Selfish Genetic Elements and Genetic Conflict
Lila Fishman and John Jaenike
OUTLINE
1. What are selfish genetic elements?
2. Diversity of selfish genetic elements
3. Selfish genetic elements and genome evolution
4. Selfish genetic elements and population variation
5. Selfish genetic elements and speciation
6. Applied uses of selfish genetic elements
While all successful genes can be said to be selfish, the term selfish genetic element (SGE) refers to heritable units that spread despite their adverse effects on individuals and populations. SGEs are remarkably abundant and diverse, ranging from mitochondrial variants to transposable elements to heritable symbiotic microorganisms. Because the spread of such elements entails a cost to other components of an organism’s genome, there can be strong selection to suppress the action of SGEs. SGEs affect several important features of organisms and populations, including genome size and structure, mutational load and mean population fitness, sex ratio, and speciation. SGEs are of applied significance, being important tools in basic research, crop development, and control of infectious diseases.
GLOSSARY
Genetic Conflict. The spread of a selfish genetic element occurs at a cost to other (nonallelic) genetic elements in the same genome or organism.
Genome Parasites. SGEs that colonize and multiply within a genome, such as mobile DNA elements.
Meiotic Drive. Historically, any process causing overtransmission of an SGE to gametes. We restrict this term to chromosomal elements that drive during meiosis rather than during gamete formation or later.
Non-Mendelian Inheritance. In a diploid nuclear locus, transmission of alternative alleles to gametes and/or progeny that deviates from the expected 1:1 ratio. Many reproductive parasites cause non-Mendelian inheritance, also known as segregation distortion or transmission ratio distortion, but other processes, such as inbreeding depression or hybrid incompatibility, can as well.
Reproductive Parasites. An SGE that spreads by altering host reproduction in ways such as meiotic drive, disabling gametes, cytoplasmic male sterility, feminization, parthenogenesis induction (PI), male killing (MK), and cytoplasmic incompatibility (CI).
Selfish Genetic Element (SGE). A heritable unit that can spread despite its adverse effects on an organism’s fitness or on other heritable elements carried by those organisms.
Somatic Parasites. SGEs that multiply within individual cells (such as petite mitochondria) or within a body of cells (such as most cancers).
1. WHAT ARE SELFISH GENETIC ELEMENTS?
For more than a century, flies of the genus Drosophila have been among the most important model organisms for studies of genetics. As in mammals, the X and Y chromosomes of Drosophila males typically obey the Mendelian law of segregation, so that males produce a 1:1 ratio of X- and Y-bearing sperm and thus equal numbers of daughters and sons; however, in some species of Drosophila, many of the females brought in from the wild produce only daughters, in clear violation of the standard rules of genetics. Chances are that such females had mated with a male that carried a “sex-ratio” gene on the X chromosome that prevents development of functional Y-bearing sperm. By incapacitating Y-bearing sperm, the sex-ratio gene—and the X chromosome on which it is located—is passed on to all of a male’s offspring. In contrast, an X chromosome lacking the sex-ratio gene plays by the rules of Mendelian segregation and is passed on to only half a male’s offspring. Therefore, by subverting the process of spermatogenesis, the sex-ratio gene gains a substantial transmission advantage and can rapidly spread through a population, even if it has negative effects on an organism’s survival and fertility, that is, its fitness. Such genes are just one example of an extraordinarily widespread, diverse, and influential group of heritable entities referred to a selfish genetic elements (SGEs).
In this example, any genes not linked to sex ratio will segregate normally and thus not experience an enhanced transmission advantage. However, they will experience the reduced fitness of occurring within an individual that carries sex ratio; therefore, most of the genome will be selected to suppress the activity of sex ratio and restore normal Mendelian segregation. Exactly this has been found in some species of Drosophila, where males from the wild appear to be genetically normal, producing offspring with 1:1 sex ratios; however, if you cross flies to get individuals carrying the X chromosome from one population and the autosomes from another (or from a closely related species), some of the resulting males will produce all-female progeny. Such a pattern indicates that a population carries a sex-ratio gene that has been suppressed by local autosomal genes, while the substitution of autosomes from another population allows the unfettered expression of the sex-ratio phenotype. This battle between the X-linked sex-ratio gene and the autosomes is an example of genetic conflict. Such conflict and coevolution between SGEs and the rest of the genome have played important roles in shaping eukaryotic genome structure, population biology, and diversification.
