1 Introduction
Independent and identically distributed residuals are a key assumption in many statistical analysis models. When analyzing spatial, i.e. geolocated data, this assumption is violated if one fails to account for the existence of spatial dependence in the modelling components. Such violation can lead to biased parameter estimates and spurious associations between the dependent variable and its covariates. Therefore, it is important to take spatial dependence into modelling consideration when it exists. Spatially dependent data arise in many research domains, for example in spatial epidemiology where researchers are interested in the relationship between disease prevalence and risk factors [9, 23], in economics where it is of interest to identify regions of housing externalities [15] or in ecological studies where species distribution needs to be mapped [10, 17]. The scales of spatial dependence also play an important role in understanding the underlying physical and biological processes [10, 17].
The assessment of spatial dependence and its scales is often done by plotting and modelling the empirical semi-variogram estimates, based on extracted residuals after first-stage statistical modelling. If the empirical semi-variogram indicates spatial dependence, then the model needs to be adjusted to account for the remaining dependence. However, semi-variogram estimates can be sensitive to outliers, choice of distance binning and sampling design. Several robust variogram estimators [4, 7] and methods to quantify uncertainty of variogram estimates [3, 5, 12] are available. One can use a maximum likelihood estimator and its uncertainty estimate to assess the spatial dependence, or use parametric bootstrap [16] by firstly fitting a variogram model and simulate new values based on the estimated model to obtain additional variogram estimates hence an uncertainty estimate. However, both approaches require a pre-defined variogram model and a sufficient sample size. An alternative approach to assess the existence of spatial dependence is to use a Monte Carlo permutation test. The permutation test is a nonparametric approach that does not make any assumptions on the distribution of residuals. Walker et al. [21] introduced a permutation test to permute the residual values across spatial locations, in order to simulate under the null hypothesis of complete spatial randomness. Diblasi and Bowman [5] compared the performance of a permutation test on their proposed test statistics based on the assumption of normally distributed residuals. Viladomat et al. [20] used a permutation method in a two-step procedure to test the correlation of two variables when they both exhibit spatial dependence.
In this paper, we propose two Monte Carlo permutation tests to assess the existence of overall spatial dependence and spatial dependence specifically at small scales, respectively. We demonstrate that our proposed methods have more accurate type I error rate compared to the standard permutation test in [21] and achieve good statistical power at the same time.
2 Assessing Spatial Dependence at Different Scales
 be a zero-mean second-order stationary spatial process that is observed at coordinates s, then the semi-variogram is defined as
be a zero-mean second-order stationary spatial process that is observed at coordinates s, then the semi-variogram is defined as

![$$[h - \delta ,h + \delta ]$$](../images/461444_1_En_45_Chapter/461444_1_En_45_Chapter_TeX_IEq2.png) . In practice, different distance binnings
. In practice, different distance binnings  are used to obtain empirical semi-variogram estimates
are used to obtain empirical semi-variogram estimates  for
for 
2.1 Permutation Test for Overall Spatial Dependence

 is the number of Monte Carlo iterations and
is the number of Monte Carlo iterations and  is the dth semi-variogram estimates from the ith permuted samples. Walker et al.
[21] compared the p-values in Eq. (3) with a type I error rate
is the dth semi-variogram estimates from the ith permuted samples. Walker et al.
[21] compared the p-values in Eq. (3) with a type I error rate  , and deemed that the null hypothesis is rejected if any p-value is below
, and deemed that the null hypothesis is rejected if any p-value is below  . This approach implicitly used the p-value combination method proposed in
[19], which takes the overall p-value
. This approach implicitly used the p-value combination method proposed in
[19], which takes the overall p-value  and compares it with
and compares it with  . When the evidence of spatial dependence is relatively strong, the p-values tend to be small. In such cases, the null hypothesis will be rejected as long as one of the p-values is smaller than
. When the evidence of spatial dependence is relatively strong, the p-values tend to be small. In such cases, the null hypothesis will be rejected as long as one of the p-values is smaller than  . However, when none of the p-values are smaller than
. However, when none of the p-values are smaller than  under settings of weak spatial dependence, the rejection region of using overall p-value
under settings of weak spatial dependence, the rejection region of using overall p-value  is smaller than using other p-value combination methods (e.g.
