Chapter 5
Cell Structure
Chapter Outline
IV. The Plasma Membrane as the Basis of Cellularity
I Introduction
Vertebrate cells (and to a degree, cells of any type) are variations on basic themes, namely the classes of substructures of which the cells are composed. While certain disciplines of biomedicine can be engaged in without recourse to cell structure, others such as pathology are vitally dependent upon this knowledge. Though this chapter, formerly entitled “Ultrastructure of Cells”, has appeared in several versions of this book, the editor and publishers have made their wishes clear, that the book is now – rather than being a “Sourcebook” – intended rather to serve as a textbook. Because of this, I have broadened the title to “Cell Structure” so as to provide a more useful context. Cell structure cannot be considered in practical terms without considering the various techniques that belong to microscopy. Since the student involved in biomedical study and practice is far more likely to have access to the so-called light (as opposed to electron) microscopes and, since the electron microscope – in addition to its cost, availability and difficulty of operation – has some severe limitations, I have included pictures of cells obtained from both sources. Since examples of many of the different cell components are present in pictures throughout the chapter (see, e.g. Fig. 5.7), only a minimal attempt has been made to make a close correlation of plates of figures with the individual sections on Nucleus, Mitochondria, etc.
II Techniques
What follows is intended to acquaint the reader with just how structure can be documented. The potential investigator will benefit wherever possible by availing him/herself of the opportunity of actually observing and, when possible, performing the procedures described below, in order to understand the process, all the better to interpret the results.
First some basic facts: light microscopes depend on a beam of light, passing through a specimen, to form an image. Depending on the source of light and other mechanical features, light microscopes include instruments known variously as “bright-field”, fluorescent, differential-interference-contrast (a.k.a. “Nomarski”) and confocal scanning microscopes, among others. Since the beam of light contains a range of wavelengths, different stains interact with the light to generate images that have different colors which correspond to different cell types and intracellular structures. Transmission-type electron microscopes use a beam of electrons that pass through the specimen, which is kept in a vacuum, to generate what amounts to a “shadowgram”, since more dense regions deflect the electrons and less dense ones allow the electrons to pass through. This results in a grayscale image, since by and large the beam of electrons has a limited wavelength and thus does not generate colors (micrographs – which is the term for pictures made with a microscope – from electron microscopes that appear in color in some magazines have been artificially colored). Those with some experience with microscopes will note that I have ignored the so-called “dissecting” microscope which, although it uses light as illumination only shows the surface of, say, an insect or an isolated organ; so too I have not spoken of the scanning electron microscope, or SEM, which produces much the same sort of picture as the dissecting light microscope (albeit, again, in shades of black and gray), since it either bounces electrons off a surface or, more commonly, generates secondary electrons just below the surface to form an image.
All that established, let us compare only microscopes that use a transmitted beam (i.e. one that passes through a specimen). It turns out that the preparation techniques for light microscopes (LMs) and transmission electron microscopes (TEMs) have a great deal in common. Though in science there are always exceptions (a phrase you commonly hear is “It’s not that simple, though”), in general, to view something in a microscope one has to: (1) preserve it (the term “fix” is usually used, implying immobilization by techniques such as freezing, immersing in chemicals, or even heat-inactivating [“cooking” for lack of a better term]); (2) dehydrate it so as to make it compatible with anhydrous materials such as paraffin or plastic; and (3) infiltrate it (“embed”) with those media. This last step is to stabilize the specimen (for our purposes, let us say a piece of brain, liver or kidney) so that it can be cut (“sectioned”) into thin slices (“sections”). Such sections range from one micrometer (abbreviated as “μm” and equivalent to roughly four one hundred-thousandths of an inch) in thickness up to 50 μm or so for LM examination, and generally in the range of 0.05–0.1 μm (known as ultrathin sections) for the TEM. Obviously, the μm to inch conversion is unwieldy; one can readily estimate sizes in microscopic images by using a couple of rules of thumb: (1) in the LM, the practical limit of resolution (meaning essentially what one can clearly distinguish as a separate structure) is about 1 μm; (2) mitochondria, when they can be resolved, are usually on the order of 1–2 μm in size (see Figs. 5.2, 5.4A, 5.5A, 5.6C, 5.7, 5.9, 5.12); also, the contractile units (“sarcomeres”: see Figs. 5.6C, 5.10) in skeletal and cardiac muscle are around 2 μm in length.
The purpose of having sections is…? …Simply to have the specimen thin enough so that the imaging beam can pass through it! This necessity leads to a basic limitation of microscopy: only a sample, a mere slice of a specimen is being examined, which can lead to erroneous conclusions about three-dimensional relationships. Also, while on the subject, interpretation of microscopic images should be recognized as an exercise in “making dynamic conclusions from static entities”. In any event, for LM specimens, paraffin – similar to the stuff some of our grandmothers and great-grandmothers may have used to seal their jars of home-made jam and jelly – is used. Specimens embedded in paraffin can be readily sectioned with a razorblade, though usually dedicated “knives” are used, designed to fit into a cutting machine – a “microtome” – that can be adjusted to cut sections of consistent thickness. These sections are floated onto glass microscope slides, usually about 1”×3” (2.54×7.62 cm) in size. To process the sections further, however, the paraffin in them must be removed with some organic solvent such as xylene, and then gradually rehydrated – brought back to water – to stain them. This deparaffinization and rehydration serves two purposes: it removes the paraffin, which would interfere with the passage of the LM’s light beam, and it renders the sectioned material – the liver, heart, kidney, whatever – compatible with a wide variety of stains (including immunohistochemical reagents), which are prepared as aqueous solutions and would not react with the material if it were still in paraffin.
TEM specimens also are cut into extremely thin slices with some sort of sharp blade, but in order to have a hard enough embedment, some sort of plastic is used to infiltrate the specimen. Most often this is an epoxy-type plastic, somewhat similar to the two-component glue (resin and catalyst) that one uses in “civilian” life. Here there is seen a substantial difference between LM and TEM preparation, though: for the TEM, the embedding plastic is not removed from the specimen, since the section – which is cut either with a knife edge made of glass or diamond, and affixed onto a 3-mm circular piece of metallic mesh known as a grid – has to withstand imaging within a vacuum chamber, through which there passes an electron beam of perhaps 100 000 volts in penetrating/imaging power. The “ultrathin” sections on their grids, rather than being stained with colored dyes, instead are contrasted with heavy metals (including osmium, uranium and lead), which enhance details of the shadowgram spoken of earlier, in particular their unit membranes (see below). The electrons are deflected or let pass at various angles, depending on the density of the parts of the section, and expose an image either on a piece of film in the microscope or, more commonly these days, the electron pattern is converted into a digital image (as also is now routinely the case with the LM).
So as a result of these processes, we can visualize either structure as seen through the LM on a glass slide, or ultrastructure (meaning literally “beyond structure”; an unfortunate term, but one that has nevertheless become firmly embedded in the literature) in the TEM, on a specimen grid. Structure and ultrastructure remain compatible, though, with limitations at both ends of the spectrum: the images in Fig. 5.1 are of the same type of structure (rat kidney glomerulus) and the specialized nature and specific appearance of the various cells can be appreciated in one way or another in each micrograph, regardless of its origin. By combining the techniques (plastic embedding and serial sectioning with a diamond-edge for both LM and TEM: Figs. 5.1A and B, respectively) bring the two microscopic images into even closer correlation. An important thing to note is that, at the enlargements (magnifications) shown, which are low for a TEM but high for an LM, the LM gives just about as much general information as the TEM! Kidney structure, which I have been studying for the past couple of decades, can be fantastically complicated when viewed in a TEM, since the various cylindrical structures, including epithelial tubules and blood vessels that make up the majority of the organ, weave in and out of section, and particularly so in ultrathin sections (Fig. 5.2). I have found that so-called “semithin” plastic sections – on the order of a quarter of a micrometer in thickness – bridge the gap between the LM and TEM nicely (Fig. 5.1). It is somewhat ironic, however, to remember that many kidney ailments depend absolutely on the TEM and thin sections for their diagnosis, since structures such as immune deposits are best appreciated there. Certainly, one must choose one’s goals carefully when choosing to do electron microscopy, but this is necessitated by the presence of many features of cells that are either poorly resolved or are practically invisible with light microscopy (Fig. 5.8 demonstrates an example of the former situation, where the TEM is required to make positive identification of autophagic bodies).
