1 Introduction
Throughout history, natural products have been used as components of traditional medicines and herbal remedies. For modern small-molecule drug development as well, natural products remain the single most productive source of inspiration [1, 2]. According to a widely cited survey of drugs approved between 1981 and 2014 [1], 6% of all small-molecule drugs are unaltered natural products, 26% are natural product derivatives, and 32% are natural product mimetics and/or contain a natural product pharmacophore.
The high importance of natural products is rooted in their evolution-based specific biological purposes, which enable them to exhibit a wide range of biological activities across different organisms. Their structural and physicochemical diversity outrivals that of modern synthetic collections [3–5], and their often high complexity with respect to molecular shape and stereochemistry [3, 6, 7] adds to their ability to modulate a significant number of targets for which no synthetic compounds are known.
Today, in addition to botanicals, natural products from bacteria, fungi, and marine life are increasingly being explored. However, developing drugs from natural products remains a challenging resource- and time-consuming task. Covalent binding, aggregate formation, decomposition, precipitation, and other chemical, physical, and biological processes pose technical barriers to assays run on crude extracts or isolated natural products [2, 8]. Apart from technical complications, the availability of material for testing remains a severe bottleneck. The sourcing process can be complex and expensive, and further complications may arise when material needs to be transferred across national boundaries [2].
Computational methods such as docking, pharmacophore modeling, and quantitative structure–activity relationship modeling can make a significant contribution to natural product-based drug discovery as they allow the selection of promising natural products for extraction, purification, (partial) synthesis, and biological testing [9]. An essential precondition for the application of in silico approaches is access to information on the molecular structure of natural products, which today is available from a large number of sources [10]. These sources can be categorized into two main classes: virtual natural product databases and physical natural product collections.
Virtual natural product databases contain the molecular structures of known natural products and vary in size, coverage, and types of information they contain for the individual compounds, among other aspects. As such, they can be further divided into encyclopedic or general, natural product databases, and specialized collections that are focused on, for example, traditional medicines, geographical regions, or bioactivities (e.g., compounds with anticancer or antimalarial activity). The majority of virtual natural product databases are accessible via online services that offer free searching and browsing functionalities. Many of them also offer an option for bulk download, thus enabling virtual screening applications, such as the Dictionary of Natural Products (DNP) [11] and Reaxys [12].
Physical natural product collections are mostly commercial offerings of in-stock natural products and natural products that are sourced or synthesized on-demand. Most vendors make the content of their collections browsable and searchable via free public web services. These web services also often include an option for bulk download. However, the download function may only be enabled after (usually free) registration for the web service.
With this contribution, we aim to provide a timely overview of natural product data sources useful for virtual screening and other applications in cheminformatics. The contribution builds on our recent analyses of virtual natural product databases and physical natural product collections [10, 13] and adds a wealth of information on the latest reported natural product data sources.
2 Virtual Natural Product Databases
Overview of virtual natural product databasesa
Data source name | Scope | Number of compoundsb | Biological datac | Free used | Bulk data access | Chemistry-aware web interface | Scientific literature | Web presence and database version | Included in the analysis published in [10] |
|---|---|---|---|---|---|---|---|---|---|
Encyclopedic and general NP databases | |||||||||
Dictionary of Natural Products (DNP) | All forms of life | >230,000 | Bioactivity data | No | Yes | Yes | – | [11] | Yes |
AntiBase | Microorganisms and higher fungi | > 43,000 | Bioactivity data (focus on antimicrobial activity) | No | No | Yes | [14] | [15] | No |
Reaxys | All forms of life | >260,000 | Bioactivity data | No | Yes | Yes | – | [12] | No |
Super Natural II | All forms of life | >325,000 | Bioactivity and toxicity data | Yes | No | Yes | [16] | [17] | No |
UNPD | All forms of life | >229,000 | None | Yes | Yes | No | [18] | Website could not be reached | Yes |
NPASS | All forms of life | ~35,000 | Bioactivity data | Yes | No | Yes | [19] | [20] | No |
CMAUP | Plants | >47,000 | Bioactivity data | Yes | Yes | Yes | [21] | [22] | No |
The Natural Products Atlas | Bacteria and fungi | >20,000 | None | Yes | Yes | Yes | – | [23] | No |
Pye et al. dataset | NPs from microorganisms and marine life published between 2012 and 2015 | >6000 | None | Yes | Yes | No | [24] | – | No |
Natural products included in the PubChem Substance Database | All forms of life | >3500 | Bioactivity data | Yes | Yes | Yes | [25] | [26] | Yes |
UEFS Natural Products | None specified | ~500 | None | Via ZINC | Via ZINC | No | – | – | Yes |
NP databases focused on traditional medicines | |||||||||
TCM database@Taiwan | Chinese medicinal herbs | >60,000 | Bioactivity data | Yes | Yes | Yes | [27] | [28] | Yes |
TCMID 2.0 | Chinese medicinal herbs | >43,000 | Bioactivity data | Yes | Yes | No | [29] | Website could not be reached | Yes |
YaTCM | Chinese medicinal herbs | >47,000 | Bioactivity data | Yes | No | Yes | [30] | [31] | No |
Chem-TCM | Chinese medicinal herbs | >12,000 | Bioactivity data | No | Yes | No | [32] | [33] | No |
HIM | Chinese medicinal herbs | ~1300 | ADME and toxicity data | Yes | Via ZINC | Via ZINC | [34] | Website could not be reached | Yes |
HIT | Chinese medicinal herbs | ~530 | Bioactivity data | Yes | Via ZINC | Via ZINC | [35] | Website could not be reached | Yes |
IMPPAT | Indian medicinal herbs | >9500 | Bioactivity data | Yes | No | Yes | [36] | [37] | No |
Databases focused on a specific habitat or geographical region | |||||||||
DMNP | Marine life | >55,000 (including NP derivatives) | Bioactivity data | No | No | Yes | – | [38] | No |
MarinLit | Marine life | >33k | Bioactivity data | No | No | Yes | – | [39] | No |
TIPdb | Taiwanese herbs | ~9000 | Bioactivity data (focus on anticancer, antiplatelet and antituberculosis activity) | Yes | Yes | No | [42] | Yes | |
NANPDB | All forms of life indigenous to North Africa | >6800 | Bioactivity data | Yes | Yes | Yes | [43] | [44] | Yes |
AfroDb | African medicinal plants | ~1000 | Bioactivity data | Yes | Yes | No | [45] | – | Yes |
SANCDB | South African plants and marine life | >700 | None | Yes | Yes | Yes | [46] | [47] | Yes |
AfroCancer | African medicinal plants with confirmed antineoplastic, cytotoxic or antiproliferative activity | ~400 | Bioactivity data (focus on anticancer activity) | Yes | Yes | No | [48] | – | Yes |
AfroMalariaDB | African plant NPs with confirmed antimalarial or antiplasmodial activity | >250 | Bioactivity data (focus on antimalarial activity) | Yes | Yes | No | [49] | – | Yes |
NuBBEDB | NPs from Brazilian plants, fungi, insects, marine organisms, and bacteria | >2200 | Bioactivity data (focus on antimicrobial activity) | Yes | Yes | Yes | [53] | Yes | |