The concepts of selfish evolution and genetic conflict have deep roots in theoretical population genetics. J.B.S. Haldane (1932) noted, “A higher plant species is at the mercy of its pollen grains. A gene which greatly accelerates pollen tube growth will spread through a species even if it causes moderately disadvantageous changes in the adult plant.” Haldane’s pollen example falls into a gray area between sexual selection, selfish evolution, and parent-offspring conflict but demonstrates a very early recognition of the multiplicity of competitive arenas experienced by genes and the conflicts that result. In the same vein, Ronald A. Fisher’s (1941) classic model demonstrating the inherent 3:2 transmission advantage of an allele causing complete self-fertilization was an explicit mathematical argument against the popular idea that evolution by natural selection always involves variation in organismal fitness and increases in population mean fitness. Thus, it has long been clear (to theoretical population geneticists, at least!) that selfish genetic elements should exist; however, it has taken most of the last century to recognize the wonderfully diverse forms taken by selfish elements, and their pervasiveness across organisms.
The history of selfish element research generally parallels the development of tools for studying genetic transmission, from microscopic observations of chromosomes in the first decade of the 1900s to the detailed analyses of molecular variation now possible. In many cases, selfish elements were described as physical phenomena (e.g., unpaired B chromosomes in bugs, visible Wolbachia endosymbionts in insect cells) or through major phenotypic effects in polymorphic populations (e.g., cytoplasmic male sterility, skewed sex ratios) decades before they were recognized as SGEs. The early exceptions were segregation distorters in Drosophila, mice, and maize, all of which are unusually tractable genetic model systems. The occurrence of SGEs in many model organisms suggests that they are far more common and diverse than generally thought. In fact, SGEs are present in every cell in our bodies and have been continuously present in our evolutionary lineage for hundreds of millions of years.
2. DIVERSITY OF SELFISH GENETIC ELEMENTS
While there is no generally accepted means by which to classify the tremendous diversity of SGEs, one way to group them is by the arena within which they compete for transmission. Genome parasites and soma and cell parasites replicate within the genome or body, respectively, of an organism. Thus, they consist of populations of elements that can multiply within individuals, and their effects on individual fitness often reflect their frequency or numbers. In contrast, most “reproductive parasites” compete via transmission to gametes or offspring; thus they act as competing alleles within the organismal population. However, cytoplasmic elements such as mitochondria and, particularly, symbionts operate in both realms, competing via both intraindividual replication ability and manipulation of transmission. We summarize the diversity of SGEs in table 1 and highlight example SGEs from each arena below.
Table 1. Examples of selfish genetic elements

Genome Parasites
Transposable elements (TEs) are segments of DNA that propagate within genomes by various “copy and paste” or “cut and paste” transposition mechanisms, thus multiplying as populations within the habitat of the host genome (figure 1). The act of transposition can result in disruptions to the regulatory or coding sequence of a host gene, thereby resulting in deleterious mutations. Despite this, TEs are extremely abundant in the genomes of plants and animals (~45% in humans, up to 70% in maize), and also occur in fungi, protists, Archaea, and Bacteria. In fact, only a handful of primarily parasitic eukaryotes appear (provisionally) to be TE free. The ubiquity of mobile elements, and their high numbers in most eukaryotes, suggests that the average per element cost is generally small. This allows their greater accumulation in taxa with smaller effective population sizes, in which selection against slightly deleterious genes is less efficient. Because individual TE insertion/deletion events can be a significant source of mutational pressure, and because repetitive tracts of TE DNA can serve as templates for recombination and rearrangement, TEs nonetheless have major impacts on genome structure and function. In addition to these impacts, phylogenetic analyses indicate that one category of TEs—retrotransposons—gave rise to retroviruses, such as HIV.
Figure 1. Retrotransposition, one of the means by which transposable elements (TEs) spread. In this case, the TE is transcribed into an RNA intermediate, which then undergoes reverse transcription to DNA, which in turn is integrated into a new position in the genome.