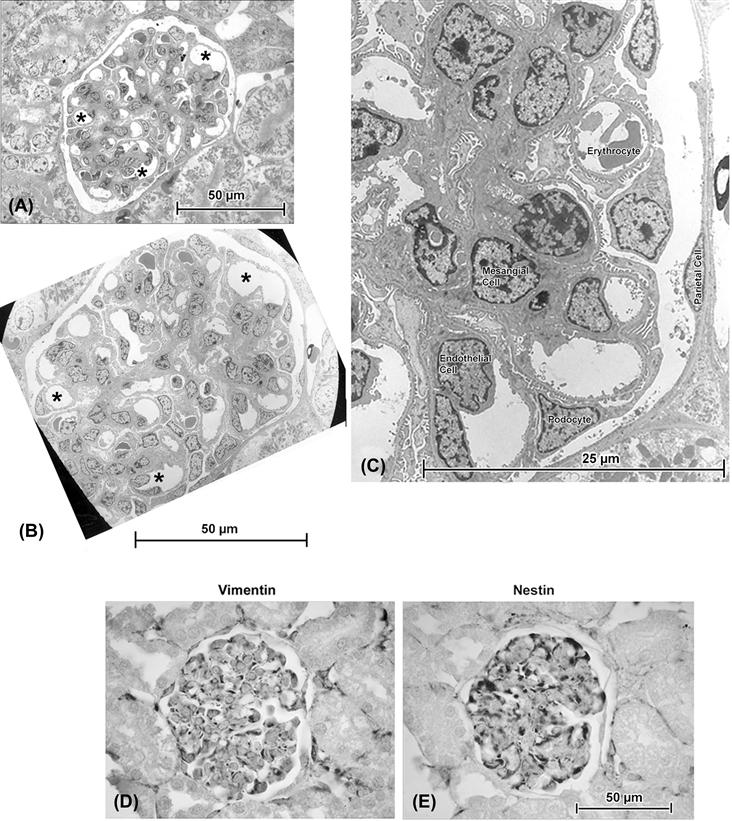
FIGURE 5.1 (A) and (B) are consecutive serial sections, taken with a diamond knife on an ultramicrotome, of a glomerulus in rat kidney. The semithin section ((A): ca. 0.25 μm thick) was cut immediately before the ultrathin section ((B): ca. 900 Å, i.e. 0.09 μm thick) and there is close correlation between such features as the pattern of capillaries (∗ in both micrographs). An enlarged view of the electron micrograph (C) identifies examples of the various cell types found in glomeruli. For most purposes, the ultrathin section – which required considerably more effort to document than the semithin – does not provide any special advantage to the investigator unless high-resolution details such as podocyte processes, endothelial fenestrations, cytoskeletal filaments or intercellular junctions are specific subjects of interest. (D) and (E) are serial consecutive paraffin sections in which immunohistochemical staining is readily accomplished: here the podocytes show reactivity (dark staining) for both vimentin, a structural protein, and nestin, a protein which under normal conditions is restricted to these cells.
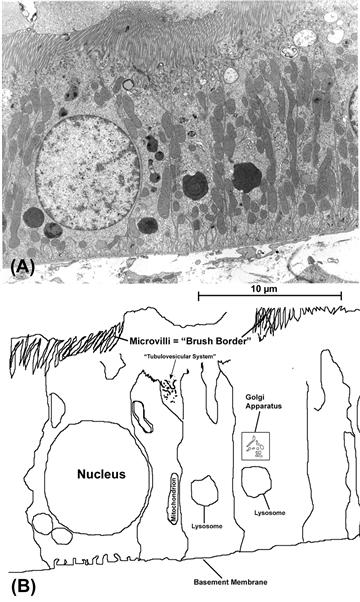
FIGURE 5.2 (A) Though a “typical” cell exists only in idealized diagrams, one type that is often used to illustrate general structural features is the epithelial cell of kidney tubules (here shown in a rat proximal tubule). In (B), a tracing of some of the cells and their organelles is shown. These cells exhibit the “polarity” that is a consistent feature of epithelial cell layers, with apical specializations (“brush border”, consisting of tightly-packed thin cell-membrane extensions known as microvilli) that provide the cell tip with a greatly enlarged surface area for absorption and release and a basal reinforcing layer of extracellular material on which the cells sit (“basement membrane”).
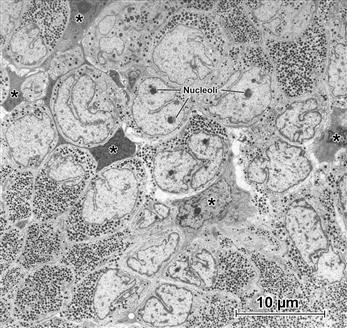
FIGURE 5.3 Pituitary gland. Low magnification (“survey”) transmission electron micrograph shows the dense concentrations of granules that contain the hormones produced by the secretory cells. Note the elaborate profiles of the nuclei (several nucleoli are indicated by Nuc). A supporting framework of “stellate” cells (examples at ∗) is recognizable because of their extended cell processes. It is instructive to point out what this picture does not show. Though the glial-like stellate cells are evident because of their opaque cytoplasm and lack of secretory granules, on the basis of their ultrastructure, the granulated cells themselves cannot be identified as to their specific hormonal content; for this, additional coordinated approaches are needed, including additional characterization and measurements of the granules at higher magnification, correlation of precise location of these cells within the whole gland and histo- and immunocytochemical staining both at the LM and TEM levels, in addition to biochemical analysis to confirm the presence of the hormones in question.
Even though I have described microscopy largely in terms of slices of tissue, it has been necessary to diverge from this in several figures (Figs. 5.4B, 5.4C, 5.5B, and 5.11B,C) to illustrate an important concept of unit membrane structure by means of the procedure known as “freeze-fracture”, in which a cutting action is retained in terms of breaking a specimen at low temperature and under vacuum, with subsequent deposition of a carbon and metal film to make a replica that demonstrates the different sides (“faces”) of these membranes that in large part define and limit many cell components (including the plasmalemma, nuclear membrane, Golgi and endoplasmic reticulum, mitochondria and lysosomes).
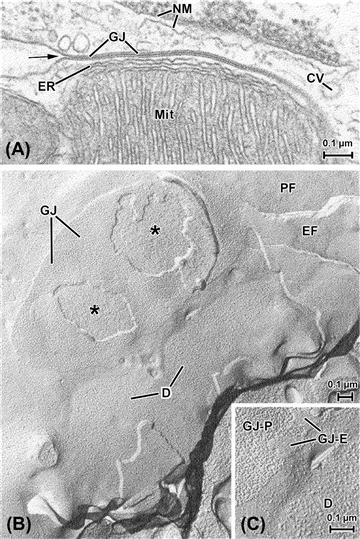
FIGURE 5.4 Unit membranes of cells. (A) Cell border formed by two cardiac muscle cells. Unit membranes are the basis of most of the structures shown here, including the double membrane of the nucleus (NM), the outer envelope and inner membrane shelves (cristae) of a mitochondrion (Mit) and several vesicular bodies, including a coated vesicle at the right of the picture (CV). The membrane-based, multilayered structure that dominates this view is a specialized adhesion known as a gap junction (GJ), formed between the two cells where the outer faces of the two cells’ plasmalemmata come into close apposition. At the arrow, the relatively unspecialized cell surface membranes are clearly continuous with the membranes that form the gap junction. Note the membrane-based tubule of endoplasmic reticulum (ER) sandwiched between the mitochondrion and the gap junction. (B) and (C) show cardiac cell membranes as revealed by the technique of freeze-fracture, in which tissue is quickly frozen and broken and a thin (ca. 2–3 nm) coating of metal is evaporated onto the specimen and stabilized with a thicker carbon film (similar techniques were used to produce the replicas depicted in Figs. 5.5B, 5.11B,C). The tissue is then digested away, leaving only the metal–carbon replica of the fractured surfaces. The fracturing process specifically cleaves unit membranes to expose their inner “faces”, thus showing the intramembranous particles (IMPs) that correspond to a variety of functionally important proteins. This also demonstrates the characteristic asymmetry inherent within a unit membrane; typically there is a greater density of larger particles on the P face, the face adjacent to the protoplasm (cytoplasm), whereas the E face (that closest to the “ectoplasm”, i.e. the extracellular space) is characteristically smoother. Compare, for example, the regions labeled PF and EF at the upper right corner of (B). This region shows the relatively unspecialized plasmalemmas of two apposed cells (cf. the cell surface membranes in (A)). The large oval body is a gap junction between the two muscle cells, broken away in two regions (∗) to expose underlying intracellular structures, probably mitochondria (a similar association is shown in the thin section in (A)). The detail shown in panel (C) shows that though this gap junction replica consists mainly of large P-face particles (GJ–P), in some areas, small regions of the junction’s E face (labeled GJ–E) have adhered, which are characterized by a finer topography of pit-like depressions, into which the P-face particles fit in the intact gap junction. In (B), two smaller ovoid membrane regions (D) appear which correspond to another type of intercellular junction, the desmosome (see also Fig. 5.11). The P-face particles belonging to desmosomes (also shown at higher magnification in (C)) are larger and more coarse-appearing than those of either the gap junction or the unspecialized plasmalemma.