BIOFACQUIM | NPs from plants, fungi, and propolis isolated and characterized in Mexico | >400 | Bioactivity data | Yes | Yes | No | [54] | [55] | No |
Databases focused on specific organisms | |||||||||
PAMDB | Pseudomonas aeruginosa | >4300 | Bioactivity data | Yes | Yes | Yes | [56] | [57] | No |
StreptomeDB 2.0 | Streptomycetes | ~4000 | Bioactivity data | Yes | Yes | Yes | [58] | [59] | Yes |
Databases focused on specific biological activities | |||||||||
NPCARE | NPs with measured anticancer activity, sourced from plants, marine species and microorganisms | >6500 from online search >1500 in bulk download | Bioactivity data (focus on anticancer activity) | Yes | Yes | No | [60] | [61] | Yes |
NPACT | NPs with measured anticancer activity, sourced from plants | >1500 | Bioactivity data (focus on anticancer activity) | Yes | Via ZINC | Yes | [62] | [63] | Yes |
InflamNat | NPs with measured anti-inflammatory activity, sourced primarily from terrestrial plants | >650 | Bioactivity data (focus on anti-inflammatory activity) | Yes | Yes | No | [64] | – | No |
Databases focused on specific NP classes | |||||||||
Carotenoids Database | Carotenoids extracted from almost 700 source organisms | >1100 | Bioactivity data | Yes | No | Yes | [65] | [66] | No |
2.1 Encyclopedic and General Natural Product Databases
2.1.1 Dictionary of Natural Products (DNP)
The Dictionary of Natural Products [11] is one of the most established encyclopedic collections of natural products available to date. The commercial database consists of more than 230k natural products, 46k of which are not covered by any of the free virtual natural product collections investigated in our recent study [10] and marked in Table 1. The molecular structures are richly annotated with compound names and synonyms, physicochemical properties (e.g., molecular weight, pKa, solubilities, and spectroscopic data), biological sources, use, and toxicity data. One particularly useful feature of this database is that the natural products are classified into 1050 structural types. Importantly, stereochemical information is stored only in Fisher-type diagrams, separate from the 2D connection tables and InChIs. The database is accessible via a web service [11] and also distributed as a CD-ROM.
2.1.2 AntiBase
AntiBase [15] is a comprehensive commercial database including more than 43k natural products collected primarily from microorganisms and higher fungi (including algae, cyanobacteria, lichens, yeasts, Ascomycetes, and Basidiomycetes). AntiBase stands out due to the large amount of spectrometric data provided (including experimental and computed 13C NMR data). The individual natural products are annotated with further physicochemical properties and biological data, such as pharmacological activities and toxicity. AntiBase is available in several software formats featuring powerful text, structure, and spectra search capabilities.
2.1.3 Reaxys
Reaxys [12] is a comprehensive resource for chemical information relevant to synthesis chemists. As such, Reaxys has no specific focus on natural products, but contains information on the molecular structures, reactions, physical properties, biological sources, and activity data for more than 260,000 natural products. Reaxys is accessible via a web interface, which features detailed search functionality. Bulk download of natural products (and other chemicals and data) is supported.
2.1.4 Super Natural II
Super Natural II [16] provides chemical information on more than 325,000 natural products and, accordingly, is currently one of the most comprehensive free data sources available. Super Natural II draws data from several preexisting databases and provides information on molecular structures (including stereochemistry annotations), suppliers, bioactivities, computed physicochemical properties, and toxicity classes. The web interface supports the download of individual structures but not bulk download.
2.1.5 Universal Natural Products Database (UNPD)
With a total of more than 229,000 entries, the Universal Natural Products Database (UNPD) [18] is currently the most comprehensive of all free and commercial resources on natural products that offer bulk download. Drawing data from a number of different sources, including the Chinese Natural Product Database (CNPD) [67], the CHDD [68] (a database of compounds of traditional Chinese medicinal herbs, previously provided by the authors of the UNPD), and the Traditional Chinese Medicines Database (TCMD) [69], the UNPD is itself a component of Super Natural II. Our recent analysis showed that approximately one-third of the natural products contained in the UNPD are not covered by any of the other investigated virtual natural product databases [13]. We also found that the UNPD covers a wide chemical space and represents all major classes of natural products. Approximately 85% of the natural products contained in the UNPD comply with Lipinski’s rule of five (here and elsewhere, statements on the compliance with Lipinski’s rule of five refer to the molecular structures of natural products after the removal of sugars and sugar-like moieties with the tool “SugarBuster” [13]). The connection tables of UNPD store 3D structures with explicit stereochemistry defined by atom coordinates (enantiomers are stored as individual entries) plus several identifiers. In recent years, significant downtimes of the web presence have been observed.
2.1.6 Natural Product Activity and Species Source (NPASS)
The Natural Product Activity and Species Source [19] is another large resource of chemical and biological information on natural products. The database currently includes more than 35,000 natural products from a total of approximately 25,000 species. Two-thirds of the natural products come from Viridiplantae; the remaining third comes primarily from Metazoa, fungi, and bacteria. Bioactivity data are recorded against approximately 3000 protein targets, more than 1300 microbial species and a similar number of cell lines. Natural Product Activity and Species Source offers a powerful, chemistry-aware web interface for browsing and searching. Data for individual natural products can easily be downloaded, but bulk download of structures and other data is not offered.
2.1.7 Collective Molecular Activities of Useful Plants (CMAUP)
Collective Molecular Activities of Useful Plants [21] is a large, new resource for information on plant natural products and their biological activities. The database stores information on over 47,000 natural products of more than 5600 plants native to greater than 150 countries and regions. The individual natural products are annotated with recorded bioactivities against more than 640 biomacromolecular targets. In addition, information on plant species, use, geographical distribution, metabolic pathways, gene ontologies, and diseases is provided. The database can be browsed and searched via a free, chemistry-aware web interface. Free bulk download of structural data (including stereochemical information) and metadata is also supported.
2.1.8 Natural Product Atlas
The Natural Product Atlas [23] has been recently introduced as a comprehensive resource of chemical information on natural products from bacteria (including cyanobacteria) and fungi (including mushrooms and lichens) reported in peer-reviewed original research articles. The current version of the database covers approximately 20,000 natural products, almost one-third of which are found in Streptomyces. Further prominent genera are Aspergillus and Penicillium, each representing approximately 10% of the data. The web service provides powerful tools for browsing, searching, and data visualization. Particularly noteworthy are the network visualization features, which allow users to obtain a solid overview of the molecular diversity and coverage of the chemical space. An option for bulk download of the database is provided.