[6, 11]). This will lead to smaller statistical power. In addition, using the minimum p-value can inflate the type I error which leads to spurious spatial dependence. To mitigate these problems, we propose a modified permutation test for overall spatial dependence.
is smaller than using other p-value combination methods (e.g.
[6, 11]). This will lead to smaller statistical power. In addition, using the minimum p-value can inflate the type I error which leads to spurious spatial dependence. To mitigate these problems, we propose a modified permutation test for overall spatial dependence.2.2 Modified Permutation Test for Overall Spatial Dependence
 -distributed test statistic. If we assume the test statistics
-distributed test statistic. If we assume the test statistics  are mutually independent, then the Fisher’s method states
are mutually independent, then the Fisher’s method states
 . For locations
. For locations  , let
, let  . The dth semi-variogram then follows a scaled
. The dth semi-variogram then follows a scaled  -distribution with
-distribution with  degrees of freedom, i.e.
degrees of freedom, i.e.
 is idempotent and has rank
is idempotent and has rank  [1]. The matrix
[1]. The matrix  is the spatial design matrix of the data at lag d
[7]. For non-gridded locations, the matrix is close to idempotent if the number of semi-variogram estimate is not too small, which yields Eq. (5) as a good approximation. For moderate to large ranks,
is the spatial design matrix of the data at lag d
[7]. For non-gridded locations, the matrix is close to idempotent if the number of semi-variogram estimate is not too small, which yields Eq. (5) as a good approximation. For moderate to large ranks,  . Therefore, we can approximate the vector of test statistics
. Therefore, we can approximate the vector of test statistics ![$$[\gamma (h_1), \gamma (h_2), \dots , \gamma (h_k)]^\top $$](../images/461444_1_En_45_Chapter/461444_1_En_45_Chapter_TeX_IEq26.png) with a multivariate normal distribution. The Brown’s method
[2] allows us to derive a scaled
with a multivariate normal distribution. The Brown’s method
[2] allows us to derive a scaled  -distribution to replace
-distribution to replace  from the Fisher’s method. The overall test statistic stays as
from the Fisher’s method. The overall test statistic stays as  ; however, the distribution under the null hypothesis becomes a scaled chi-squared distribution
; however, the distribution under the null hypothesis becomes a scaled chi-squared distribution  , with
, with
 and
and  [13]. The latter has been shown to be more robust compared to polynomial approximation when there is deviation from normality in the test statistics. In our modified permutation test, we use the empirical Brown’s method to combine the p-values generated by Monte Carlo permutations into an overall test statistic and compare it with
[13]. The latter has been shown to be more robust compared to polynomial approximation when there is deviation from normality in the test statistics. In our modified permutation test, we use the empirical Brown’s method to combine the p-values generated by Monte Carlo permutations into an overall test statistic and compare it with  .
.2.3 Permutation Test for Spatial Dependence at Small Scales
Sometimes the existence of scale-specific spatial dependence is a more meaningful hypothesis to test against. We propose a permutation test to permute the residuals in a way such that only small-scale dependence is destroyed.
Instead of randomly permuting the residuals over all spatial locations, we first apply a clustering algorithm on the locations to divide them into small clusters. The clustering algorithm should not result in clusters that have a high variance in size. Popular algorithms such as k-means or hierarchical clustering can be used, or simply hex-binning when the locations are evenly distributed over the spatial domain. After clusters are defined, the residuals are randomly permuted only within each cluster. The null hypothesis then concerns only the first few semi-variogram estimates at small scale. The clustering algorithm should be tuned depending on the scale of interest. Since there are still correlations among the pointwise p-values, we use the empirical Brown’s method to combine them to get an overall test statistic.
