1 Introduction
 and
and  of size
of size  and
and  , respectively, coming from some non-negative discrete exponential family with natural parameter
, respectively, coming from some non-negative discrete exponential family with natural parameter  , base measure
, base measure  and normalising constant
and normalising constant 

 of size
of size  to test
to test
 [10]. Moreover, T is a sufficient statistic for the nuisance parameter of the test, the population constant
[10]. Moreover, T is a sufficient statistic for the nuisance parameter of the test, the population constant  , if we assume the standard one-way ANOVA model for the means
, if we assume the standard one-way ANOVA model for the means  [11].
[11]. and its conditional distribution given T under
and its conditional distribution given T under  in (1) is
in (1) is
 the set of non-negative integer vectors of length n with sum of entries equal to x.
the set of non-negative integer vectors of length n with sum of entries equal to x.In order to perform the test (1), we can either find the critical values for any given risk of type I error or, alternatively, compute the p-value corresponding to the observed value  of U. Unfortunately, the distribution (2) can rarely be computed in closed form. In most cases, it is necessary to approximate (2) through Markov Chain Monte Carlo (MCMC).
of U. Unfortunately, the distribution (2) can rarely be computed in closed form. In most cases, it is necessary to approximate (2) through Markov Chain Monte Carlo (MCMC).
MCMC sampling methods suffer two major drawbacks in the discrete setting: the construction of the Markov basis needed to build the chain is computationally expensive and the chain may mix slowly [8]. The idea of speeding-up the MCMC sampling is therefore not new in the Algebraic Statistics literature. In [7], the first drawback is addressed in the case of bounded contingency tables, instead of computing the entire Markov basis in an initial step, sets of local moves that connect each table in the reference set with a set of neighbouring tables are studied. A similar approach for bounded two-way contingency tables under the independence model with positive bounds is presented in [13]. A hybrid scheme using MCMC and sequential importance sampling able to address both drawbacks has been proposed in [8]. We propose a strategy that exploits the structure of the sample space for UMPU tests and does not require sequential importance sampling but only standard independent Monte Carlo sampling.
2 Markov Chain Monte Carlo Samplings
 , the sample space to be inspected under
, the sample space to be inspected under  , is the fibre of non-negative integer vectors of size
, is the fibre of non-negative integer vectors of size  and with entries which add up to t
and with entries which add up to t
 is the vector of length N with all entries equal to 1.
is the vector of length N with all entries equal to 1. given
given  under
under  as specified in (1)
as specified in (1)
 and
and  is 1 if
is 1 if  and 0 otherwise and
and 0 otherwise and  with a slight abuse of notation.
with a slight abuse of notation. . The first one samples vectors
. The first one samples vectors  , while the second one samples orbits of permutations
, while the second one samples orbits of permutations  . Both MCMC algorithms make use of a Markov basis, a set of moves allowing to build a connected Markov chain over
. Both MCMC algorithms make use of a Markov basis, a set of moves allowing to build a connected Markov chain over  using only simple additions/subtractions
[6]. Specifically, a Markov basis for a matrix A is a finite set of moves
using only simple additions/subtractions
[6]. Specifically, a Markov basis for a matrix A is a finite set of moves  such that
such that - 1.
 belongs to the integer kernel of A,
belongs to the integer kernel of A,  ;
; - 2.
every pair of elements
 in
in  is connected by a path formed by a sequence
is connected by a path formed by a sequence  of moves
of moves  and signs
and signs  , and this path is contained in
, and this path is contained in  .
.
Markov bases can be found analytically [5, 6] or using the algebraic software 4ti2 [1].
2.1 MCMC—Vector-Based
 .