2.1.9 Pye et al. Dataset
As part of a comprehensive survey of natural products discovered between 1941 and 2015, Pye et al. have recently published a dataset of almost 6300 natural products that have been published between 2012 and 2015 [24]. As such, the dataset provides a good overview of the chemical space of natural products discovered in recent years. All structures are available as isomeric SMILES (simplified molecular input line entry specification) from the supporting information.
2.1.10 Natural Products Included in the PubChem Substance Database
The PubChem database [70] contains structures of more than 3500 natural products, which can be retrieved using the query “MLSMR [SRC] AND NP[CMT]” [25]. Most compounds are annotated with bioactivity data, covering a total of more than 650 biomolecular targets. Approximately 40% of all compounds are not covered by any other resource investigated in our recent study [13]. More than 95% of all natural products of this dataset comply with Lipinski’s rule of five; greater than half of all compounds are alkaloids. All structures are downloadable and include stereochemical information.
2.1.11 UEFS Natural Products
Researchers from the State University of Feira de Santana (UEFS) in Brazil have deposited a dataset of approximately 500 natural products for download at the ZINC database [71, 72]. The natural products have been compiled from papers that the authors and collaborators have published separately. Noteworthy is the relatively high proportion of flavonoids in the dataset [13].
2.2 Databases Focused on Traditional Medicines
2.2.1 Traditional Chinese Medicine Database@Taiwan
The TCM Database@Taiwan [27] is the most comprehensive free resource for molecular structures of natural products related to TCM. It has been compiled from Chinese medical texts and various dictionaries, and contains the structures of more than 60,000 natural products from over 450 herb, animal, and mineral product TCMs. Important features of this database include the organization of the data into 22 TCM usage classes, such as “digestant medicinal”, and comprehensive ingredient-to-TCM mapping. We found that 38% of all natural products of the TCM Database@Taiwan are alkaloids, which is one of the highest percentages observed among all investigated databases [13]. The database also stands out due to its large proportion of high molecular weight natural products, among which polyphenols and basic alkaloids are particularly prominent. In contrast to the previously discussed natural product databases, the proportion of natural products in compliance with Lipinski’s rule of five is only 51%. The web interface of the TCM Database@Taiwan offers advanced search functionalities based on molecular structures and physicochemical properties. Bulk download of all molecular structures including stereochemical information is supported.
2.2.2 Traditional Chinese Medicine Integrated Database (TCMID 2.0)
The TCMID 2.0 [29] is a large database of natural products that links traditional Chinese with modern western medicine by incorporating data on drugs, targets, and diseases. The database integrates data on herbal ingredients from, among many other sources, the TCM Database@Taiwan, TCM-ID [73], and the Encyclopedia of Traditional Chinese Medicines [74]. Since its initial release in 2013, the database has been substantially expanded, with the latest release counting more than 43k compounds. As major additions to the latest release, almost 4k mass spectra of natural products and over 176,000 protein-protein interactions have been added. The TCMID 2.0 web interface offers, among many other features, a tool for visualizing ingredient-target-drug-disease networks and herb-target-disease networks. This enables users, for example, to browse the natural products of a herb of interest, the targets of these natural products and how they are linked to diseases. As such, the platform can provide valuable information on multi-target effects and molecular mechanisms. Download of molecular structures (including stereochemical information) and associated data is possible in principle. At the time of writing, the online presence of this database could not be confirmed.
2.2.3 Yet Another Traditional Chinese Medicine Database (YaTCM)
The YaTCM database [30] is a further recently introduced database on natural products from Chinese medicinal herbs. The database currently holds more than 47,000 records of natural products found in over 6200 herbs. Like TCMID 2.0 (which is integrated into YaTCM), the chemical data are supplemented with a wealth of information on targets (approximately 3500 therapeutic targets are covered), pathways, and diseases. The web service offers chemistry-aware browsing and search functionality. The website also features an in silico model for target prediction and tools for visualizing networks of TCM recipes, herbs, natural products, known and predicted protein targets, pathways, and diseases. Bulk download of chemical information is not supported.
2.2.4 Chemical Database of Traditional Chinese Medicine (Chem-TCM)
Chem-TCM [33] is a commercial resource that holds more than 12,000 records on natural products from approximately 350 herbs used in TCM. The database provides rich chemical information, including molecular structures with stereochemical information, names and identifiers, molecular scaffold types, and natural product classes. The botanical information includes Latin binomial botanical names, pharmaceutical names, and Chinese herb names. Chem-TCM seeks to link TCM to western medicine by including activities against 41 drug targets predicted with a random forest model [32]. In addition, the database includes estimated affinities of molecular activities according to 28 traditional Chinese herbal medicine categories. Chem-TCM is provided via a chemistry-aware software application and as SD files.
2.2.5 Herbal Ingredients In Vivo Metabolism Database (HIM)
The Herbal Ingredients In Vivo Metabolism (HIM) [34] consists of around 1300 natural products richly annotated with absorption, distribution, metabolism, and excretion (ADME) data and information on compound toxicity. Most natural products of HIM comply with Lipinski’s rule of five, and approximately one-third of the natural products in this database are not available from any of the other resources that we investigated recently [13].
At the time of writing, the online presence of this database could not be confirmed. The molecular structures of HIM can, however, be accessed via the ZINC database and include stereochemical information.
2.2.6 Herbal Ingredients’ Targets Database (HIT)
The Herbal Ingredients’ Targets (HIT) database [35] is a collection of more than 530 active ingredients from herbs. Most natural products of HIT comply with Lipinski’s rule of five [13]. As for HIM, the web presence of HIT could not be confirmed at the time of writing, but the molecular structures (including stereochemical information) are available via the ZINC database. The natural products stored in HIT are covered to a large extent by other databases [13].
2.2.7 Indian Medicinal Plants, Phytochemistry, and Therapeutics Database (IMPPAT)
The Indian Medicinal Plants, Phytochemistry, and Therapeutics (IMPPAT) database [36] is a rich resource of chemical, biological, and botanical information on Indian medicinal plants, covering more than 9500 natural products from more than 1700 species. The chemistry-aware web interface allows browsing and searching. A network visualization tool allows the investigation of plant-natural product associations, plant-therapeutic use associations, and plant-formulation associations. Bulk download of molecular structures is not supported.
2.3 Databases Focused on a Specific Habitat or Geographic Region
2.3.1 Dictionary of Marine Natural Products (DMNP)
The Dictionary of Marine Natural Products [38] is a subset of the Dictionary of Natural Products (DNP) containing more than 55,000 marine natural products and their derivatives. This commercial resource is provided as a web service (with similar capacities as that of the DNP) and is also distributed as a combination of a book and CD-ROM.
