Chapter 3. NLP on Apache Spark
It’s no longer news that there is a data deluge. Every day, people and devices are creating huge amounts of data. Text data is definitely one of the main kinds of data that humans produce. People write millions of comments, product reviews, Reddit messages, and tweets per day. This data is incredibly valuable—for both research and commerce. Because of the scale at which this data is created, our approach to working with it has changed.
Most of the original research in NLP was done on small data sets with hundreds or thousands of documents. You may think that it would be easier to build NLP applications now that we have so much more text data with which to build better models. However, these pieces of text have different pragmatics and are of different varieties, so leveraging them is more complicated from a data-science perspective. From the software engineering perspective, big data introduces many challenges. Structured data has predictable size and organization, which makes it easier to store and distribute efficiently. Text data is much less consistent. This makes parallelizing and distributing work more important and potentially more complex. Distributed computing frameworks like Spark help us manage these challenges and complexities.
In this chapter, we will discuss the Apache Spark and Spark NLP. First, we will cover some basic concepts that will help us understand distributed computing. Then, we will talk briefly about the history of distributed computing. We will talk about some important modules in Spark—Spark SQL and MLlib. This will give us the background and context needed to talk about Spark NLP in technical detail.
Now, we’ll cover some technical concepts that will be helpful in understanding how Spark works. The explanations will be high level. If you are interested in maximizing performance, I suggest looking more into these topics. For the general audience, I hope this material will give you the intuition necessary to help make decisions when designing and building Spark-based applications.
Parallelism, Concurrency, Distributing Computation
Let’s start by defining some terms. A process can be thought of as a running program. A process executes its code using an allotted portion of memory also known as a memory space. A thread is a sequence of execution steps within a process that the operating system can schedule. Sharing data between processes generally requires copying data between the different memory spaces. When a Java or Scala program is run, the Java Virtual Machine (JVM) is the process. The threads of a process share access to the same memory space, which they access concurrently.
Concurrent access of data can be tricky. For example, let’s say we want to generate word counts. If two threads are working on this process, it’s possible for us to get the wrong count. Consider the following program (written in pseudo-Python). In this program, we will use a thread pool. A thread pool is a way to separate partitioning work from scheduling. We allocate a certain number of threads, and then we go through our data asking for threads for the pool. The operating system can then schedule the work.
0: def word_count(tokenized_documents): # list of lists of tokens
1: word_counts = {}
2: thread_pool = ThreadPool()
3: i = 0
4: for thread in thread_pool
5: run thread:
6: while i < len(tokenized_documents):
7: doc = tokenized_documents[i]
8: i += 1
9: for token in doc:
10: old_count = word_counts.get(token, 0)
11: word_counts[token] = old_count + 1
12: return word_counts
This looks reasonable, but we see that the code under run thread references data in the shared memory space like i and word_counts. Table 3-1 shows the execution of this program with two threads in the ThreadPool starting at line 6.
| time | thread1 | thread2 | i | valid_state |
|---|---|---|---|---|
| 0 |
while i < len(tokenized_documents) |
0 | yes | |
| 1 |
while i < len(tokenized_documents) |
0 | yes | |
| 2 |
doc = tokenized_documents[i] |
0 | yes | |
| 3 |
doc = tokenized_documents[i] |
0 | NO |
At time 3, thread2 will be retrieving tokenized_documents[0], while thread1 is already set to work on the first document. This program has a race condition, in which we can get incorrect results depending on the sequence of operations done in the different threads. Avoiding these problems generally involves writing code that is safe for concurrent access. For example, we can pause thread2 until thread1 is finished updating by locking on tokenized_documents. If you look at the code, there is another race condition, on i. If thread1 takes the last document, tokenized_documents[N-1], thread2 starts its while-loop check, thread1 updates i, then thread2 uses i. We will be accessing tokenized_documents[N], which doesn’t exist. So let’s lock on i.