.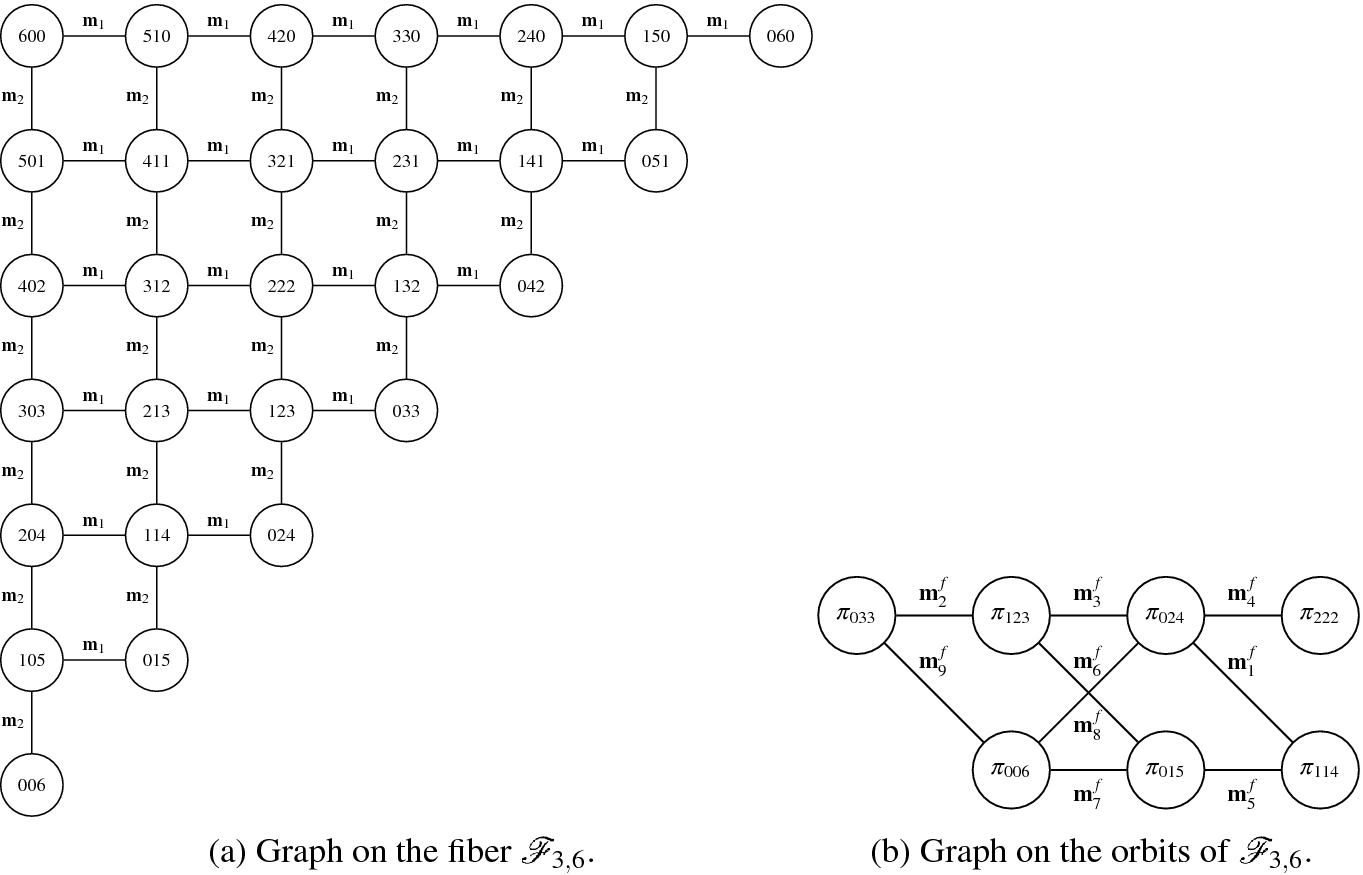
Vector-based and orbit-based parametrisation of the fibre 
2.2 MCMC—Orbit-Based
The second algorithm is built by exploiting the link of  with orbits of permutations
with orbits of permutations  . Clearly, if
. Clearly, if  , every permutation of
, every permutation of  is an element of
is an element of  too. Moreover, different orbits do not intersect. Therefore the orbits of permutations
too. Moreover, different orbits do not intersect. Therefore the orbits of permutations  form a partition of
form a partition of  .
.
This partition is particularly interesting, as the elements which belong to the same orbit have the same probability of being sampled from  ,
[12].
,
[12].
- Step 1:
Sample one orbit
 from the set of orbits
from the set of orbits  .
.- Step 2:
Sample uniformly from
 .
.
 in orbit
in orbit 

 .
. contained in the fibre is given by
contained in the fibre is given by  [5], with
[5], with  defined in
[9, 14]. The values of the partition function can be computed using the recurrence
defined in
[9, 14]. The values of the partition function can be computed using the recurrence
To perform Step 1, we parametrise the fibre  in such a way that all the vectors in the same permutation orbit are mapped to the same element. To do so, we consider a frequency-based representation. In this representation the orbit
in such a way that all the vectors in the same permutation orbit are mapped to the same element. To do so, we consider a frequency-based representation. In this representation the orbit  is mapped into
is mapped into  . In this notation, vectors (0, 4, 2) and (2, 0, 4), which belong to the same orbit, correspond to the same frequency vector.
. In this notation, vectors (0, 4, 2) and (2, 0, 4), which belong to the same orbit, correspond to the same frequency vector.