2.3.2 MarinLit Database
The MarinLit database [39] is a large database of marine natural products collected from journal articles. The commercial resource currently lists more than 33,000 natural products, richly annotated with bibliographic information, molecular structure, names, biological sources, physicochemical properties, and identifiers. MarinLit’s web interface provides powerful search functionalities and features for the dereplication of natural products.
2.3.3 Taiwan Indigenous Plant Database (TIPdb)
The TIPdb database [40] provides information on the anticancer, antituberculosis, and antiplatelet activity of more than 9000 natural products of plants indigenous to Taiwan. Noteworthy are the rather high percentage of natural products with sugars and sugar-like moieties (25%) and a rather low percentage of alkaloids (14%) [13]. The web service offers basic browsing and searching functionality, and the molecular structures of all natural products can be downloaded in bulk.
2.3.4 Northern African Natural Products Database (NANPDB)
With more than 6800 natural products records, NANPDB [43] is the largest database of natural products isolated from species native to Northern Africa, primarily plants but also endophytes, animals, fungi, and bacteria. This freely accessible database has been compiled from many different sources, including articles published in natural product journals as well as Ph.D. theses. The database provides information on source organisms, biological activities, and activity types (e.g., antimalarial, cancer-related). We have shown that the chemical space covered by NANPDB is similar to that of approved drugs, with more than 90% of all compounds complying with Lipinski’s rule of five [13]. Noteworthy is the high proportion of natural products containing sugars and sugar-like moieties (28%). The Northern African Natural Products Database is provided via a chemistry-aware web interface [44] and can be downloaded in SMILES and SD file format (including stereochemical information).
2.3.5 AfroDb Database
The AfroDb database [45] is a diverse collection of natural products found in African medicinal plants. Worth mentioning is the high percentage of phenols and phenol ethers in this database (61%), which is approximately double of that of the DNP [13]. The molecular structures (including stereochemical information) are freely available in the supplementary information of the original publication and via the ZINC database.
2.3.6 South African Natural Compound Database (SANCDB)
The SANCDB [46] is composed of more than 700 natural products from plants and marine life native to South Africa. The database has been compiled manually from the literature and contains information on molecular structure (including stereochemistry information), name, structural class, source organism, and physicochemical properties. A free, chemistry-aware web interface for searching and browsing is provided. The resource is also accessible via a representational state transfer application programming interface (REST API).
2.3.7 African Anticancer Natural Products Library (AfroCancer)
AfroCancer [48] focuses on natural products from African medicinal plants with confirmed antineoplastic, cytotoxic, or antiproliferative activity. The database contains a high percentage of phenols and phenolic compounds (57%) [13]. The molecular structures (including stereochemical information) are freely available in the supplementary information of the original publication.
2.3.8 African Antimalarial Natural Products Library (AfroMalariaDB)
The AfroMalariaDB [49] is focused on natural products with antimalarial or antiplasmodial activity confirmed by in vitro and/or in vivo experiments. It consists of approximately 250 natural products collected from more than 130 African plants. Like AfroDb and AfroCancer, AfroMalariaDB is rich in phenols and phenolic compounds [13]. The database is available for download in the supplementary information of the original publication.
2.3.9 Nuclei of Bioassays, Biosynthesis, and Ecophysiology of Natural Products Database (NuBBEDB)
The NuBBE database [50, 51] lists more than 2200 natural products of mainly plants but also fungi, insects, marine organisms, and bacteria native to Brazil. In addition to chemical information, pharmacological and toxicological data are provided. Most of the natural products contained in NuBBEDB are drug-like [50]. Compared to other sources, a low proportion of alkaloids (9%) is observed [50]. The chemistry-aware web interface allows the search for compounds according to structure, spectroscopic information, physicochemical properties, and biological source. Bulk download of structures in MOL2 file format is available.
2.3.10 BIOFACQUIM Database
The BIOFACQUIM database [54] is a manually compiled dataset of natural products isolated and characterized in Mexico. Approximately three-quarters of the 400 natural products currently listed in this database are from plants and 23% are from fungi. The web service offers basic searching functionality and bulk download of all data (molecular structures including stereochemical information).
2.4 Databases Focused on Specific Organisms
2.4.1 Pseudomonas aeruginosa Metabolome Database (PAMDB)
The PAMDB [56] is a rich resource of natural products found in Pseudomonas aeruginosa. The database contains more than 4300 natural products linked to ontology, reaction, and pathway data. The database also provides information on the physicochemical properties of natural products and cross-links to external resources. The PAMDB can be browsed and searched via a chemistry-aware web interface [57]. The web service also offers bulk download of data in various formats.
2.4.2 StreptomeDB 2.0
StreptomeDB 2.0 [58] is a comprehensive database of about 4000 natural products produced by Streptomycetes. The database has been compiled from the literature, the Novel Antibiotics Database [75], and KNApSAcK [76, 77]. The individual molecular structures (including stereochemical information) are annotated with names, Streptomyces species, biological activities, and key physicochemical properties. Approximately one-third of the natural products recorded in StreptomeDB2.0 are not available from any of the other resources that we investigated recently [13]. StreptomeDB2.0 stands out by having one of the largest proportions of natural products containing sugars and sugar-like moieties (25%). Although most of the natural products of StreptomeDB2.0 cover areas in chemical space that are also densely populated with approved drugs, only a relatively small portion of the natural products in this database comply with Lipinski’s rule of five (70%). Noteworthy are a high proportion of alkaloids (47%), although only relatively few of these contain a basic nitrogen (19%). The database can be freely searched and browsed via a chemistry-aware web interface. Bulk download of the data in SD file format with chirality flags is supported.
2.5 Databases Focused on Specific Biological Activities
2.5.1 Database of Natural Products for Cancer Gene Regulation (NPCARE)
The NPCARE database [60] contains more than 6500 natural products with potential anticancer activity measured for a total of approximately 1100 cell lines for 34 cancer types. The natural products in NPCARE originate from more than 2000 plants, marine species, and microorganisms. The provided data include chemical information (including molecular structures with stereochemistry annotations) and information on modulated genes and proteins. The molecular structures of a subset of more than 1500 compounds are available for bulk download (the SMILES notations do not include stereochemical information; however, this information can be retrieved using the PubChem compound identifiers provided).
2.5.2 Naturally Occurring Plant-Based Anti-cancer Compound-Activity-Target Database (NPACT)
The NPACT database [62] is focused on plant-derived natural products with experimentally confirmed cancer-inhibitory activity. The database lists more than 1500 compounds annotated with approximately 5200 compound-cell line and 2000 compound-target interactions. Cross-links with other resources such as the HIT database and PubChem are also provided. The chemistry-aware web interface allows browsing and searching. The molecular structures including stereochemical information can be downloaded from the ZINC database.