 . In this case, spatial dependence only exists at small scales. The 95% confidence band shown in light blue is based on random permutation where all of the spatial dependence is destroyed. The 95% confidence band in red is based on cluster permutation where only the small-scale spatial dependence is destroyed. The latter allows more powerful hypothesis testing focusing only on a small scale, as we will show in the simulation results.
. In this case, spatial dependence only exists at small scales. The 95% confidence band shown in light blue is based on random permutation where all of the spatial dependence is destroyed. The 95% confidence band in red is based on cluster permutation where only the small-scale spatial dependence is destroyed. The latter allows more powerful hypothesis testing focusing only on a small scale, as we will show in the simulation results.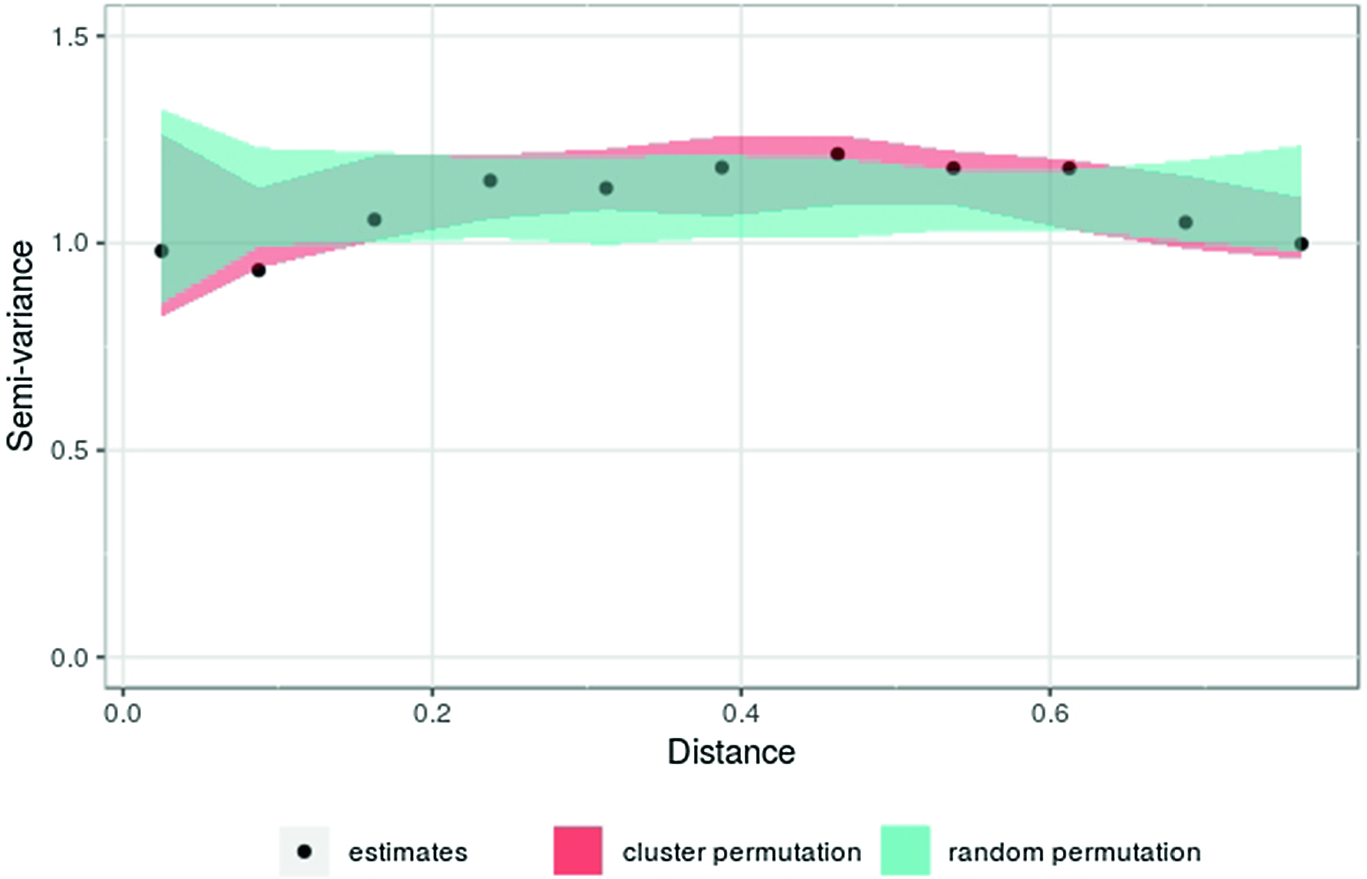
Semi-variogram estimates based on simulated residuals. Ribbons indicate 95% confidence band of permutation samples. The original and modified permutation test for overall spatial dependence provided p-values of 0.001 and 0.078, respectively, while the cluster permutation method provided an overall p-value of 0.027
3 Simulation Study
We conduct a simulation study to compare the permutation tests in terms of type I error rate and statistical power of detecting spatial dependence. The null hypothesis of random permutation is that there is no spatial dependence, while the null hypothesis of cluster permutation is that there is no spatial dependence at small scales.