Soma and Cell Parasites
Somatic mutations can cause cancer by promoting uncontrolled cell division. Cancerous cell lineages spread within individuals, usurping resources and shortening life span, and thus reduce the fitness of all genes within the germ line. Because every cell division has the potential to lead to a cancerous mutation, species with the most total cell divisions—those that are large and/or long lived, like humans—are the most vulnerable. Animals have evolved a variety of mechanisms to effectively detect and control rogue cancerous cells during the reproductive life span of the individual. The rapid propagation of aberrant somatic cells does not generally increase transmission to the next generation, so such lineages cannot spread within populations. Interestingly, cancers are relatively rare in plants, despite their potential for large size and long life span; however, because plant tissues are not segregated into soma and germ lines, there is much greater potential for genetic transmission of a cancer-causing mutation. Thus, selection has probably favored strong anticancer mechanisms in plants.
Petite colonies in yeast are similarly caused by repeated mutation of short-lived selfish elements. Yeast mitochondria with mutations that reduce respiratory activity can replicate more rapidly than normal mitochondria and thus spread to high frequency within a cell; however, this leads to slow growth by the host cells, producing the “petite” phenotype, and low colony fitness. It has been argued that the uniparental inheritance of mitochondria, which has evolved independently in numerous eukaryotic lineages, is in part a strategy to minimize opportunities for interorganelle competition within individuals and forestall the spread of such selfish mutants via sexual reproduction.
Reproductive Parasites
A wide variety of SGEs spread through manipulation of various components of an organism’s reproduction to promote their own transmission. These reproductive parasites can be divided into female meiotic drivers, allelic killers, and cytoplasmic distorters.
Female Meiotic Drivers
In most plants and animals, female meiosis is asymmetrical, with only one of the four products becoming the oocyte (or megagametophyte in plants). Thus, any chromosomal element that preferentially segregates to the egg pole gains a transmission advantage. Such a process is termed meiotic drive. But such drive can entail significant costs: chromosomal competition can cause errors in segregation resulting in chromosome loss and gamete sterility, and deleterious alleles can accumulate in the driving regions of chromosomes, which tend to be areas of unusually low recombination.
The best-characterized example of meiotic drive is the Ab10/knob neocentromere system in maize, in which an aberrantly long arm of chromosome 10 (Ab10) locks together large heterochromatic regions of repetitive DNA (knobs) and a number of genes that cause the knobs to speed to the outer spindle poles ahead of the centromeres. In heterozygotes, this neocentromeric behavior can gain the Ab10 variant up to 70 percent transmission (vs. 50% expected). Ab10 is found at generally low frequencies (<10%) in populations of maize and its wild relative teosinte, indicating that correspondingly strong costs must oppose its spread. In populations with Ab10 present, additional knobs accumulate on the arms of other chromosomes and also preferentially segregate to the egg in Ab10 carriers, acting as secondary parasites on the system.
Paradoxically, the usual mediators of Mendelian chromosomal segregation—centromeres—may themselves act as selfish elements. It has been argued that any centromeric variant that could bias its own segregation toward the egg pole should spread, leading to repeated rounds of centromere turnover and conflict or coevolution between the DNA and protein components of the centromere. Though logically compelling, this model remains controversial in the absence of evidence of driving centromeres.
Allelic Killers
In contrast to female meiotic drive elements, which subvert equal meiosis to enhance their absolute representation in a heterozygote’s gametes, allelic killers increase their relative fitness by disabling products of meiosis or offspring that carry alternative alleles. Although such processes are often referred to as meiotic drive or segregation distortion, allelic killers act postmeiotically.
The best-studied autosomal allelic killers are t haplotype in mice, Segregation Distorter (SD) in Drosophila melanogaster, and Sporekiller in Neurospora fungi. Each of these systems comprises, minimally, a killer locus with killer/nonkiller alleles and a target or responder locus with sensitive/insensitive alleles. These loci are locked together in region of low recombination, with the killer/insensitive and nonkiller/sensitive associations predominating, as a killer/sensitive combination would be suicidal. Thus, tight linkage of killer and insensitive alleles is essential for maintaining the observed polymorphisms in sperm/spore killer systems.