2.5.3 InflamNat Database
The InflamNat database [64] contains 665 natural products with experimentally confirmed anti-inflammatory activity. Most natural products (86%) originate from terrestrial plants; a minority comes from marine life, terrestrial fungi, and bacteria. The InflamNat database is rich in flavonoids and triterpenoids. Cross-linking with the PubChem Bioassay database provides information on the biomolecular targets of the natural products. All structures are provided in the supporting information of the publication on InflamNat.
2.6 Databases Focused on Specific Natural Product Classes
2.6.1 Carotenoids Database
The Carotenoids Database [65] contains over 1100 natural carotenoids extracted from almost 700 source organisms. The resource was compiled from the primary literature. The web interface provides access to molecular structures, source organisms, and biological function of the individual carotenoids. The structures of individual carotenoids can be downloaded in various formats (including stereochemical information) but only one molecule at a time.
3 Physical Natural Product Collections
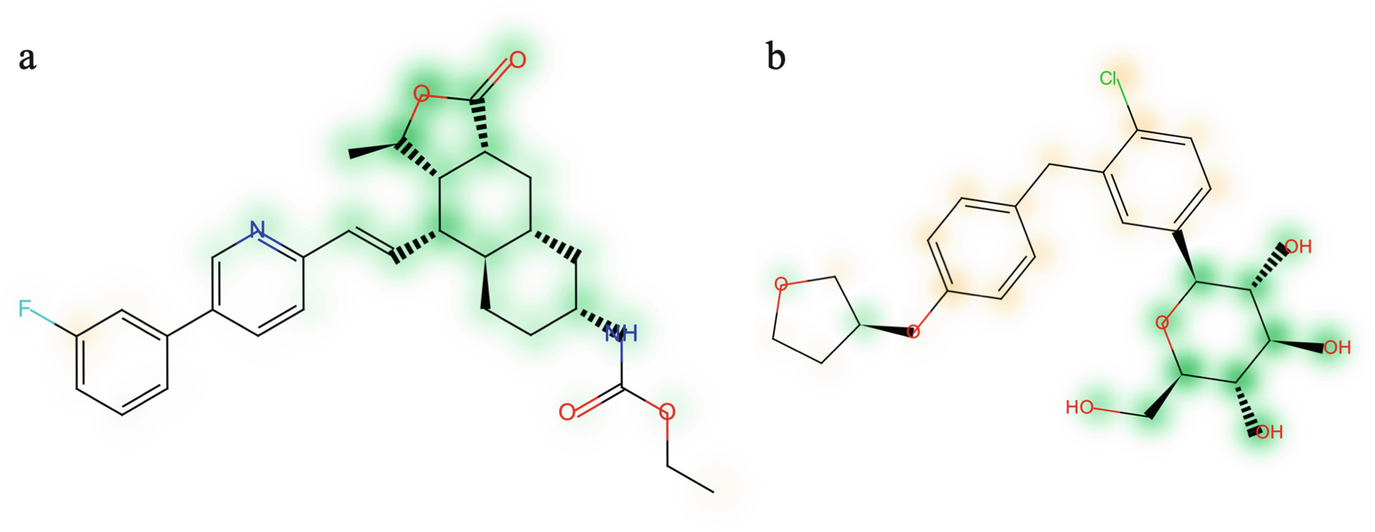
Similarity maps of (a) vorapaxar and (b) empagliflozin. Green-highlighted atoms contribute to the classification of a molecule as a natural product; orange-highlighted atoms contribute to the classification of a molecule as a synthetic compound. Adapted from [78] (CC BY 4.0; https://creativecommons.org/licenses/by/4.0)
Physical natural product collectionsa
Supplier name | (Sub-)set name | Number of compounds | Collection composition | Molecular structures provided free of charge | Web presence |
|---|---|---|---|---|---|
Ambinter and Greenpharma | Natural products | >8000; plated collection of 480 NPs | NPs only | Yes | |
Ambinter and Greenpharma | Natural product derivatives | >11,000 | (Semi-) synthetic compounds | Yes | |
AnalytiCon Discovery | MEGx—Purified natural products of microbial and plant origin | ~5000 | NPs only | Yes | [81] |
AnalytiCon Discovery | NATx—Semi-synthetic natural product-derived compounds | >29,000 | NPs and (semi-) synthetic compounds | Yes | [81] |
AnalytiCon Discovery | MACROx—Next generation macrocycles | >2000 | Semisynthetic compounds based on nine scaffolds | Yes | [81] |
AnalytiCon Discovery | FRGx—Fragments from Nature | >200 | NPs and (semi-) synthetic compounds | Yes | [81] |
Chengdu Biopurify Phytochemicals | TCM Compounds Library | >4600 | NPs and (semi-) synthetic compounds | Yes | [82] |
Selleck Chemicals | Natural Products | ~1600 (plated) | NPs only | Yes | [83] |
TargetMol | Natural Compound Library | >1500 (plated) | NPs only | Yes | [84] |
MedChem Express | Natural Product Library | >1500; plated collection of >900 NPs | NPs only | Yes | [85] |
InterBioScreen | Natural Compound (NC) Collection | >1300 natural compounds and 66,000 derivatives and analogs | NPs and (semi-) synthetic compounds; distinguishable by tags | Yes | [86] |
InterBioScreen | Building Blocks | >13,000 | NPs and (semi-) synthetic compounds | Yes | [86] |
InterBioScreen | Natural Scaffold Libraries | >500 | NPs and (semi-) synthetic compounds | Yes | [86] |
TimTec | Natural Product Library (NPL) | ~800 | NPs only | No | [87] |
TimTec | Natural Derivatives Library (NDL) | ~3000 | NPs and (semi-) synthetic compounds | Yes | [87] |
TimTec | Flavonoids Collection | ~500 | NPs and (semi-) synthetic compounds | Yes | [87] |
TimTec | Flavonoid Derivatives Extended Collection | >4000 | NPs and (semi-) synthetic compounds | Yes | [87] |
TimTec | Gossypol Derivatives Collection | ~100 | NPs and (semi-) synthetic compounds | Yes | [87] |
AK Scientific | Natural Products | ~500 | NPs only | Yes | [88] |
Developmental Therapeutic Program (DTP) of NCI NIH | Natural Products Set IV | ~400 | NPs only | Yes | [89] |
INDOFINE Chemical | Natural Products, Flavonoids, Coumarins, etc. | >4000 | NPs and (semi-) synthetic compounds | Yes | [90] |
Pharmeks | Screening Compounds | >360,000 (>2800 NPs and NP derivatives) | NPs and (semi-) synthetic compounds; distinguishable by tags | Yes | [91] |
Pharmeks | Building Blocks | >12,000 | NPs and (semi-) synthetic compounds | Yes | [91] |
Princeton BioMolecular Research | Macrocycles | >1500 | NPs and (semi-) synthetic compounds | Yes | [92] |
MicroSource Discovery Systems | Natural Products Collection (NatProd) | ~800 | NPs and (semi-) synthetic compounds | Yes | [93] |
Specs | Natural Products | >600 | NPs and (semi-) synthetic compounds | Yes | [94] |
3.1 Pure Natural Product Collections
In this section, we list offerings of pure natural product collections and mixed collections in which genuine natural products are clearly marked and can hence be distinguished from other compounds.