3.1 Simulation Setup
We simulate a Gaussian process on n uniformly distributed locations within a ![$$[0,1]\times [0,1]$$](../images/461444_1_En_45_Chapter/461444_1_En_45_Chapter_TeX_IEq35.png) domain. Without loss of generality, we assume the Gaussian process has an exponential covariance function from the Matérn family, i.e.
domain. Without loss of generality, we assume the Gaussian process has an exponential covariance function from the Matérn family, i.e.  , where
, where  denotes the Euclidean distance between locations
denotes the Euclidean distance between locations  and
and  . The magnitude of spatial covariance is represented by
. The magnitude of spatial covariance is represented by  , the magnitude of noise is
, the magnitude of noise is  . The spatial range
. The spatial range  controls the covariance decay over distance, which corresponds to an effective range of
controls the covariance decay over distance, which corresponds to an effective range of  for the exponential covariance function. Hence, 95% of the spatial correlation disappears at distance
for the exponential covariance function. Hence, 95% of the spatial correlation disappears at distance  .
.
We simulate a total of 10 different spatial dependence structures each with 5 different sample sizes, where  ,
,  and
and  . Different
. Different  represents differing strengths of spatial dependence. We choose
represents differing strengths of spatial dependence. We choose  and repeat each scenario 1000 times. For the cluster permutation, we use k-means clustering with 5 clusters when
and repeat each scenario 1000 times. For the cluster permutation, we use k-means clustering with 5 clusters when  , and with 10 clusters for all other sample sizes. The null hypothesis is that the first two semi-variogram estimates are 0, which corresponds to no spatial dependence at a distance smaller than approximately 0.1. All the simulation results are obtained in R version 3.5
[14].
, and with 10 clusters for all other sample sizes. The null hypothesis is that the first two semi-variogram estimates are 0, which corresponds to no spatial dependence at a distance smaller than approximately 0.1. All the simulation results are obtained in R version 3.5
[14].
3.2 Simulation Results
Figure 2 shows the simulation results. The permutation test for overall spatial dependence (denoted as random+min) using minimum p-value is shown in solid lines; the modified permutation test for overall spatial dependence with empirical Brown’s method (denoted as random+ebm) is shown in dotted lines; the permutation test for spatial dependence at small scales with empirical Brown’s method (denoted as cluster+ebm) is shown in dashed lines.
 , the power of the clustering-based permutation test is higher than the modified permutation test.