III Cell Theory
The appreciation of the very existence of individual cells did not come about in the form of a linear, smooth progression. Despite Robert Hooke’s often-cited observations (ca. 1665) of individual units (actually cell walls; the living cells were long gone) in thin sheets of cork, aided by one of the earliest microscopes (he likened the units to cubicles [L. “cellula: small rooms”] occupied by monks or other sorts of prisoners, thence the term “cells”), centuries later the cellular nature of some tissues was still questioned. Though in the late 1830s Matthias Schleiden and Theodor Schwann had proposed that most tissues were composed of individual cells, toward the end of the 19th century the reticular theory of Joseph von Gerlach and Camillo Golgi came to stand as an exception. This posited that nervous tissue was a syncytium, i.e. a system possessing multiple nuclei but having its cell substance continuous through its entire network. Opposed to reticular theory was the neuronal theory of Santiago Ramon y Cahal, whose exquisite staining techniques and careful microscopic renderings demonstrated neurons to be individual entities (the accuracy of his drawings has in fact been confirmed in modern times, by examination of his original preparations and three-dimensional reconstruction from them by the confocal scanning light microscope).
While the battle over nerve structure was carried out in Europe, an international controversy developed in the case of the heart, with cellular theory being espoused by the camp of Zimmerman in Germany (among others) and the contrasting view – namely that heart muscle was a true syncytium – being defended by Harvey Ernest Jordan in America (Jordan, incidentally, was the first dean of the University of Virginia School of Medicine, where this review is being written). In truth, both nervous tissue and cardiac muscle behave as if they were true syncytia, with impulses being passed from cell to cell; in the case of nerve this most often happens through the intercellular contacts known as synapses (see Fig. 5.12C), whereas in heart, the muscle cells are held together via elaborate arrays of intercellular connections known collectively as intercalated disks, components of which are the basis of cell-to-cell transmission (see Fig. 5.11D).
Ultimately refinements to microscopic equipment – in particular the invention of electron microscopes – carried the day for the establishment of cell theory as valid for both brain and heart, since the actual separations between adjacent cells could finally be visualized. This did not happen until the 1950s, however, and even in the face of definitive electron microscopic evidence, some clung to the syncytial theory of heart organization into the following decade (for further review of this, see Forbes and Sperelakis, 1985).
IV The Plasma Membrane as the Basis of Cellularity
To understand the fundamental mechanisms that constitute the normal activities of any organism, it is essential to consider the structural correlates that underlie these activities. Once cell theory was established, there arose another important question which revolved around the nature of the barriers that separate cells from one another and from the surrounding environment. A major breakthrough in resolving this question was made independently by Gorter and Grendel in the 1920s and Davson and Danielli in the 1930s. The latter authors examined the behavior of phospholipids in aqueous environments and determined that the most favorable energetic configuration was a bilayer having the hydrophilic polar headgroups located at the water interface and the hydrophobic non-polar tails apposed to each other. Such a cell membrane (“plasmalemma”), formed by a phospholipid bilayer, would be impermeable to fluids and charged species, such as ions. Thus it would isolate the cell contents from the extracellular environment. It should be understood that those cell contents, specifically organelles including the Golgi complex, mitochondria and endoplasmic reticulum, are themselves composed in large part of similarly-constructed unit membranes.
Proteins were recognized as being closely associated with the phospholipid bilayer and early models proposed that they form a layer on either side of the membrane (i.e. a protein–lipid sandwich). This model remained hypothetical until the introduction and refinement of the transmission electron microscope (TEM) in the 1930s and 1940s; in the TEM, resolution was so much improved that, in properly-prepared “ultrathin” sections, all cells were bounded by a trilaminar membrane, 70 Å (=7 nm) thick that to some observers resembled a “railroad track” consisting of two darker lines separated by a relatively clear inner region (Fig. 5.4A). This structure was termed the unit membrane by J.D. Robertson in the early 1950s. It had already been recognized by Davson and Danielli that many of the proteins associated with the cell membrane were tightly attached, since they were not extracted by changing the ionic strength of the bathing solutions or even by mild detergent treatment. Thus, the notion of a phospholipid bilayer with a coating of protein had evolved, by the 1960s, into a “fluid mosaic” model proposed by Singer and Nicholson. In this model, there were both peripheral (or extrinsic) and integral (or intrinsic) membrane proteins. Peripheral proteins are associated with the inner and outer surfaces of the membrane, but are not anchored in it. Integral membrane proteins are actually embedded in at least one leaflet of the phospholipid bilayer (Figs. 5.4B,C) and some traverse the entire thickness of the membrane.
The fluid mosaic model was more consistent with functional studies, which clearly demonstrated that cell membranes were not totally impermeable, but rather were semipermeable. The integral membrane proteins included ion channels that acted as low-resistance pores, as well as energy-dependent pumps. This model better accounted for the establishment of ionic gradients and the influx or efflux of ions under various conditions.
As mentioned, the extrinsic proteins are membrane-associated rather than actually embedded in the lipid bilayer. The extracellular matrix exists on the outer cell surface as a layer of varying thickness, known as the basement membrane or basal lamina, depending on the cell type (see Figs. 5.2 and 5.7). Historically, this coating was considered to be an inert structural framework composed of collagen and elastic fibrils. Not only can the extracellular matrix act as a filtration device but, in addition, recent evidence points to a more dynamic role, with proteins such as fibronectin and laminin active in stimulating a number of cellular activities. These functions are likely mediated through interactions with intrinsic membrane proteins which, in turn, are connected to and exert influences on the extrinsic membrane proteins that form subplasmalemmal networks on the inner (cytoplasmic) surface of the cell membrane. Ultimately, signals from outside the cell may be communicated deep within the cell via the interior membrane systems and elements of the cytoskeleton and may even influence gene activity in the nucleus. In this sense, the unit membranes can be considered to represent the “nervous system” of the cell.
The contents of the cell enclosed by the plasmalemma are collectively known as the cytoplasm. The cytoplasm is not an amorphous soup, but in fact contains a number of distinct structural elements known as organelles (by analogy to the different organs of the body). Figure 5.2A is an electron micrograph of kidney tubule epithelial cells, often considered to be a characteristic type of animal cell, with a diagrammatic representation of the same micrograph in Fig. 5.2B, where some of the organelles are shown; these serve to compartmentalize different activities of the cell, just as the organs perform different functions within the body.
V Nucleus
The most prominent organelle within the cell is readily visible in the light microscope and, since it is most often located at the cell center, it came to be called the nucleus. The nucleus contains most of the cell’s DNA and its genetic apparatus as well. Because it is the locus of the genetic programming that directs cytoplasmic synthetic activity, as well as the site at which cell division is initiated, the nucleus can be considered as the organelle that corresponds in the cell in part to the reproductive system of the body.
In most cells, the nucleus is an ovoid body, but it can assume a variety of shapes and sizes (see Figs. 5.1, 5.2, 5.3 and 5.5). Multiple nuclei are characteristic of certain cell types (e.g. skeletal myocytes). Early EM observations discovered the nucleus to be bounded by the nuclear envelope, essentially a double unit membrane (see Fig. 5.4A) and noted as well a dense intranuclear body, the nucleolus (see Figs 5.3, 5.5). The nucleus was characterized in addition by darker material (heterochromatin), found interspersed with the lighter euchromatin (see Fig. 5.2). Beyond these initial descriptions, the regional specializations of the nucleoplasm remain rather poorly understood. Recent studies with antibody localization of nuclear-specific proteins demonstrate the presence of domains within the nucleoplasm; these are not readily apparent even with ultrastructural inspection, but appear to be organized through attachment to the filamentous proteins of the nuclear matrix. On the basis of compartmentation of DNA polymerases and RNA splicing factors, it has been suggested that discrete domains are involved in different processes, such as transcription and translation of the genetic material.
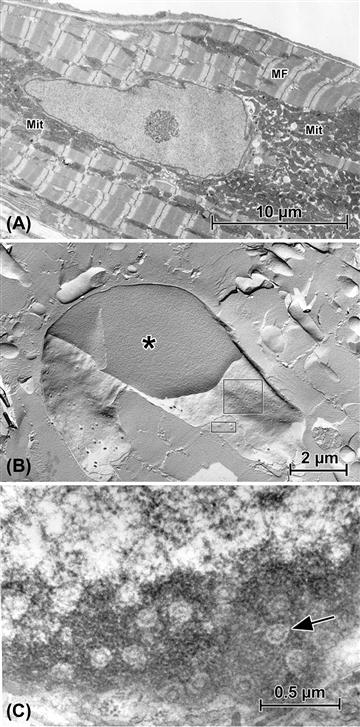
FIGURE 5.5 Aspects of nuclear structure as seen in cardiac muscle cells (“cardiomyocytes”). (A) The nucleolus is prominent within this nuclear profile. Additional characteristics of the cardiomyocyte are the interspersed arrays of myofibrils (MF) and mitochondria (Mit). (B) Freeze-fracture replica of a nucleus. The plane of fracture reveals cytoplasmic myofibrils and mitochondria (see panel (A)) and breaks through the nucleus to show the homogeneous nucleoplasm (∗), then passes into the outer nuclear membrane leaflet (larger rectangle) to show nuclear pores as depressions, then into the inner nuclear membrane (small rectangle), where the pores appear as corresponding protuberances. (C) Grazing thin section of nucleus, passing through the peripheral dark heterochromatin to reveal the complex nuclear pores’ substructure (arrow).