3.1.1 Ambinter and Greenpharma
With more than 8000 listed compounds, the physical natural product collection of Ambinter and Greenpharma [79] is one of the largest offerings available to date. As we have shown previously [13], approximately half of all these natural products are available exclusively from these providers. The collection stands out due to the well-balanced representation of all major natural product classes, which is comparable to that observed for the DNP [13]. Ambinter and Greenpharma also offer a collection of more than 11,000 purchasable natural product derivatives and a preformatted collection of 480 diverse natural products.
3.1.2 AnalytiCon Discovery
AnalytiCon Discovery [81] offers a continuously growing collection of purchasable natural products (“MEGx”). The collection consists of approximately 5000 compounds, the majority of which are available exclusively from this provider [13]. Among the offered compounds are many microbial natural products. The MEGx has the highest proportion of natural products containing sugar or sugar-like fragments among all natural product collections we investigated previously. In contrast, the percentage of alkaloids in this collection is low (14%). AnalytiCon also offers collections of more than 29,000 semisynthetic compounds derived from natural products (“NATx”), over 2000 macrocycles (“MACROx”), and more than 200 fragments from Nature (“FRGx”).
3.1.3 Chengdu Biopurify Phytochemicals
Chengdu Biopurify Phytochemicals [82] offers a collection of over 4600 compounds related to TCM. The collection is rich in flavonoids, alkaloids, phenols, and terpenoids. Many of the natural products are offered exclusively by this provider.
3.1.4 Selleck Chemicals
Selleck Chemicals [83] offers a plated collection of over 1600 natural products for screening. The collection is rich in flavonoids and phenolic natural products, and more than three-quarters of the natural products in this collection comply with Lipinski’s rule of five [13].
3.1.5 TargetMol Collection
TargetMol [84] offers a plated collection of more than 1500 natural products for screening. The compounds originate from plants, animals, microorganisms, and other organisms. Many of the natural products of this collection are active on pharmaceutically relevant proteins.
3.1.6 MedChem Express Collection
The MedChem Express collection [85] offers a diverse ensemble of more than 1500 natural products, including 216 alkaloids, 189 terpenoids and glycosides, 183 acids and aldehydes, 156 flavonoids, and 88 saccharides and glycosides. The company also offers a plated collection of more than 900 natural products for screening.
3.1.7 InterBioScreen Collection
InterBioScreen [86] offers the Natural Compound (NC) collection of purchasable compounds, which contains over 1300 genuine natural products plus 66,000 natural product derivatives (the labels allow the discrimination of genuine natural products from natural product analogs and derivatives). The vast majority of natural products contained in this collection originate from plants, 5 to 10% are isolated from microbes, and another 5% from marine species. The NC collection includes uncommon compounds as well, such as certain classes of phytoalexins, allelopathic agents, and specific sex attractants. In our recent studies, we found that the NC collection features the highest rate of steroids among all investigated natural product databases [13]. Approximately 95% of all compounds of the natural product collection comply with Lipinski’s rule of five. InterBioScreen also offers a collection of over 13,000 building blocks that are partly related to natural products, plus more than 500 natural product scaffolds for compound synthesis.
3.1.8 TimTec Collection
The Natural Product Library (NPL) from TimTec [87] consistes of approximately 800 genuine natural products. These natural products originate primarily from plants, but some have animal, bacterial, or fungal origins. In addition, TimTec offers the Natural Derivatives Library (NDL), which is composed of more than 3000 natural product derivatives, natural product analogs, and semi-natural compounds. A subset of 500 flavonoid derivatives based on nine core flavonoid scaffolds is available, as are an extended collection of over 4000 flavonoid derivatives and a small collection of gossypol derivatives.
3.1.9 AK Scientific Collection
AK Scientific [88] offers a collection of approximately 500 natural products including alkaloids, flavonoids, stilbenoids, terpenoids, and terpenes. The company also provides a subset of synthetic compounds and additives, containing over 100 flavonoids, food preservatives/additives, and vitamins.
3.1.10 Natural Products Set IV of the National Cancer Institute’s Developmental Therapeutic Program (DTP)
The Developmental Therapeutic Program of the National Cancer Institute, National Institutes of Health, provides a plated collection of approximately 400 natural products for experimental screening. These natural products have been selected from 140,000 compounds available from the DTP Open Repository based on compound diversity, availability, and purity. According to our previous analysis [13], more than 60% of these compounds are available exclusively from this source. Approximately 80% comply with Lipinski’s rule of five, which is the lowest among all investigated physical collections. Noteworthy is the high proportion of alkaloids (42%).
3.2 Mixed Collections of Natural Products, Semisynthetic, and Synthetic Compounds
More than 100 vendors offer natural products for experimental testing today, as will be discussed in the next section. However, only a rather small number of vendors explicitly mention the presence of natural products in their mixed physical collections. One of them is INDOFINE Chemical [90], which offers around 4000 natural products and semisynthetic compounds including flavones, isoflavones, flavanones, coumarins, chromones, chalcones, and lipids. The company also has a broad portfolio of synthetic compounds.
Pharmeks [91] offers a diverse, mostly heterocyclic collection of 360,000 organic molecules, 2800 of which are natural products or natural product derivatives. In addition, Pharmeks also offers more than 12,000 building blocks of both synthetic compounds and natural products.
Princeton BioMolecular Research [92] provides a collection of over 1500 macrocyclic natural products, natural product derivatives, and synthetic compounds. MicroSource Discovery Systems [93] offers its Natural Products Collection (“NatProd”), which is composed of 800 natural products and natural product derivatives originating from plant, animal, and microbial sources. Specs [94] offers a collection of over 600 isolated or synthesized natural products and natural product derivatives originating from fungi, bacteria, plants, marine species, and other organisms.
4 Coverage and Reach of Molecular Structures Deposited in Natural Product Collections
As part of one of our previous studies [10], we have analyzed the coverage and reach of 18 virtual natural product databases (marked in Table 1 as included in the analysis published in Ref. [10]) and several physical natural product collections. The number of unique compounds contained in the individual datasets was determined by counting unique InChIs (without stereochemistry and fixed hydrogen layers) derived from neutralized molecules (i.e., counter-ions of salts removed and compounds neutralized with the Wash function in the Molecular Operating Environment (MOE) [95]). Summarized here are some of the most relevant findings of this study.