, the power of the clustering-based permutation test is higher than the modified permutation test.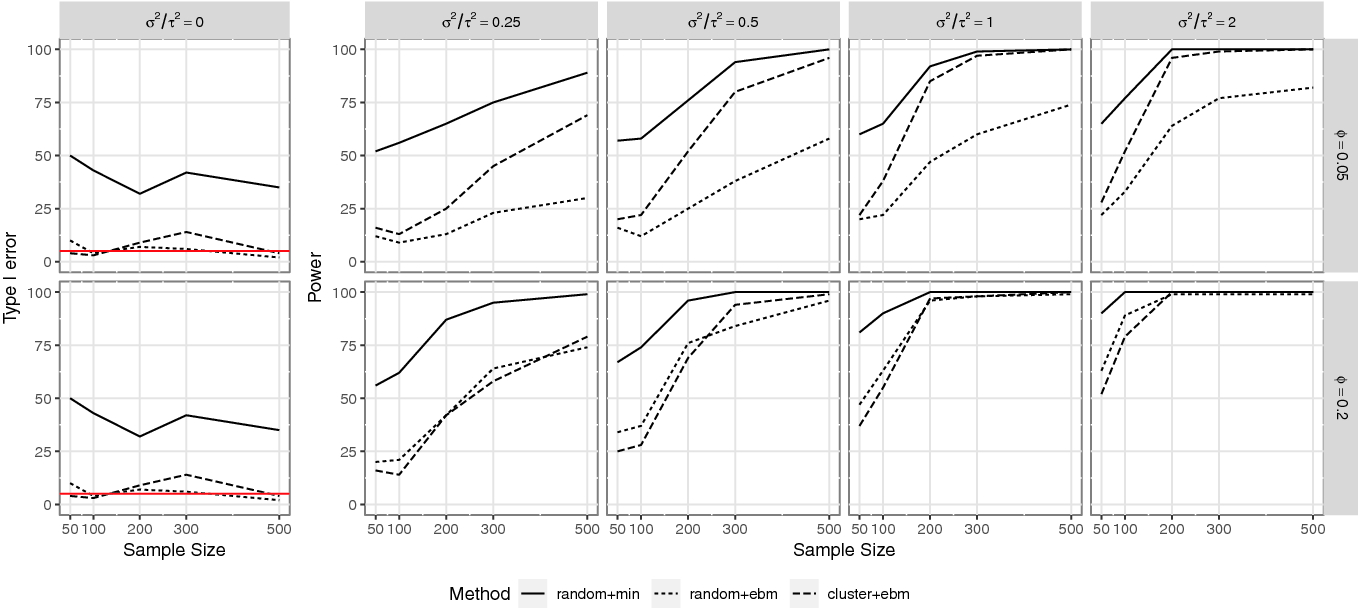
Simulation results showing type I error rate (first column) and statistical power (other columns) of three different permutation tests for different scenarios. The horizontal red line in the first column shows the nominal type I error rate at 5%
4 Discussion and Outlook
This paper presents two new approaches of testing spatial dependence using Monte Carlo permutation tests. The first is a modified version of the permutation test for overall spatial dependence. Instead of using the minimum p-values at different distance binnings as the overall p-value, we propose a modified version that uses empirical Brown’s method to combine the p-values into a new test statistic. The second is a clustering-based permutation test for spatial dependence at small scales. Instead of the null hypothesis of complete spatial randomness, sometimes it is of interest to focus only on the existence of spatial dependence at small scales. In such a situation, our proposed approach can improve the statistical power compared to using an overall permutation test. Both approaches are implemented in the open-source software package variosig [20] available on the Comprehensive R Archive Network (CRAN, https://cloud.r-project.org/).
Our simulation study shows that the type I error rate is maintained by the modified permutation test for overall spatial dependence and the clustering-based permutation test for spatial dependence at small scales. The clustering-based permutation test has increased statistical power compared to the modified permutation test when the sample size is not too small. When the interest is spatial dependence at small scales, the clustering-based permutation test should be used.
In addition to permutation tests, our proposed clustering-based permutation method can also be used in conjunction with a functional boxplot [18] to obtain a visual inspection of the spatial dependence at small scales. The permutation tests can also be applied to large spatial datasets, since it is computationally efficient and can be tuned using the number of permutations. Finally, the result of our permutation tests can help to inform about subsequent analysis such as spatial interpolation, scale decomposition and regression modelling.
This work was supported by the Swiss National Science Foundation (grant no. 175529).