 being the number of distinct elements in the orbit
being the number of distinct elements in the orbit  .
. , i.e. the usual permutation cdf,
, i.e. the usual permutation cdf,
3 Comparison of Vector-Based and Orbit-Based MCMC
 , which is faster than performing an MCMC sampling. On the other hand, Step 1 performs an MCMC sampling over the set of orbits
, which is faster than performing an MCMC sampling. On the other hand, Step 1 performs an MCMC sampling over the set of orbits  contained in
contained in  , whose cardinality is smaller than that of the set of vectors in
, whose cardinality is smaller than that of the set of vectors in  :
:
 and the cardinality of
and the cardinality of  for values of N and t between 5 and 100. Even for moderately sized samples, the number of orbits contained in
for values of N and t between 5 and 100. Even for moderately sized samples, the number of orbits contained in  is about two orders of magnitude smaller than the number of vectors (e.g. for
is about two orders of magnitude smaller than the number of vectors (e.g. for  and
and 
 ).
). the number of distinct vectors in the orbit sampled at iteration i.
the number of distinct vectors in the orbit sampled at iteration i.Ratio between the number of orbits  and the number of vectors
and the number of vectors  in
in  for several values of N and t
for several values of N and t
| 5 | 10 | 15 | 20 | 30 | 50 | 100 |
|---|---|---|---|---|---|---|---|
5 | 0.056 | 0.030 | 0.022 | 0.018 | 0.015 | 0.012 | 0.010 |
10 | 0.004 |
|
|
|
|
|
|
15 |
|
|
|
|
|
|
|
20 |
|
|
|
|
|
|
|
50 |
|
|
|
|
|
|
|
We show how the reduction in the dimension of the space explored by the MCMC algorithm improves convergence and accuracy with respect to the truth (4) through the simulation study in the next section.
4 Simulation Study
 and
and  are Poisson distributed with mean
are Poisson distributed with mean  and
and  , respectively. In this case, the exact distribution (4) under
, respectively. In this case, the exact distribution (4) under  is the binomial distribution
is the binomial distribution
 [10, 11].
[10, 11].We compare the exact conditional cdf above with the approximated cdfs given by the vector-based algorithm, the orbit-based algorithm and the standard permutation cdf over the orbit of the observed pooled vector (this is the limit case of the orbit-based algorithm when only the observed orbit is sampled). A preliminary simulation study is presented in [4].
We consider 9 scenarios built taking three sample sizes  and, for each sample size, three different couples of population means
and, for each sample size, three different couples of population means  .
.
The comparisons performed are two: first, we check the convergence behaviour on a fixed runtime (15 s) for both algorithms; then we compare their accuracy through the mean squared error (MSE).
4.1 Convergence Comparison
 for each scenario above and we run both algorithms for 15 s. Figure 2 shows four examples of the behaviour of the two MCMC procedures which are representative of the nine scenarios.