4.1 Coverage of Free and Commercial Virtual Natural Product Collections
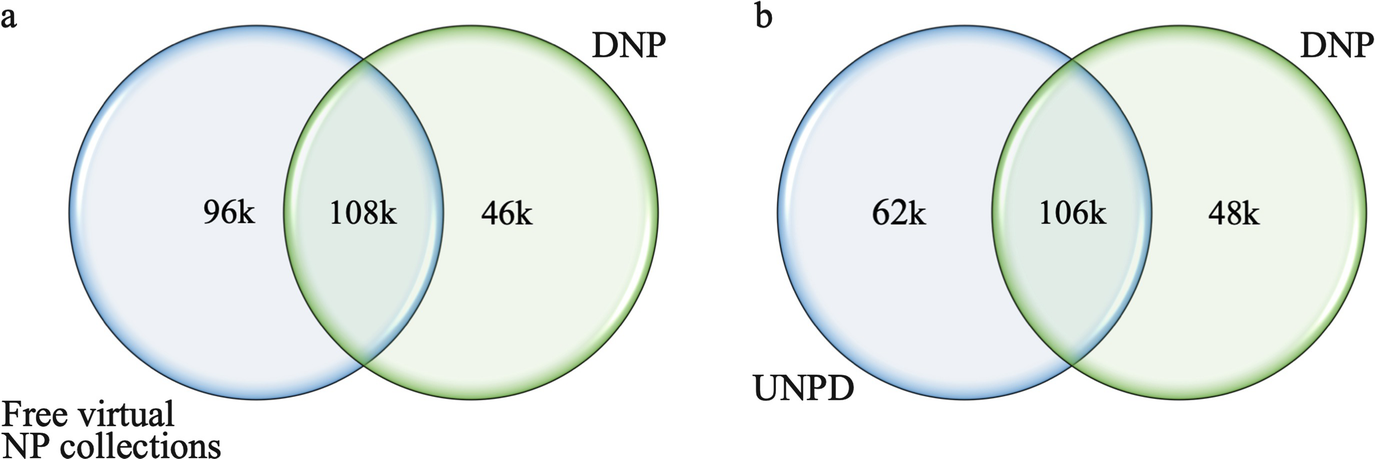
The overlap between the Dictionary of Natural Products (DNP) and (a) the freely accessible virtual natural product collections or (b) the Universal Natural Products Database (UNPD). Reprinted with permission from [10]. Copyright 2017 American Chemical Society
4.2 Readily Obtainable Natural Products and Derivatives
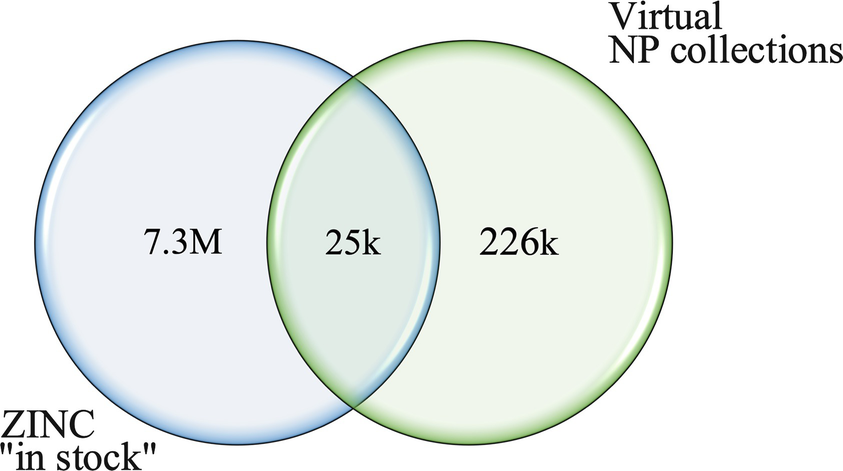
Comparison of the content of virtual natural product collections and the ZINC “in-stock” subset. Reprinted with permission from [10]. Copyright 2017 American Chemical Society
Numbers of natural products readily purchasable from suppliersa
Number of readily purchasable NPs | Suppliers |
|---|---|
>5000 | Molport, TimTec, AK Scientific, Tetrahedron Scientific, BOC Sciences, FineTech Industry, Sigma Aldrich, Specs, National Cancer Institute (NCI) |
3000 to 5000 | Fluorochem, Nanjing Kaimubo Pharmatech Company, Hong Kong Chemhere, Oxchem Corporation, BePharm, Zelinsky Institute, Combi-Blocks, Debye Scientific, Matrix Scientific, WuXi AppTec, Ark Pharm, Bide Pharmatech, BioSynth, InterBioScreen, Labseeker, StruChem, Alfa-Aesar |
2000 to 3000 | AstaTech, Enamine, Oakwood Chemical, Frontier Scientific Services, Alfa Chemistry, Key Organics, Apollo Scientific, W&J PharmaChem, AnalytiCon Discovery, Acros Organics, Shanghai Pi Chemicals, Syntharise Chemical |
1000 to 2000 | Toronto Research Chemicals, Capot Chemical, Rostar, INDOFINE Chemical, Alinda, Pharmeks, Innovapharm, Synthon-Lab, Vesino Industrial, Life Chemicals, Bosche Scientific, Chem-Impex International, Vitas-M Laboratory, Biopurify Phytochemicals, Otava Chemicals, A2Z Synthesis, Cayman Chemical, Accela ChemBio, Molepedia, Curpys Chemicals, ChemDiv, AsisChem |
100 to 1000 | Boerchem Pharmatech, AbovChem, Ryan Scientific, Hangzhou Yuhao Chemical Technology, TargetMol, APExBIO, Princeton BioMolecular Research, EDASA Scientific, ChemBridge, Maybridge, MolMall, HDH Pharma, UORSY, Chemik, Bachem, Creative Peptides, MedChem Express, Aronis, Heteroz, Selleck Chemicals, Tocris, Frinton Laboratories, Asinex, Synchem, EndoTherm Life Science Molecules, Coresyn, SpiroChem, Advanced ChemBlock |
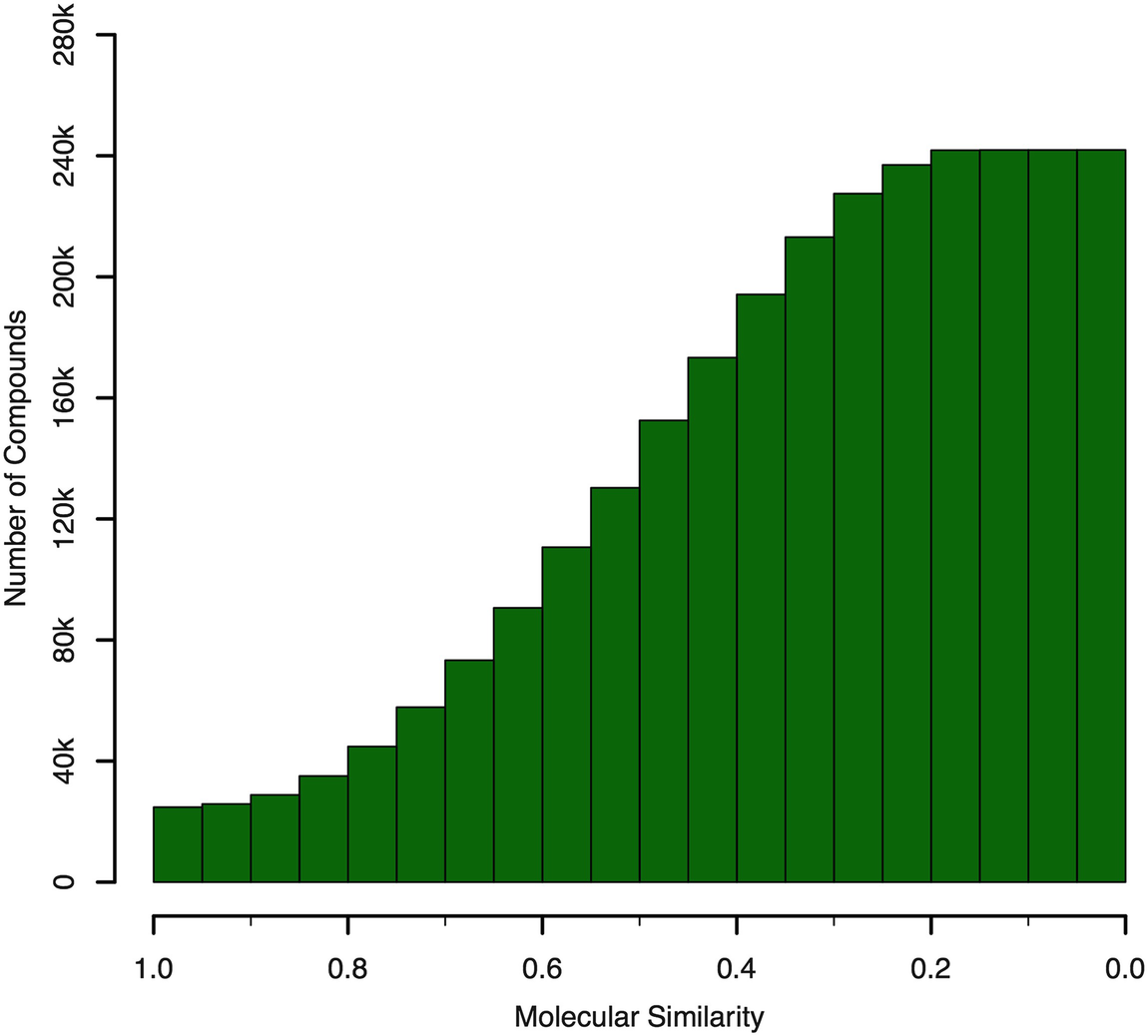
Cumulative histogram of maximum molecular similarity (Tanimoto coefficient) for the compounds in virtual natural product libraries compared to the ZINC “in-stock” subset. The bars in the histogram represent the number of known natural products with a maximum molecular similarity greater than or equal to the bin threshold. Reprinted with permission from [10]. Copyright 2017 American Chemical Society
Macrocycles have gained significant interest in the context of drug discovery in recent years. Due to their conformational constraints, macrocycles can provide advantages in entropic binding and specificity [98]. Our analysis has shown that approximately 14% (35,000) of all 250,000 known natural products contain rings formed by more than seven atoms. However, only approximately 800 genuine natural products with a ring size larger than seven atoms are readily obtainable (note that, e.g., AnalytiCon offers more than 2000 semisynthetic, macrocyclic compounds based on nine scaffolds).
5 Resources for Biological Data on Natural Products
The majority of virtual natural product databases provide biological information in addition to chemical data (Table 1). Most of this information is in the form of bioactivities measured for organisms, cells, or individual biomacromolecules. Several resources provide information on pathways, diseases, and ADME properties.