When competition between siblings is strong, a parental allele can spread by killing offspring that do not carry it. Examples include maternal effect Medea loci in Tribolium beetles, several maternal effect killers in mice, a paternal effect killer in the nematode Caenorhabditis elegans, and it has been speculated, restriction-modification systems in bacteria. All of these systems appear to have a killer + target structure similar to that of gamete killers.
Conflicts between gametes or offspring can be especially acute in organisms with chromosomal sex determination, as gametes or offspring carry either the X or the Y from a given parent, and there is essentially no recombination between these chromosomes. Sex chromosome drive is particularly common in flies and mosquitoes, which have XY (male) versus XX (female) sex determination. As discussed above, in males carrying a “sex ratio” X chromosome, sperm carrying the Y fail to differentiate or function properly, resulting in strongly female-biased offspring sex ratios (figure 2). Males with a nondriving X produce equal numbers male and female offspring. Mechanistically, this X chromosome drive entails an interaction between loci on the X and Y chromosomes. In population genetic terms, however, the competition is between the sex-ratio and standard X chromosomes. When females mate with only a single male, the X of a sex-ratio male is passed on to all the offspring, while the X of a standard male is passed on to only half of them; thus, a sex-ratio X chromosome can potentially spread to the point of causing extinction of a population or species.
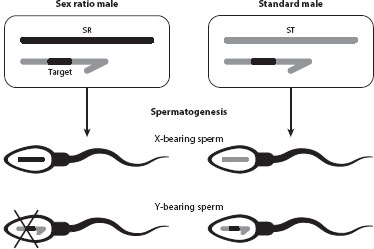
Figure 2. X drive in male flies. Males that carry a sex-ratio X chromosome (SR) produce only X-bearing sperm, whereas males with a standard X chromosome (ST) produce equal numbers of X- and Y-bearing sperm.
Cytoplasmic Distorters
Uniparentally inherited organelles and symbionts can spread by increasing the frequency of the carrier sex at the expense of the noncarrier sex. The two best examples of this are mitochondria that cause cytoplasmic male sterility (CMS) in flowering plants and Wolbachia (and other maternally transmitted microbial symbionts) in arthropods, both of which are remarkably widespread and have far-reaching effects on organismal variation.
Cytoplasmic male sterility. Maternally transmitted organelles (mitochondria and chloroplasts) have a strong interest in increasing female fertility. Most flowering plants are hermaphroditic, producing both male and female gametes within each flower. This generates a conflict over floral development between organellar (only transmitted in seeds) and nuclear genes (transmitted in both pollen and seeds). (For simplicity, we use the term seed to refer to a plant megagametophyte.) Mitochondrial CMS mutants are associated with increased resource allocation to seeds at the expense of pollen production and thus have a significant transmission advantage (figure 3). However, as such a CMS variant spreads, nuclear mutations that restore pollen fertility are favored, both to regain lost male fitness and to take advantage of the increasingly female-biased population sex ratio. This coevolution is predicted, under most conditions, to result in rapid fixation of both the CMS mitochondria and its matched restorer. However, under some conditions, polymorphism of CMS and restorer may be maintained, resulting in a mixture of female and hermaphroditic plants in the population (gynodioecy). About 7 percent of plant species are gynodioecious, suggesting that CMS-restorer interactions are common and contribute to plant mating system diversification. Male sterility due to CMS-restorer mismatch is common in inter- and intraspecific hybrids, indicating that many currently hermaphroditic populations have histories of CMS-restorer coevolution. Rapid evolution under positive selection of the gene family that includes most known restorer alleles also supports the hypothesis of a widespread and ongoing history of cytonuclear conflict in angiosperms.
Figure 3. Cytoplasmic male sterility (CMS), found in numerous plants. A specific mitochondrial haplotype results in male sterility and failure to produce pollen, resulting in availability of resources for production of additional eggs, through which mitochondria are transmitted.