As noted earlier, the nuclear envelope is a double bilayer. The outer membrane, often decorated with spherical RNA-containing particles called ribosomes, is often continuous in places with a network of cytoplasmic membranes known collectively as the endoplasmic reticulum (ER) (e.g. Fig. 5.6B). The nuclear envelope has a proteinaceous lamina at its inner surface, which is composed of cytoskeletal proteins called lamins; the major portion of the envelope is the prominent double membrane itself (see Fig. 5.4A), which is interrupted at periodic intervals by circular discontinuities, the nuclear pore complexes (see Fig. 5.5B,C). Detailed studies have demonstrated that the nuclear pore complexes are complicated structures indeed, incorporating an eight-subunit annular structure centered around an aqueous channel that allows passive diffusion of small molecules and ions. The nuclear envelope is not a complete barrier, because it is necessary for RNA, proteins and ions to pass in and out of the nucleus; the system of nuclear pores regulates the trafficking of these substances.
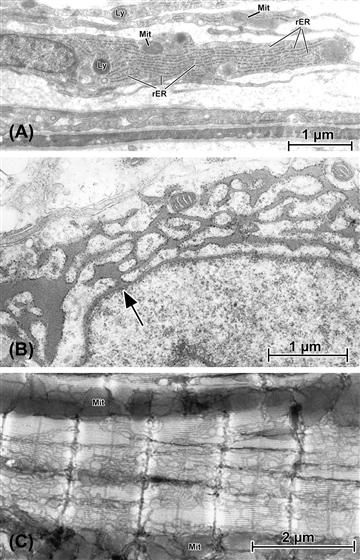
FIGURE 5.6 The variable appearances of endoplasmic reticulum (“ER”). (A) Fibroblastic cells that form the capsule layer of kidney. Each of these elongated cells is characterized by a corresponding flattened nucleus, along with limited numbers of lysosomes (Ly) and mitochondria (Mit). Filling the cytoplasm of these collagen-producing cells are parallel arrays of rough ER (rER). (B) Pituitary cell (thyrotroph) in which synthetic activity has been stimulated by removal of the thyroid gland. This induces a dramatic increase in production of thyroid-stimulating hormone (TSH) and, instead of generating storage granules (see Fig. 5.3), the rough ER expands into interconnected networks of dilated tubules and cisternae filled with freshly-synthesized hormone. Note (at arrow) the connection between the nuclear envelope and rER. (C) Atrial tissue from mouse in which the smooth endoplasmic reticulum (known as “SR” in muscle, is specifically stained by an osmium impregnation technique related to those used by Golgi and Cajal (see Cell Theory section) to contrast neurons. In this application, only the SR is darkened; by cutting relatively thick (ca. 2 μm) plastic sections for the TEM, the extent, distribution and elaborate patterning of the SR where it enwraps the banded myofibrils can be appreciated. Linear masses of mitochondria (Mit) are seen running between some myofibrils.
VI Endoplasmic Reticulum
The ER was originally described by light microscopists in the late 1800s, who used special preparative procedures that selectively infiltrated this organelle system with a variety of staining moieties (see Fig. 5.6C). Understanding of the structure of the ER was greatly advanced in the late 1940s and early 1950s by the elegant EM work of Porter and Palade, who demonstrated that the ER was an extensive network of interconnecting tubules and cisterns.
Ribosomes are compact rounded particles, approximately 300 nm in diameter, that dissociate in the presence of low Mg2+ into two smaller units. They consist in large part of ribosomal RNA but, in eukaryotes, as much as 50% of the ribosomal mass can be composed of associated proteins. Ribosomes are often found organized into strands or rosettes called polyribosomes or simply “polysomes” (Fig. 5.7). These are aggregations of ribosomes active in the messenger RNA-directed linkage of amino acids to form peptide chains. Protein synthesis is initiated on ribosomes, which may be free in the cytoplasm or bound to the ER to form a complex known as rough endoplasmic reticulum (see Figs. 5.6A,B, 5.7). The rough ER is presumed to be involved virtually exclusively in protein synthesis. In contrast, ER that does not have attached ribosomes is known as smooth endoplasmic reticulum (see Figs. 5.6C, 5.7) and has been implicated in a number of different functions depending on the particular cell type. In steroid hormone-secreting cells, for example, smooth ER is associated with the production of secretory products. In liver cells, the smooth ER has been shown to be active in glycogen metabolism. In cardiac, skeletal and smooth muscle, the smooth ER is better known as the sarcoplasmic reticulum (SR) (see Fig. 5.6C); SR membrane proteins are responsible for the uptake, sequestration, and release of Ca2+ during the cycles of excitation–contraction coupling and relaxation. ER in non-muscle cells may also be involved in Ca2+ metabolism.
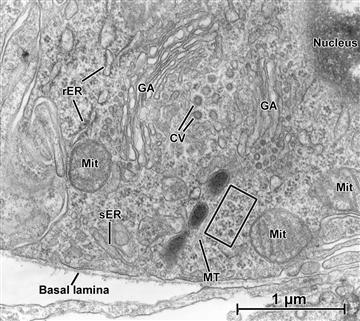
FIGURE 5.7 Transmission electron micrograph of a “juxtaglomerular cell” in kidney blood vessel. In this limited area, examples of many of the organelles described in this chapter are seen in close proximity to one another. In particular, the saccules, cisternae and vesicles that make up the Golgi system (GA) are clearly defined. In this cell, which is the source of the vasoactive hormone renin, polyribosomes are abundant (examples within the rectangle), along with smooth ER (sER), rough ER (rER), mitochondria (Mit), and a microtubule (MT).
VII Golgi Apparatus
Synthesis of membrane proteins and secretory products is completed in another organelle, which is at least functionally connected with the ER and is called the Golgi apparatus (see Fig. 5.7). This organelle was named for Camillo Golgi, a 19th-century microscopist-histologist who developed numerous staining techniques and described many of the light-microscopic features of cells. In the electron microscope, this organelle was found to be composed of flattened disk- or saucer-shaped membranous elements known as cisternae. In each Golgi apparatus, numbers of these cisternae are organized into stacks, with individual cisternae interconnected through tubular channels. As secretory products pass through the Golgi apparatus, they undergo chemical modifications such as glycosylation, proteolytic cleavage, phosphorylation and sulfation. Other functions performed by the Golgi apparatus include carbohydrate metabolism, targeting of plasmalemmal proteins (pumps, channels and receptors) and the condensation of secretory materials into opaque “secretory granules” (see Fig. 5.3).
Although a variety of terminologies has been used to identify different regions of the Golgi apparatus, four morphological subsections are generally recognized. The cisternae are slightly dished in conformation, which confers both concave and convex faces to the Golgi complexes. The convex or cis face is also known as the forming or immature face, since it is at this level that newly synthesized proteins enter the Golgi. The medial or intermediate region comprises the varying numbers of cisternae in the middle of the stack. The concave or trans face is also called the mature or secretory face, since it is the site from which large secretory vesicles bud off after their contents have been modified for export. The trans-Golgi network is a reticulum of tubules emanating from the trans face and is thought to be associated with the lysosomal system.
This general scheme is applicable to all Golgi complexes and transport within the Golgi is generally vectorial, going from the cis to the trans face. However, more recent evidence suggests that maturation and pinching off is not totally restricted to the trans face. There is likely much more intra-Golgi shuttling back and forth of material than was originally believed. The numerous vesicles of various shapes and sizes in the immediate vicinity of the Golgi complex (see Fig. 5.7) may be transporting proteins along the pathway during maturation and processing. There is also evidence for some retrograde transport from the Golgi apparatus to the ER.