for each scenario above and we run both algorithms for 15 s. Figure 2 shows four examples of the behaviour of the two MCMC procedures which are representative of the nine scenarios.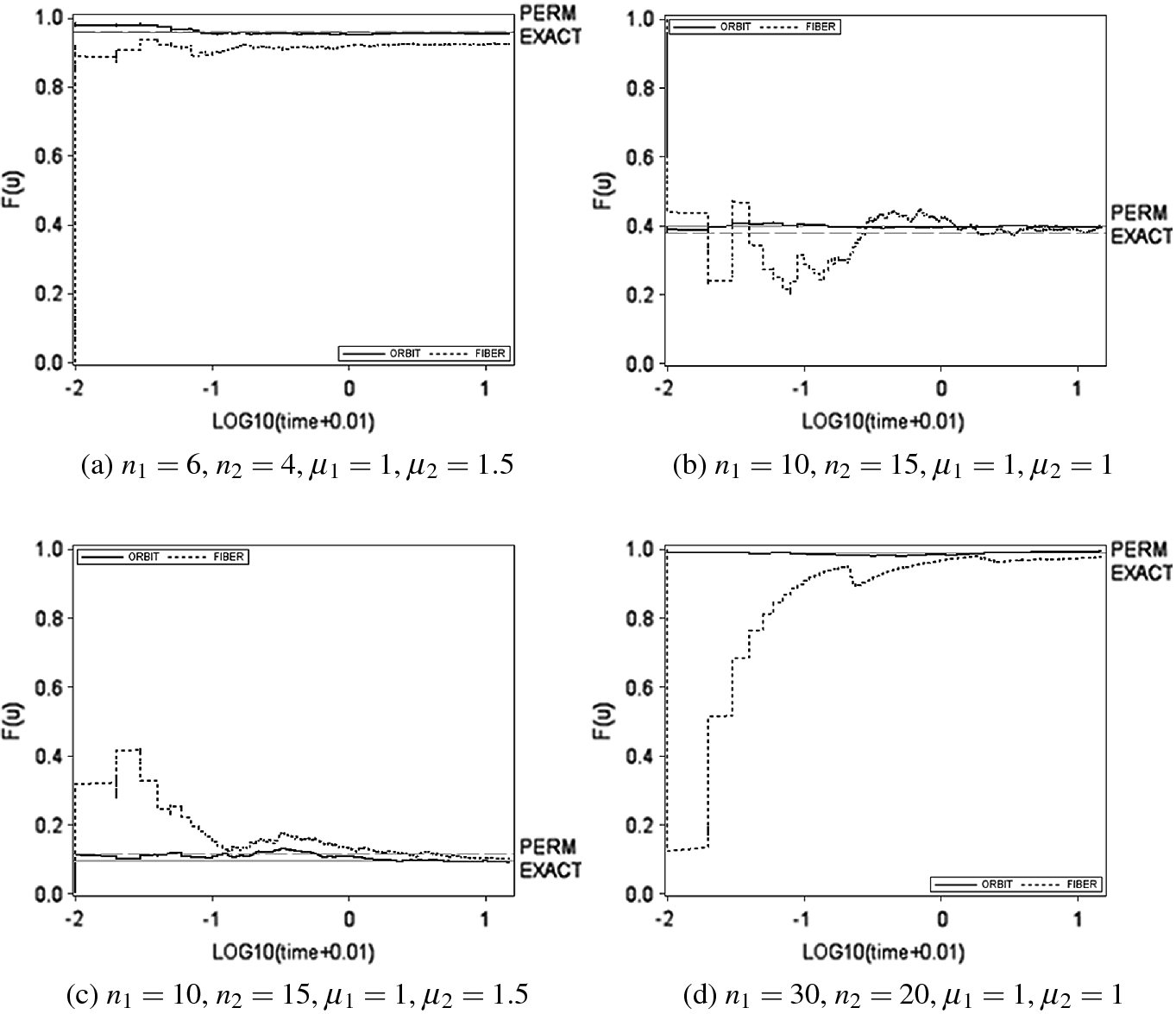
Comparison of the convergence to the exact value (solid horizontal line) in 15 s for the vector-based algorithm (dashed line) and the orbit-based algorithm (solid line). The Monte Carlo permutation estimate of  (dashed horizontal line) is reported too. The number of Monte Carlo permutations per orbit is 5, 000. The plots show the estimates achieved as functions of the log-time
(dashed horizontal line) is reported too. The number of Monte Carlo permutations per orbit is 5, 000. The plots show the estimates achieved as functions of the log-time
The orbit-based algorithm is very fast and achieves good convergence in  seconds. On the contrary, the vector-based algorithm is much less efficient, in fact, its convergence to the exact value is not always satisfactory even after 15 s (Fig. 2a, b).
seconds. On the contrary, the vector-based algorithm is much less efficient, in fact, its convergence to the exact value is not always satisfactory even after 15 s (Fig. 2a, b).
It would be possible to further reduce the computational time required by the orbit-based algorithm by exploiting one of the key features of this new approach, namely the possibility of sampling from each orbit independently. The Monte Carlo samplings in Step 2 could be made in parallel: once the chain reaches an orbit  the Monte Carlo sampling over
the Monte Carlo sampling over  can be performed while the chain keeps on moving on the set of orbits.
can be performed while the chain keeps on moving on the set of orbits.
4.2 Accuracy Comparison

Mean Squared Error (MSE) for the vector-based, the orbit-based and the Monte Carlo permutation sampling. Both MCMC algorithms were run for 15 s with no burn-in steps
|
|
|
| Orbit-based | Vector-based | Permutation |
|---|---|---|---|---|---|---|
6 | 4 | 1 | 1 | 0.00012 | 0.0016 | 0.00284 |
6 | 4 | 1 | 1.5 | 0.00012 | 0.00083 | 0.00212 |
6 | 4 | 1 | 2 | 0.00016 | 0.00043 | 0.00221 |
10 | 15 | 1 | 1 | 0.00034 | 0.00131 | 0.00077 |
10 | 15 | 1 | 1.5 | 0.00009 | 0.00046 | 0.00074 |
10 | 15 | 1 | 2 | 0.00007 | 0.00017 | 0.00057 |
20 | 30 | 1 | 1 | 0.00069 | 0.00132 | 0.00036 |
20 | 30 | 1 | 1.5 | 0.00006 | 0.00053 | 0.00027 |
20 | 30 | 1 | 2 | 0.00001 | 0.00011 | 0.00009 |
 ,
,  ,
,  ,
,  , where the standard Monte Carlo permutation sampling has the smallest MSE. Table 3 shows the ratio between the MSE of the vector-based algorithm and the MSE of the orbit-based algorithm (column 5) and the ratio between the MSE of the standard Monte Carlo permutation sampling and the MSE of the orbit-based algorithm (column 6). The MSE of the vector-based algorithm is at least 1.9 times bigger than that of the orbit-based algorithm, while the MSE of the standard Monte Carlo permutation sampling can be 22.7 times bigger than that of the orbit-based algorithm (scenario 1).