Wolbachia and other symbionts. Insects and other arthropods are infected with a variety of maternally transmitted microbial symbionts, some of which spread by mechanisms involving manipulation of host reproduction. Such selfish symbionts include bacteria belonging to the genera Wolbachia and Rickettsia (Alphaproteobacteria), Arsenophonus (Gammaproteobacteria), Spiroplasma (Firmicutes), and Cardinium (Flavobacteria), and microsporidian protozoa. Of these, Wolbachia is by far the most common, infecting perhaps two-thirds of all insect species. Wolbachia can manipulate host reproduction in a variety of ways, including feminization (developmental conversion of a genetic male into an egg-producing female), male killing (embryonic death of the males, giving their infected sisters access to more resources), parthenogenesis induction (forgoing production of males altogether), and cytoplasmic incompatibility (in which matings between infected males and uninfected females lead to high levels of offspring mortality). Phylogenetic analyses indicate that Wolbachia do not infect insect lineages for long evolutionary periods, implying very high rates of colonization of new host species by Wolbachia and extinction from currently infected species. Recent studies have shown that some strains of Wolbachia that are reproductive parasites can also protect their insect hosts from the adverse effects of RNA virus infection. Thus, Wolbachia can function both as a selfish reproductive parasite and as a context-dependent mutualist.
3. SELFISH GENETIC ELEMENTS AND GENOME EVOLUTION
A major fraction of the DNA of many eukaryotes (including humans) consists of transposable elements, making transposable element numbers a primary determinant of genome size. Although TEs make up nearly half the human genome, they contribute relatively little to current mutational variation, as most are dead (missing key components) or otherwise inactivated. In contrast, TEs are extremely abundant and active in many flowering plants. Much of this variation is unexplained, but some undoubtedly reflects aspects of the organismal ecology, such as mating system, that influence the evolutionary dynamics of TEs. For example, small effective population sizes in the highly inbreeding annual plant Arabidopsis thaliana may reduce the efficiency of selection against TE-caused mutations and explain its higher load of TEs relative to closely related outcrosser A. lyrata. On the other hand, long-term self-fertilization or asexuality reduces opportunities for TE movement among lineages and may even select for TEs with “self-control” by favoring organismal lineages with few or weaker TEs. The latter prediction is borne out by the complete lack of several otherwise-widespread families of transposons in the long-term asexual bdelloid rotifers, despite evidence of TE acquisition by horizontal gene transfer. Under conditions of asexual reproduction or long-term selfing, the interests of all genetic elements within an organism are aligned, thus effectively eliminating conflict and the problems of SGEs.
Mobile elements also affect the physical structure of genomes. TEs are major components of the heterochromatic centromeric and telomeric regions of most chromosomes, contributing to the function of these regions in ways that are only beginning to be understood. Telomeres are repetitive DNA regions at chromosome ends that are eroded during replication and thus must be actively maintained (generally by a telomerase) for cell division to occur properly. In Drosophila, end-directed transposition of three types of retrotransposon actually maintains telomere length, an important function for the individual that indicates domestication of a formerly selfish element.
The accumulation of closely related TE sequences throughout a genome provides substrates for recombination between nonhomologous chromosomal regions; it is thus a major source of inversions, translocations, and other rearrangements. Such rearrangements contribute to rapid evolution of chromosome structure in some lineages, particularly flowering plants. If rearrangements vary in their transmission through female meiosis, as has been demonstrated in mice, humans, chickens, and flies, meiotic drive may also contribute to the fixation of such variants. Chromosomal rearrangements are implicated in the evolution of species barriers, both as direct causes of low hybrid fitness and as suppressors of recombination. Thus, the incidence and distribution of TEs across lineages, as well as the opportunities for drive during meiosis, may influence mechanisms and patterns of species diversification.
Other kinds of selfish genetic elements also influence genome- or chromosome-level processes. Most notably, reproductive SGEs may be an important source of selection on recombination rates. A mutation (such as a chromosomal rearrangement) that suppresses recombination in the vicinity of a drive allele will reap the benefits of the linked driver’s transmission advantage. Linked mutations that enhance drive will also be favored, favoring low recombination over increasingly larger regions as additional enhancers accumulate. There is abundant evidence for both chromosomal and allelic suppression of recombination around active drive loci. For example, both the maize Ab10/knob system and a centromere-associated female meiotic driver in yellow monkeyflower (Mimulus guttatus) encompass vast chromosomal regions locked together by inversions. The rapid spread of selfish nuclear elements along with reduced recombination can result in the spread of linked deleterious alleles. Unlinked loci (that is, most of the genome) should favor greater recombination in the vicinity of a driving element, both to increase the efficiency of selection against deleterious hitchhikers and to break up associations between killer and enhancer/insensitive alleles. For example, the higher recombination rates generally observed near centromeres in female versus male mammals may have evolved to suppress centromere drive in females.