VIII Lysosomes
In addition to packaging and modifying secretory products, the Golgi apparatus plays a role in the formation of another important cellular organelle, the lysosome (see Figs. 5.2, 5.6A, 5.8A,B). In fact, the functional continuity of the ER, the Golgi apparatus and the lysosomes has led to the concept of the GERL (Golgi–endoplasmic reticulum–lysosome) system. Lysosomes are a heterogeneous population of membrane-limited vesicles, differing in size and density, that were identified and characterized by De Duve in the mid-1950s. Lysosomes contain hydrolytic enzymes that can break down virtually every form of biological material and thus these organelles act, in part, as the cell’s digestive system. Many of the lysosomal enzymes are more effective at low pH and the lumen of the lysosome accordingly is more acidic than the cytoplasm. The lysosome acts to break down and recycle intracellular components such as cytoskeletal proteins and defunct organelles, such as mitochondria (by a process known as autophagy), as well as to digest extracellular material that has been trapped in phagocytic vesicles formed by internalized plasmalemma (heterophagy). Viewed in the electron microscope, newly formed “primary” lysosomes are more uniform in size, with an amorphous electron-opaque content. After fusing with phagocytic vesicles or other cell contents, they form “secondary” lysosomes, whose size and internal density is more variable (Fig. 5.8). Lysosomes containing indigestible material often remain in the cytoplasm as residual bodies, also variously known as aging pigment or lipofuscin.
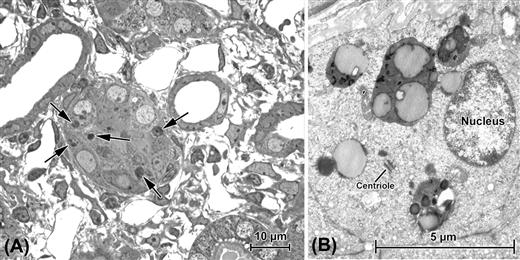
FIGURE 5.8 Autophagy in kidney proximal tubules following obstruction of the ureter. (A) “Semithin” (ca. 0.25 μm) section of tissue embedded in plastic. Use of such sections allows survey viewing in the light microscope for selection of areas to be more closely examined in the TEM, as seen in (B). In this case, the structures of interest are the dark bodies indicated in (A) by arrows; for all practical purposes, their substructure is below the resolution of the light microscope and it is only in the TEM that they are revealed to be autolysosomes, the complex products of a multistep process (described in text) that isolates defunct or excess cell contents for digestion. Nuclei in cells undergoing autophagy often appear normal; a centriole is also present in one cell.
Cell death mechanisms are now of particular interest. necrosis, in which the semipermeable properties of the plasmalemma are compromised, has been well known for many decades and the term apoptosis (a.k.a. “type I programmed cell death”) was coined in the 1970s (apoptosis is discussed further in the section on Mitochondria). More recently, however, autophagy has attracted special interest. Though lysosomes were well-established as fundamental cellular organelles, their role in autophagy (literally “self-eating”; also known as type II programmed cell death) is only now being detailed. The sequestration of cellular materials into lysosomes to form secondary lysosomes has now been found, largely on the basis of electron microscopic studies, to be more complex. Autophagy is now considered to be initiated by the formation of a double-membrane structure, the “isolation membrane” (likely derived from the ER, but this is still not completely clear), which develops into a body known as an autophagosome (or autophagic vacuole), which is the entity that envelops the target organelle or substance. The autophagosome with its contents then joins (“docks and fuses”) with the lysosome proper, forming the secondary lysosome, now known as the autolysosome. Autophagy is not entirely an ineluctable path to cell death, however, and in some cases – such as starvation – promotes cell survival.
Peroxisomes, while similar in overall appearance to lysosomes, are considered to be a separate class of organelle. These bodies are abundant in liver, kidney and heart (among others) and contain both peroxidase and catalase, through the action of which fatty acids are broken down and the resulting, potentially harmful hydrogen peroxidase is dissociated into water and oxygen. Endogenous peroxidatic activity is of technical interest when one is performing immunohistochemistry; peroxisomes and erythrocytes (which also have peroxidatic activity) require “quenching” with hydrogen peroxidase to neutralize endogenous peroxidase, since certain steps of standard immunohistochemical procedures utilize exogenous peroxidase in generating the colored compounds (chromogens) that mark the sites of antigens (see Fig. 5.1D,E).
IX Mitochondria
Aside from the nucleus, the most obvious feature of the majority of cells is the collection of organelles called mitochondria (Fig. 5.9). The term “mitochondrion” (literally “thread-grain”) was introduced at the turn of the 20th century based on the appearance of these bodies, under the light microscope, as elongated granules. In the TEM, each mitochondrion is seen to be bounded by a double bilayer membrane (see Fig. 5.4A). The outer bilayer is smooth in contour and forms the boundary around the entire mitochondrion. The inner membrane has a variable number of infoldings, called cristae, which increase the internal surface area substantially (see Figs. 5.4A, 5.9).
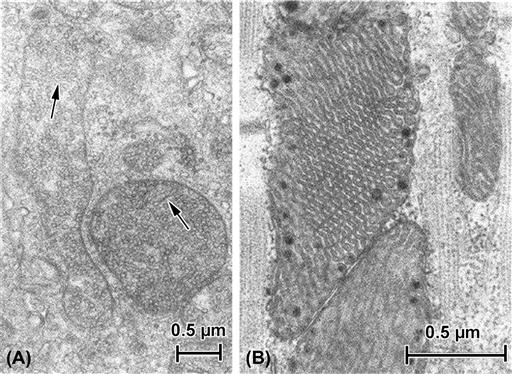
FIGURE 5.9 Mitochondria. (A) Mitochondria in cell from adrenal cortex. Steroid-secreting cells often are characterized by spheroidal or elongate mitochondria packed with internal membranes (cristae) that take the form of fine tubules (arrows) or chains of tiny vesicles. (B) Mitochondria in cardiac muscle. In addition to being tightly sandwiched between the myofibrils (collections of contractile filaments; see also Figs 5.6C, 5.10A), cardiac mitochondria are often densely populated with elaborate pleated shelves of internal cristae. Opaque granules often are present within the mitochondrial matrix.
It is now clear that the mitochondria are sites of cellular respiration and thus are the center of energy production in the cell. The inner mitochondrial membrane contains abundant transport proteins responsible for establishing a proton gradient between the intermembrane space and the matrix. The proton motive force created by this gradient is essential for driving many of the reactions of oxidative phosphorylation, a fundamental metabolic process occurring within the inner compartment of the mitochondrion. Oxidative phosphorylation generates much of the cell’s adenosine triphosphate (ATP), the high-energy compound that is the source of energy for most cellular activities. The enzymes responsible for the tricarboxylic acid cycle, one component of oxidative phosphorylation, are located in the mitochondrial matrix and the enzymes involved in electron transport, another major function of the oxidative phosphorylation process, are associated with the inner mitochondrial membrane.
As would be expected, mitochondria concentrate at sites within the cell that require high energy utilization, as well as being more abundant in cells that exhibit a high degree of metabolic activity. For example, heart muscle tissue has one of the higher measured metabolic rates; accordingly, in myocardial cells, mitochondria constitute up to 40% of the total cell volume, and are sandwiched in between the collections of contractile proteins (myofibrils) (see Figs. 5.5A, 5.6C).
Apoptosis, as mentioned earlier, is a mechanism of “programmed cell death”. Morphologically, it is most clearly evident in changes in nuclear appearance (condensed chromatin, nuclear fragmentation). However, the impetus for the apoptotic process is found in the mitochondrion where, under certain circumstances, there is a change in its membrane permeability and thence its electrical potential. “Apoptogenic factors” such as cytochrome c and proteases are then released from the mitochondria and lead to the so-called “caspase cascade” and eventually to the nuclear alterations that are obvious in the microscope. Like autophagy, however, apoptosis can serve beneficial purposes. Apoptosis is a necessary process, particularly in development; e.g. it is thought to bring about the establishment of microvessel lumina in kidney glomeruli (Fierlbeck et al., 2003).
X Cytoskeleton
The cytoplasm is not just a watery bag in which organelles move about at will. In fact, cell contents are to one degree or another compartmentalized, with certain cytoplasmic regions specialized to subserve particular functions. Underlying this compartmentalization is the framework of fibrillar structures that make up the cytoskeleton of each cell. Normally, 20–35% of the total protein of a cell is tied up in the cytoskeleton, although this proportion can vary, being considerably greater in muscle, where cytoskeletal proteins also form part of the extensive contractile apparatus. In all cells, cytoskeletal elements are involved in intracellular motility (such as migration of chromosomes during mitosis, translocation of organelles and cytoplasmic streaming), cell locomotion, and maintenance of cell shape. More specialized functions, such as muscle contraction and ciliary/flagellar movement, also are supported by cytoskeletal elements.
XA Filaments
Filaments are composed of backbones of single proteins, polymerized to form long, slender fibrillar structures. Such filaments are both helical and polar and frequently interact with one another. There are three general filament categories in cells: microfilaments, often called “thin filaments”, which are primarily composed of the protein actin and ranging from 6 to 8 nm in diameter; thick filaments, consisting mainly of myosin and 12–15 nm in average diameter; and intermediate filaments, so called because their customary 10 nm average diameter is intermediate between that of thin and thick filaments (thus also known as 100 Å or 10 nm filaments). Intermediate filaments may be composed of a variety of related proteins, depending on cell type.