The ChEMBL [99] database is one of the most comprehensive sources of measured biological activities of small molecules. The database is manually compiled primarily from scientific publications. It also draws information from other sources such as the PubChem Bioassay database [100, 101]. The latest version of the ChEMBL database counts over 1.8 million distinct compounds annotated with more than 15.2 million activity records on a total of more than 12,000 targets. In our recent analysis, we found that approximately 16% (40,000) of known natural products are contained in ChEMBL [10].
6 Resources for Structural Data on Natural Products
The Cambridge Structural Database (CSD) [102] provides a wealth of information on the three-dimensional structures of small-molecule organic and metal-organic compounds. Currently, the database is approaching the milestone of storing 1 million structures derived by X-ray and neutron diffraction analysis.
Structural information of natural products bound to their biomacromolecular targets is available from the Protein Data Bank (PDB) [103] but remains sparse. We found that for approximately 2000 natural products at least one X-ray crystal structure in complex with a biomacromolecule is deposited in the PDB [13]. A small number of structures of protein-bound macrocyclic natural products are also available [104].
7 Conclusions
During the last few years, the chemical, biological, and structural information available on natural products has increased substantially. Today, the molecular structures of several hundred thousand natural products are available from a large number of different sources. In particular, natural products from botanical sources are to a large part covered by subscription-free resources that permit bulk export or download of data, allowing an array of different cheminformatics methods to be employed in the context of drug discovery. It is important to mention that the quality and quantity of the information provided by the individual sources vary substantially. For example, not all sources provide information on stereochemical properties, which in fact are often incomplete or inaccurate for natural products anyway. To the best of our knowledge, there have been no systematic studies on the quality of the data provided by natural product databases. This would, of course, be an important aspect to examine further.
Measured data on biological activities and ADME properties are becoming increasingly available, whereas structural information on natural products bound to their biomacromolecular target remain sparse. The bottleneck for drug discovery continues to be the availability of material for experimental testing. It is estimated that only about 10% (25,000) of all known natural products are readily obtainable from commercial and other sources. However, a substantially higher number of natural product-like compounds are readily obtainable.
In the coming years, we expect a further increased growth rate for chemical, biological, and structural data on natural products. In particular, we expect resources providing free access and bulk data download to play an ever more important role. One major challenge is to develop strategies for the sustainability of such valuable sources. What is seen today, unfortunately, is that many databases are no longer maintained after they have been reported in the scientific literature, and there are many examples of resources that go offline even within 1 year after their launch. This phenomenon is, of course, not specific to natural product databases but part of a general and largely unsolved problem.
Despite the remaining challenges, the large amount of data on natural products available today enables investigators to effectively employ computational methods and make substantial contributions to natural product-based drug discovery.
Funding Sources
Ya Chen is supported by the China Scholarship Council (201606010345). Johannes Kirchmair is supported by the Bergen Research Foundation (BFS)—grant no. BFS2017TMT01.