4. SELFISH GENETIC ELEMENTS AND POPULATION VARIATION
SGEs interact with other components of population biology in diverse ways, acting as everything from sources of mutation and expression variation to selective factors in the evolution of mating systems and phenotypic diversification.
As TEs jump around, they often land in functional genes, thus disrupting their expression or function. In taxa with a large percentage of active transposable elements, TEs are a significant source of (primarily deleterious) mutational pressure. For example, in Drosophila, TEs are responsible for up to 80 percent of visible newly arising visible mutations. TE insertions into regulatory (nonprotein-coding) regions may affect gene expression in potentially adaptive ways. In D. melanogaster, genome scans suggest that a small number of TEs in regulatory regions may have been involved in adaptation to temperate conditions during its recent range expansion out of Africa. Over the longer term, there are numerous individual cases in which TE sequences have been co-opted for other functions. For example, the V(D)J recombination mechanism, which underlies the ability of the vertebrate immune system to recognize an extraordinary diversity of antigens, relies in part on an enzyme derived from a TE transposase gene. These integrations of TEs into the “normal” machinery of an organism do not explain their evolutionary proliferation, but they do beautifully illustrate how evolution by natural selection works with whatever raw materials are at hand.
Reproductive parasites also contribute to standing variation in individual fitness within populations, but through balancing selection on the selfish element (and/or linked loci) rather than mutation. Allelic killers can reduce the fertility of carriers, and segregation distorters can become associated with linked deleterious alleles and thus influence diverse fitness traits. For example, the driving t haplotype in mice negatively affects male territorial behavior and female fertility, in addition to its direct effects on male fertility. In Drosophila melanogaster in Africa, the driving SD locus has carried along a single linked haplotype representing nearly 40 percent of chromosome 2. Given that both these SGEs were first identified through segregation distortion of genetic markers in thoroughly studied lab crosses, such major effects in wild populations suggest that undiscovered selfish elements may frequently contribute to natural fitness variation in less genetically tractable taxa.
If the spread of an SGE alters the properties of a population, this can bring about selection on morphological, behavioral, or life history traits. Some SGEs bias offspring sex ratios (X and Y drive elements, some endosymbionts) or the sex expression of adult individuals (CMS, some endosymbionts), thus affecting the population-level sex ratio. In sexually dimorphic stalk-eyed flies (Cyrtodiopsis), for example, high frequencies of a driving X chromosome lead to extremely female-biased sex ratios. Females that mate with males carrying a nondriving X are favored, as they produce male offspring, which being rare in the population, have exceptionally high mean fitness. Stalk-eyed fly females exhibit a preference for males with long eye stalks, which are genetically associated with the nondriving X. Evidence of selfish elements influencing sexual selection, particularly female choice, has also been documented in guppies, mice, butterflies, and fruit flies. In flowering plants, cytoplasmic male sterility similarly alters selection on reproductive allocation, and females and hermaphrodites in gynodioecious species tend to be dimorphic for a variety of floral characters in addition to pollen production. Given the direct effects of many selfish elements on individual fertility and population sex ratios, they are likely to be generally important factors in sexual selection and mating system evolution in a wide variety of taxa.
5. SELFISH GENETIC ELEMENTS AND SPECIATION
Reproductive parasites and their suppressors tend to evolve rapidly, interact epistatically, and negatively affect fertility when mismatched. Theoretically, joint fixation of driver and suppressor is often more likely than polymorphism, and evidence from X-linked drivers in flies and CMS in plants suggests that hidden drive systems are indeed common. Molecular analyses of hybrid incompatibilities in flies, mice, plants, and yeast indicate that SGEs contribute significantly to the evolution of species barriers.
The endosymbiont Wolbachia causes cytoplasmic incompatibility (CI) in a wide variety of insects in a Wolbachia strain-specific manner. Therefore, if two populations are fixed for different Wolbachia strains, there can be bidirectional CI, a potentially significant source component of postzygotic isolation between populations. While CI may not be sufficient to cause speciation, it probably does contribute to overall reproductive isolation between incipient species and thus contributes to speciation and diversification.