XA1 Microfilaments
As noted above, the backbone of microfilaments is composed of the cytoskeletal protein actin; in most cell types, however, additional regulatory proteins (e.g. troponin) and structural proteins (e.g. tropomyosin) are intimately associated with the actin filaments. G (globular)-actin polymerizes to form filaments and in this form is known as F (filamentous)-actin. A number of associated proteins may regulate initiation and growth of a filament, as well as its final length. Tropomyosin, for example, stabilizes thin filaments of muscle cells.
In different cells, and in different tissues, thin filaments are associated with a variety of recognizable intracellular formations. Bundles of filaments, all having the same polarity, form the cores of microvilli in epithelial cells; actin filaments are also evident in smooth muscle and striated muscle cells as intrinsic parts of the contractile apparatus (Fig. 5.10). Bundles of thin filaments having opposite polarities are also found, located on opposing sides of the Z disks in skeletal and cardiac muscle, across from one another at dense bodies in smooth muscle, and within the stress fibers of non-muscle cells. Three-dimensional networks of filaments are found in subplasmalemmal (cortical) arrays in many cells.
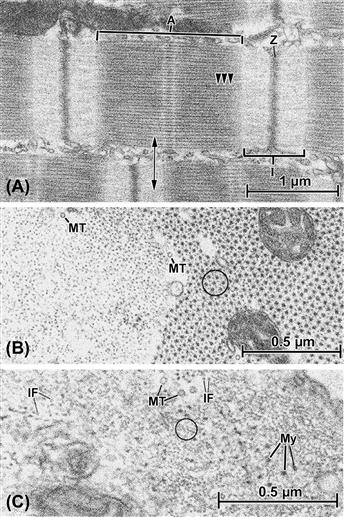
FIGURE 5.10 Fibrillar elements in muscle cells. (A) Longitudinal section through a cardiac muscle cell, showing the characteristic banded pattern of the ordered masses of contractile filaments known as myofibrils. Between myofibrils, other cell organelles, such as mitochondria and endoplasmic (sarcoplasmic) reticulum, are sandwiched. The units of the banding pattern are known as sarcomeres and the major filament categories that contribute to their formation are thin (actin) filaments and thick (myosin) filaments. The so-called A band (A) is a composite of interdigitated actin and myosin and the subtle cross-hatching pattern derives from portions of the myosin filaments that form cross-bridges (several shown by arrowheads) with the adjacent actin filaments. The I band (I) is occupied only by thin filaments, which insert into the dense material of the Z bands (Z), which are primarily composed of the protein α-actinin. Adjacent Z bands define a sarcomere unit, each of which covers a distance of ca. 2.2 μm in heart. Note in this panel that adjacent myofibrils are not always aligned with one another in terms of the levels of their sarcomere patterns. (B) A transverse section through two cardiac myofibrils, its plane of sectioning corresponding to the double-headed arrow in (A). At the left of the picture, the section passes through the I-band region and therefore reveals only actin filaments, whereas at the right the A band is cut, showing the characteristic hexagonally arrayed myosin filaments, each surrounded by actin filaments, usually in the “six-around-one” pattern circled. Mitochondria are enmeshed by the filaments in one myofibril, and additional fibrillar components, namely microtubules (MT), appear in the same orientation as the muscle filaments. (C) Cross-section through a vascular smooth muscle cell in the wall of a coronary artery. Like the cardiac muscle shown in the other panels, the smooth muscle contains examples of actin (circled) and myosin (My), but a geometric arrangement similar to that of cardiac or skeletal muscle is not evident. Cytoskeletal fibrils, including microtubules (MT) and intermediate filaments (IF), also are present in the same orientation as the contractile actin and myosin.
XA2 Thick Filaments
The most detailed structural information regarding myosin-containing filaments comes from the study of striated muscle thick filaments (see Fig. 5.10A,B). Smooth muscle cells clearly contain thick filaments (see Fig. 5.10C), but the exact organization of the molecules within the filaments is still controversial. It is likely that non-muscle cells also contain thick filaments, but that they are shorter and less ultrastructurally obvious.
It is generally accepted that thick filaments are required for movement and motility, which are carried out through interactions with actin-containing thin filaments.
XA3 Intermediate Filaments
Intermediate filaments (IFs) actually comprise a related family of proteins found in virtually all cell types; lamins are IF proteins that form the internal lamina of cell nuclei, for example. However, there are also tissue-specific IF proteins. Vimentin is found in mesenchymal tissue (connective tissue, bone, blood, cartilage); desmin is localized in muscle; neurofilament protein, or neurofilamin (NF), is found in neurons; GFAP (for “glial fibrillar acidic protein”) is found in glial cells of the nervous system. Finally, keratins are IF proteins that are abundant in epithelial cells. Additional IF proteins do not form filaments (e.g. nestin: see Fig. 5.1E), but help to stabilize the filamentous forms.
IFs are often observed as loose three-dimensional networks, intermixed with other cellular components (see Fig. 5.10C), but in astrocytes and other cells can form dense skeins that consist primarily of 10 nm filaments. As pointed out above, IFs form the framework of the inner nuclear membrane, the nuclear lamina. IFs are also associated with subplasmalemmal plaques adjacent to the plasma membrane at specialized cell–cell contacts (desmosomes) (see Fig. 5.11D, F) and are disposed in muscle in transverse meshworks that link and align myofibrils to one another, to the sarcolemma, and perhaps also to the nucleus. Thus, IFs are an internal scaffolding that can form networks to link peripheral and central components of the cell as a mechanically integrated complex.
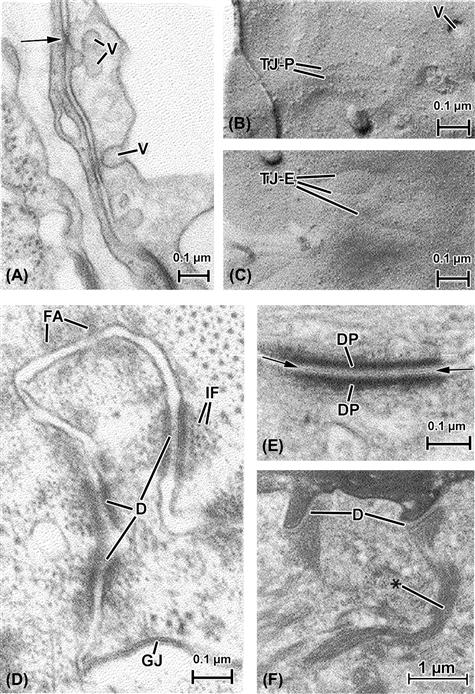
FIGURE 5.11 Cell-to-cell appositions, also called intercellular junctions. (A), (B) and (C) show endocardium (the endothelial lining of the heart) both in thin section (A) and freeze-fracture replicas (B) and (C). (A) The plane of section passes vertically through two overlapping endocardial cells, whose plasmalemmas bear cytoplasmic vesicles (V) that support transport of materials in and out of these cells. The plasma membranes of the two endocardial cells come into close proximity at several points, at one appearing to touch or fuse (arrow). Such junctions are known generally as “tight junctions” and form barriers to the penetration of certain materials past them in the intercellular spaces. (B) A freeze-fracture replica of the endocardial membrane’s P face (see Fig. 5.4B) shows vesicular openings (V) as well as linear arrays of particles that correspond to the tight junction (TJ-P). (C) These correspond in replicas of the E face to linear indentations (TJ–E) into which the corresponding P-face particles fit to form an interlocking junctional barrier. (D) and (E) show junctions from the myocardial cell-to-cell junctional complex (the “intercalated disk”), which consists of interdigitated muscle cell tips that bear a variety of interspersed types of junctions, including gap junctions (GJ, see also Fig. 5.4). (D) At the top of the micrograph there appears a zone of fascia adherens (FA), the points at which thin actin filaments of the myofibrils insert into the intercalated disk. The adherens-type junctions are characterized primarily by accumulations of intracellular dense material that face one another across the intercellular space, but which incorporate no particular extracellular specializations. In contrast, the adhesive junctions known as desmosomes (three of which appear in this picture, D) not only bear intensely opaque intracellular densities closely associated with intermediate filaments (IF), but also contain a distinct component located in the extracellular space (see also (E)). (E) A desmosome is shown at high magnification. The intracellular material is concentrated into desmosomal “plaques” (DP) which, together with the associated plasmalemmal unit membranes, are arranged in a striking parallel array. The extracellular material, known as a “central lamella” (between arrows), is likely responsible for maintaining the register of the apposed membranes, having toothed projections from a center linear component that anchor in the adjacent plasma membrane leaflets. (F) Keratinocytes in the epithelium of skin. Here the major filament type is keratin, considered to be one of the family of intermediate filaments. Keratin is concentrated into fibrous cords (∗) that insert onto the intracellular plaque material of the numerous desmosomes (D) that connect the cells of this layer of the skin.