, where the standard Monte Carlo permutation sampling has the smallest MSE. Table 3 shows the ratio between the MSE of the vector-based algorithm and the MSE of the orbit-based algorithm (column 5) and the ratio between the MSE of the standard Monte Carlo permutation sampling and the MSE of the orbit-based algorithm (column 6). The MSE of the vector-based algorithm is at least 1.9 times bigger than that of the orbit-based algorithm, while the MSE of the standard Monte Carlo permutation sampling can be 22.7 times bigger than that of the orbit-based algorithm (scenario 1).Ratio between the MSE of the vector-based and the MSE of the orbit-based algorithm (column 5) and ratio between the MSE of the standard Monte Carlo permutation sampling and the MSE of the orbit-based algorithm (column 6).
|
|
|
| MSE vector/MSE orbit | MSE perm/MSE orbit |
|---|---|---|---|---|---|
6 | 4 | 1 | 1 | 12.82 | 22.7 |
6 | 4 | 1 | 1.5 | 6.98 | 17.79 |
6 | 4 | 1 | 2 | 2.67 | 13.82 |
10 | 15 | 1 | 1 | 3.9 | 2.31 |
10 | 15 | 1 | 1.5 | 4.96 | 8 |
10 | 15 | 1 | 2 | 2.45 | 8.27 |
20 | 30 | 1 | 1 | 1.9 | 0.52 |
20 | 30 | 1 | 1.5 | 9.03 | 4.58 |
20 | 30 | 1 | 2 | 15.9 | 12.77 |
Number of iterations for 15 s
Scenario | N. iterations | Ratio | ||||
|---|---|---|---|---|---|---|
|
|
|
| Orbit | Vector | Vector/Orbit |
6 | 4 | 1 | 1 | 23,977 | 53,842 | 2.25 |
6 | 4 | 1 | 1.5 | 24,169 | 53,210 | 2.20 |
6 | 4 | 1 | 2 | 24,560 | 57,382 | 2.34 |
10 | 15 | 1 | 1 | 11,950 | 52,504 | 4.39 |
10 | 15 | 1 | 1.5 | 11,564 | 54,836 | 4.74 |
10 | 15 | 1 | 2 | 7326 | 53,492 | 7.30 |
20 | 30 | 1 | 1 | 4675 | 45,576 | 9.75 |
20 | 30 | 1 | 1.5 | 3174 | 44,817 | 14.12 |
20 | 30 | 1 | 2 | 2572 | 48,003 | 18.66 |
5 Conclusions
The orbit-based algorithm grants a faster convergence to the exact distribution if compared to the standard MCMC algorithm proposed in [6]. At the same time, it gives more reliable estimates by decreasing the MSE. This simulation-based observation can be proved by comparing the variance of the estimators used by the two algorithms (the indicator function in (4) and the permutation cdf (5) respectively) [5].
When permutation-invariant statistics are used, the orbit-based algorithm is dramatically simplified. In this case, it is only necessary to walk among orbits of permutations without performing the second-step sampling and thus the reduction in computational time is significant.
Finally, it is worth noting that the MCMC sampling procedure based on orbits of permutations establishes a link between standard permutation and algebraic-statistics-based sampling that, to the best of our knowledge, has not been previously noted.
A preliminary version of this work has been presented at the 4th ISNPS conference in June 2018 in Salerno, Italy. Extensions of the present work include hypothesis testing for  groups and data fitting
[5].
groups and data fitting
[5].
R. Fontana acknowledges that the present research has been partially supported by MIUR grant Dipartimenti di Eccellenza 2018–2022 (E11G18000350001). The authors thank the anonymous referee for his/her helpful comments.

















