6. APPLIED USES OF SELFISH GENETIC ELEMENTS
Selfish elements have been surprisingly useful to humans, and a variety of novel applications are in development. Both transposable elements and restriction-modification systems are used extensively as molecular genetic tools, allowing targeted manipulation of DNA sequences. Because male sterility facilitates the generation of F1 hybrid seed, CMS is an important tool in agriculture. Research has recently begun on using reproductive parasites to control the insect vectors of human diseases such as malaria and dengue fever. These approaches take advantage of the potentially rapid spread of SGEs to manipulate the genetics or population biology of wild populations in a cost-effective manner. In principle, such approaches could be used either to knock down populations of vectors or to bring about the spread of factors that reduce vector effectiveness (e.g., malarial resistance). For example, experimental introduction of Wolbachia into the mosquito Aedes aegypti results in the expression of CI; thus, Wolbachia has the potential to spread through populations of these mosquitoes. Remarkably, these Wolbachia also render the mosquitoes unsuitable as vectors for dengue virus; therefore, as Wolbachia spreads through mosquito populations, the incidence of dengue fever in humans is expected to decline. Because such applications involve the release of biologically modified organisms, these approaches must contend with practical, ethical, and social hurdles prior to implementation. Understanding the evolutionary dynamics of naturally occurring SGEs is key to evaluating the costs and benefits of selfish-element–driven vector control.
FURTHER READING
Burt, A. 2003. Site-specific selfish genes as tools for the control and genetic engineering of natural populations. Proceedings of the Royal Society B 270: 921–928.
Burt, A., and R. Trivers. 2006. Genes in Conflict: The Biology of Selfish Genetic Elements. Cambridge, MA: Harvard University Press.
Chase, C. D. 2007. Cytoplasmic male sterility: A window to the world of plant mitochondrial-nuclear interactions. Trends in Genetics 23: 81–90.
Dawkins, R. 1976. The Selfish Gene. Oxford: Oxford University Press.
Doolittle, W. F., and C. Sapienza. 1980. Selfish genes, the phenotype paradigm and genome evolution. Nature 284: 601–603.
Haig, D., and A. Grafen. 1991. Genetic scrambling as a defence against meiotic drive. Journal of Theoretical Biology 153: 531–558.
Henikoff, S., K. Ahmad, and H. S. Malik. 2001. The centromere paradox: Stable inheritance with rapidly evolving DNA. Science 293: 1098–1102.
Hurst, G.D.D., and J. H. Werren. 2001. The role of selfish genetic elements in eukaryotic evolution. Nature Reviews Genetics 2: 597–606.
Hurst, L. D., A. Atlan, and B. O. Bengtsson. 1996. Genetic conflicts. Quarterly Review of Biology 71: 317–364.
Jones, R. N., M. Gonzalez-Sanchez, M. Gonzalez-Garcia, J. M. Vega, and M. J. Puertas. 2008. Chromosomes with a life of their own. Cytogenetic Genome Research 120: 265–280.
Kazazian, H. H. 2004. Mobile elements: Drivers of genome evolution. Science 303: 1626–1632.
Lyttle, T. W. 1991. Segregation distorters. Annual Review of Genetics 25: 511–557.
Merrill, C., L. Bayraktaroglu, A. Kusano, and B. Ganetzky. 1999. Truncated RanGAP encoded by the segregation distorter locus of Drosophila. Science 283: 1742–1745.
Naito, T., K. Kusano, and I Kobayashi. 1995. Selfish behavior of restriction-modification systems. Science 267: 897–899.
Orgel, L. E., and Crick, F.H.C. 1980. Selfish DNA: The ultimate parasite. Nature 284: 604–607.
Pardo-Manuel de Villena, F., and C. Sapienza. 2001. Nonrandom segregation during meiosis: The unfairness of females. Mammalian Genome 12: 331–339.
Turelli, M., and A. A. Hoffmann. 1991. Rapid spread of an inherited incompatibility factor in California Drosophila. Nature 353: 440–442.
Werren, J. H., L. Baldo, and M. E. Clark. 2008. Wolbachia: Master manipulators of invertebrate biology. Nature Reviews Microbiology 6: 741–751.