XB Microtubules
Microtubules are elongate, hollow cylinders of notably larger diameter (ca. 25 nm) than other cytoskeletal fibrils (see Figs. 5.7, 5.10B,C) and are composed of the protein tubulin. The walls of the microtubules are formed from protofilaments aligned parallel to the long axis of the microtubule. The number of protofilaments that make up the microtubule cross-section varies, but 13 is the most common complement. The protofilaments are polar structures and are assembled so that they confer polarity to the microtubule.
There are several different microtubule-associated proteins (MAPs), including MAPI, MAP2 and tau (aberrant formations composed of tau are responsible for the formation of “tangles” within neurons in Alzheimer’s disease). The MAPs serve to stabilize the microtubules and are the targets of regulatory signals. One of the most functionally important MAPs is dynein, which forms side arms on the microtubules found in the cores of cilia and flagella. These side arms enable microtubules to slide past one another, allowing bending of the whole structure and accounting for their basic motility. In addition to being the fibrillar component of cilia and flagella, microtubules are the basis of the so-called MTOCs (microtubule-organizing centers). Microtubules are involved in movement of chromosomes during cell division and perform a role in translocation of organelles within the cytoplasm. In some cases, microtubules also can form the framework on which some of the cell compartments are organized and it has even been shown that the orientation of IFs depends, in some instances, upon the integrity of the microtubules in the same cell.
XI Cell Junctions
Where they are located in tissues and organs, cells do not exist in isolation, but rather exert multiple influences on one another. When they reside in different parts of the organism, the influences are usually realized through an intermediary milieu such as the bloodstream, but where neighboring cells come into contact with one another, specialized structures known as intermembranous junctions connect them. The electron microscope has proven a particularly apt tool for detecting such connections, since they usually incorporate some form of modification of the apposed cell plasmalemmas that is best visualized in the TEM.
Four types of intercellular junctions are commonly encountered in vertebrate tissue: (1) tight junctions; (2) gap junctions; (3) intermediate or adherens junctions; and (4) desmosomes. Epithelial layers such as seen in small intestine display a rather stylized “junctional complex” that consists of the four junctional types arranged in a sequence from the luminal surface down to the basal surface; in order, the complex goes tight junction/adherens junction/desmosome/gap junction.
Because of the extreme thinness of sections used in the electron microscope, the views of cells and their constituents are essentially two-dimensional; in such views, the casual observer can easily fail to appreciate the true shape and extent of any particular profile. This is certainly the case for specialized intercellular junctions. More stringent observations, including examination of multiple sections of cells in various orientations, as well as use of serial sections, has shown, in epithelium as well as other tissues, that some categories of junctions occupy continuous bands of plasmalemma (zonulae) that form rings about the lateral cell surface. Other junctions are more strictly delimited, in three dimensions occupying round, ovoid, or sometimes irregular patches of plasmalemma in each of the joined cells (see Fig. 5.4B).
Tight junctions, viewed in thin sections, are identified, as the name would imply, by plasmalemmal appositions that come into such close contact as to obliterate the intercellular space between cells. Often multiple points of leaflet fusion are visible along short stretches of apposed membranes (Fig. 5.11A). Replication of membrane surfaces by freeze-fracture techniques (Figs. 5.11B,C) has revealed that tight junctions actually are composed of intramembranous particles arranged to form networks of “tongue-and-groove” interdigitations, which provide a barrier largely impenetrable to most particles and fluids. Tight junctional arrays prevent direct diffusion of various products through the extracellular fluid space, thus forcing their passage “through proper channels,” so to speak, namely via mechanisms such as vesicular transport through the cytoplasm. Tight junctions are also found in abundance in pancreatic acini, as well as between endothelial cells in certain segments of blood vessels. To some degree, tight junctions are likely responsible for maintenance of the blood–brain barrier.
Gap junctions, also known as nexuses or maculae communicantes, were in early studies mistaken for tight junctions, but it was realized, through the use of special staining procedures and particle tracer techniques, that these complexes were different in their overall thickness and, furthermore, allowed particles below a certain size to penetrate the extracellular space at their level. Gap junctions appear in thin EM sections to be composed of seven layers (see Figs. 5.4A, 5.11D), the middle one appearing as a 2–4 nm “gap”, explaining the common name for this type of junction. Closer examination has revealed that the gap junction is an assemblage of hexagonally packed “connexons” (as their individual intramembranous particles are called; see Fig. 5.4B,C), each of which contains a tiny pore at its center. It is thought that such pores are aligned to form thin channels, allowing intracytoplasmic passage of ions (e.g. Ca2+) and other substances (such as ATP and amino acids) directly from the cytoplasm of one cell into the interior of the other, a distance calculated to be only about 2.5 nm. In this way, it is postulated, neighboring cells can be “coupled” both electrically and metabolically.
Adherens junctions, though often descriptively linked with desmosomes, can be distinguished from them structurally on the basis of the adherens junctions being more diffuse-appearing and more widely distributed than desmosomes (see Fig. 5.11D). In addition to this morphological disparity are fundamental differences in the species of proteins associated with and composing the two types of junctions.
Adherens junctions are frequently associated with microfilaments, whereas desmosomes instead are accompanied by intermediate filaments. A well-described example of this dichotomy is found in cardiac muscle in the intercalated disks, the zones of extensive junctional attachment between adjacent cardiac muscle cells (see Fig. 5.11D,E). Here at the cell tips, the actin filaments of the myofibrils terminate in the amorphous dense zones of the fascia adherens portion of the disk. In contrast, myofibrils do not come into contact with desmosomes. Furthermore, desmosomes exhibit a far more distinct and layered ultrastructure (see Fig. 5.11E), including a central lamella lying in the extracellular space between cells and intensely opaque intracellular, subplasmalemmal plaques, within whose substance profiles of intermediate filaments can often be detected. In epithelial cells, both desmosomes and hemidesmosomes – which are essentially “half” desmosomes confined to the cells’ basal regions – contain subplasmalemmal plaques that are the insertion points for cords of keratin-type intermediate filaments (see Fig. 5.11F). As mentioned, distinctly different types of proteins are specific to each type of junction, and so, according to the philosophy of form following function, their structural resemblance to one another is related largely to their similar adhesive roles in cells.
XII Special Tissues, Specialized Ultrastructure
Although a chapter such as this one necessarily deals in generalities, in reality few researchers work on generic “cells”. Instead, certain types of cells, for one or many reasons, are of special interest to individual investigators. It is therefore useful to view cell ultrastructure as it is specifically arranged to form a cell type that is particularly appealing to physiologists: the nerve cell or neuron. This cell type not only generates and transfers electrochemical signals, it is also heavily devoted to synthetic activity. Though neurons vary considerably in size and shape in their various locations in the central, peripheral and enteric nervous systems, as a group they are characterized by their ability to exercise their functions at points far removed from the central cell region that contains the nucleus. This action-at-a-distance ability can be directly correlated with ultrastructural features of neurons. The cell body or soma, which contains a single prominent nucleus, also houses the bulk of the synthetic machinery of the cell (Fig. 5.12A), including extensive, often multiple Golgi apparatus and groupings of rough endoplasmic reticulum cisternal profiles, known collectively as “Nissl substance”, because of the staining properties of their ribonucleoprotein. “Free” ribosomes (i.e. not attached to membranes) and polysomes abound as well within the neuronal soma.
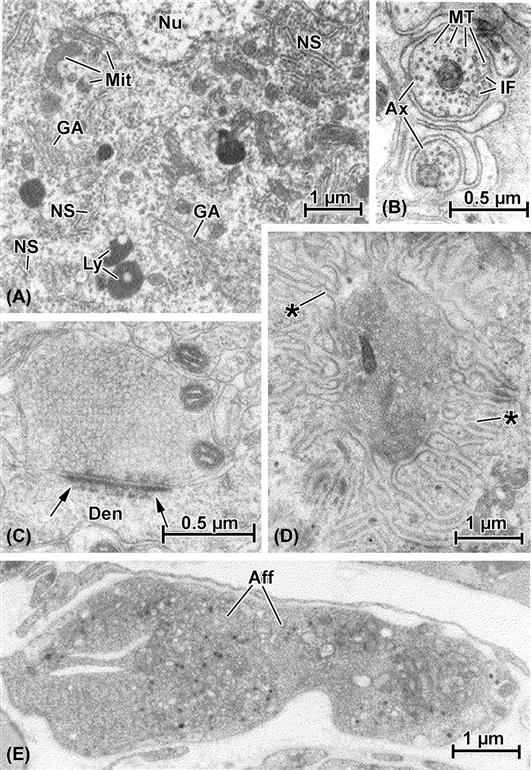
FIGURE 5.12 Electron micrographs illustrating specialized ultrastructure of neurons. (A) A portion of the cell body, or soma, that contains the nucleus (Nu). The cytoplasm in this region is filled with membrane-based synthetic organelles, including multiple profiles of the Golgi apparatus (GA) and cytoplasmic islands of rough endoplasmic reticulum and polyribosomes (known collectively as “Nissl substance,” NS). Numerous small mitochondria are scattered throughout the soma and other examples of typical cell organelles such as lysosomes (Ly) are represented as well. (B) High-magnification view of axons (Ax), the elongated cytoplasmic processes of neurons. These are cut transversely and contain individual mitochondria (Mi) and numerous cross-sectioned cytoskeletal elements, including both microtubules (MT) and intermediate filaments (IF). (C) Axodendritic synapse (i.e. a nerve-to-nerve contact made by an axon with a dendrite, Den). The presynaptic axon terminal is packed with neurosecretory vesicles and also contains mitochondria. The hallmark of this complex is the synaptic density (arrows) which both attaches the two apposed nerve elements and delineates the region in which transfer of neurotransmitter will occur. (D) Motor end-plate, a contact made by an axon terminal of the peripheral nervous system with a skeletal muscle fiber. Although much larger than the CNS terminal (see scale bars in (B), (C), (D) and (E)), like it, the end-plate terminal is filled with vesicles; instead of a dense plaque at the point of contact, however, there is a deep depression in the skeletal muscle into which the terminal nestles, together with elaborate secondary folds formed by the muscle cell membrane (∗). (E) An afferent or sensory nerve terminal (Aff) in heart. This axon termination is considerably larger than most efferent terminals (see (C) and (D)) and contains a variety of inclusions such as mitochondria, clear and dense vesicles and other structures.
It is the cytoplasmic processes of neurons, their dendrites and axons, that most effectively attest to their specialized functions. While dendrites, which generally bring signals toward the cell body, are usually limited to no more than several micrometers in length, the points at which the axons terminate may be located several millimeters away (or even farther, depending upon such factors as the size of the animal, type of neuron and its particular location in the body). Both dendrites and axons, as would be expected, are well endowed with cytoskeletal elements, oriented preferentially along the processes in a configuration that best supports their attenuated shapes. Electron microscopic observation demonstrates that microtubules and intermediate filaments are the most prominent constituents of axons (see Fig. 5.12B).
As pointed out, neurons, though organized along certain unified architectural lines, can vary markedly in their ultrastructure. Nowhere is this more pronounced than at their most distal portions, the tips of the axons, or axon terminals. These termini may have as their target other neurons, or may abut contractile cells, such as smooth muscle (both in viscera and blood vessels), cardiac muscle cells or skeletal myocytes. Axon terminals are usually characterized by concentrations of vesicles of one type or another. Where these axon terminals occur, they may lack any specialized structure that attaches them to the target cell. Often, though, where neurons terminate on one another, a distinct appositional complex known as the synapse is found.
The axon terminal (also known as a “bouton”) constitutes the presynaptic element, and may terminate on a neuronal soma or another axon but, in the central nervous system (CNS), more often is found in contact with a dendrite (see Fig. 5.12C). In addition to the neurosecretory vesicles that fill the axonal bouton, there is a prominent synaptic density whose function appears similar to that of an adherens or desmosomal type of adhesive junction. The synapse functions through a process of vesicle fusion with the presynaptic membrane, with subsequent exocytosis of the transmitter chemical substance (which can be either excitatory or inhibitory) into the synaptic cleft, where it acts on the postsynaptic membrane side of the complex (i.e. the dendritic membrane in Fig. 5.12C), changing its electrical potential and thus generating a signal (thence the term “chemoelectric” for such an event). Where no synapse is formed, as for example in axon terminations near blood vessel walls, diffusion of neurotransmitter is sufficient to modulate the contractile activity of the vascular smooth muscle cells there.
In the case of vertebrate motor neuron axons, which terminate on skeletal muscle fibers, considerable specialization characterizes both the axon terminal and the adjacent muscle cell. This sort of “synapse” is better known as a motor end-plate (see Fig. 5.12D). The axon tip here is considerably larger than the typical boutons found in the CNS and, accordingly, contains a considerably greater number of neurosecretory vesicles. The underlying muscle cell plasmalemma (“sarcolemma”), furthermore, has itself become specialized, containing concentrations of membrane-associated receptors (usually for acetylcholine) and forming both a large depression into which the axon fits (the synaptic “gutter”) and numerous secondary infoldings that increase the amount of excitable muscle membrane available to the released neurotransmitter.
The terminal axons discussed thus far are efferent, i.e. they convey outbound signals from the nerve cell body to its distant target. Also found are afferent, or sensory, terminals, which function to send stimuli such as pressure or pain back to the soma. The contents of such terminals usually greatly vary, including not only vesicles of different shapes and sizes, but also numerous mitochondria, granules, and whorled membranous bodies (see Fig. 5.12E).
Acknowledgments
Over a 40-year career in microscopy, I have been privileged to have trained by and collaborated with many excellent scientists, among them Professors James Norman Dent, W. Gerald Robison, Nick Sperelakis and Lennart Heimer. During this time, my research projects were greatly aided by Ms Jan Redick, Miss Barbara Ann Plantholt, Ms Susan Purdy-Ramos, Mr Lawrence A. Hawkey, Ms Ellen van Niel and Ms Barbara Thornhill and many others too numerous to list. Special thanks goes to Dr Robert L. Chevalier, in whose laboratory I have worked for the past decade, and in which this latest version of the present chapter was prepared.
BIBLIOGRAPHY
1. Andre J. Mitochondria. Biol Cell. 1994;80:103–106.
2. Bershadsky AD, Vasiliev JM. Cytoskeleton. New York: Plenum Press; 1988.
3. Clermont Y, Rambourg A, Hermo L. Trans-Golgi network (TGN) of different cell types: three-dimensional structural characteristics and variability. Anat Rec. 1995;24:289–301.
4. Davis LI. The nuclear pore complex. Annu Rev Biochem. 1995;64:865–896.
5. Dessev GN. Nuclear envelope structure. Curr Opin Cell Biol. 1992;4:430–435.
6. Di Fiore MSH. Atlas of Normal Histology. 6th ed. Philadelphia: Lea and Febiger; 1974.
7. Fawcett DW. The Cell. 2nd ed. Philadelphia: W.B. Saunders; 1981.
8. Fawcett DW. Bloom and Fawcett: A Textbook of Histology. 12th ed. New York: Hodder Arnold; 1997.
9. Fierlbeck W, Liu A, Coyle R, Ballermann BJ. Endothelial cell apoptosis during glomerular capillary lumen formation in vivo. J Am Soc Nephrol. 2003;14:1349–1354.
10. Forbes MS, Sperelakis N. Intercalated discs of mammalian heart: a review of structure and function. Tissue Cell. 1985;17:605–648.
11. Gerace L. Molecular trafficking across the nuclear pore complex. Curr Opin Cell Biol. 1992;4:637–645.
12. Gonatas NK. Contributions to the physiology and pathology of the Golgi apparatus. Am J Pathol. 1994;145:751–761.
13. Hayat MA. Principles and Techniques of Electron Microscopy. 3rd ed. Boca Raton: CRC Press; 1989.
14. Hotchkiss RS, Strasser A, McDunn JE, Swanson PE. Cell death. New Engl J Med. 2009;361:1570–1583.
15. Lodish H, Berk A, Kaiser CA, et al. Molecular Cell Biology. 6th ed. New York: W.H. Freeman; 2007.
16. Novikoff AB. The endoplasmic reticulum: a cytochemist’s view. Proc Natl Acad Sci USA. 1976;73:2781–2786.
17. Nunnari J, Walter P. Protein targeting to and translocation across the membrane of the endoplasmic reticulum. Curr Opin Cell Biol. 1992;4:573–580.
18. Palade GE. The endoplasmic reticulum. J Biophys Biochem Cytol. 1956;2:85–98.
19. Pante N, Aebi U. Molecular dissection of the nuclear pore complex. Crit Rev Biochem Mol Biol. 1996;31:153–199.
20. Peters A, Palay SL, Webster HdeF. The Fine Structure of the Nervous System Neurons and their Supporting Cells. 3rd ed. New York: Oxford University Press; 1991.
21. Porter KR, Bonneville MA. Fine Structure of Cells and Tissue. Philadelphia: Lea and Febiger; 1973.
22. Rhodin JAG. Histology A Text and Atlas. New York: Oxford University Press; 1974.
23. Shay JW. Cell and Molecular Biology of the Cytoskeleton. New York: Plenum Press; 1986.