0: def word_count(tokenized_documents): # list of lists of tokens
1: word_counts = {}
2: thread_pool = ThreadPool()
3: i = 0
4: for thread in thread_pool
5: run thread:
6: while True:
7: lock i:
8: if i < len(tokenized_documents)
9: doc = tokenized_documents[i]
10: i += 1
11: else:
12: break
13: for token in doc:
14: lock word_counts:
15: old_count = word_counts.get(token, 0)
16: word_counts[token] = old_count + 1
17: return word_counts
Now, we are locking on i and checking i in the loop. We also lock on word_counts so that if two threads want to update the counts of the same word, they won’t accidentally pull a stale value for old_count. Table 3-2 shows the execution of this program now from line 7.
| time | thread1 | thread2 | i | valid state |
|---|---|---|---|---|
| 0 |
lock i |
0 | yes | |
| 1 |
if i < len(tokenized_documents) |
blocked | 0 | yes |
| 2 |
doc = tokenized_documents[i] |
blocked | 0 | yes |
| 3 |
i += 1 |
blocked | 0 | yes |
| 4 |
lock i |
1 | yes | |
| 5 |
lock word_counts |
1 | yes | |
| 6 |
if i < len(tokenized_documents) |
1 | yes | |
| 7 |
old_count = word_counts.get(token, 0) |
1 | yes | |
| 8 |
doc = tokenized_documents[i] |
1 | yes | |
| 9 |
word_counts[token] = old_count + 1 |
1 | yes | |
| 10 |
i += 1 |
1 | yes | |
| 11 |
lock word_counts |
2 | yes | |
| 12 |
old_count = word_counts.get(token, 0) |
blocked | 2 | yes |
| 13 |
word_counts[token] = old_count + 1 |
blocked | 2 | yes |
| 14 |
lock word_counts |
2 | yes | |
| 15 | blocked |
old_count = word_counts.get(token, 0) |
2 | yes |
| 16 | blocked |
word_counts[token] = old_count + 1 |
2 | yes |
We fixed the problem, but at the cost of frequently blocking one of the threads. This means that we are getting less advantage of the parallelism. It would be better to design our algorithm so that the threads don’t share state. We will see an example of this when we talk about MapReduce.
Sometimes, parallelizing on one machine is not sufficient, so we distribute the work across many machines grouped together in a cluster. When all the work is done on a machine, we are bringing the data (in memory or on disk) to the code, but when distributing work we are bringing the code to the data. Distributing the work of a program across a cluster means we have new concerns. We don’t have access to a shared memory space, so we need to be more thoughtful in how we design our algorithms. Although processes on different machines don’t share a common memory space, we still need to consider concurrency because the threads of the process on a given machine of the cluster still share common (local) memory space. Fortunately, modern frameworks like Spark mostly take care of these concerns, but it’s still good to keep this in mind when designing your programs.
Programs that work on text data often find some form of parallelization helpful because processing the text into structured data is often the most time-consuming stage of a program. Most NLP pipelines ultimately output structured numeric data, which means that the data that’s loaded, the text, can often be much larger than the data that is output. Unfortunately, because of the complexity of NLP algorithms, text processing in distributed frameworks is generally limited to basic techniques. Fortunately, we have Spark NLP, which we will discuss shortly.
Parallelization Before Apache Hadoop
HTCondor is a framework developed at the University of Wisconsin–Madison starting in 1988. It boasts an impressive catalog of uses. It was used by NASA, the Human Genome Project, and the Large Hadron Collider. Technically, it’s not just a framework for distributing computation—it also can manage resources. In fact, it can be used with other frameworks for distributing computation. It was built with the idea that machines in the cluster may be owned by different users, so work can be scheduled based on available resources. This is from a time when clusters of computers were not as available.
GNU parallel and pexec are UNIX tools that can be used to parallelize work on a single machine, as well as across machines. This requires that the distributable portions of work be run from the command line. These tools allow us to utilize resources across machines, but it doesn’t help with parallelizing our algorithms.
MapReduce and Apache Hadoop
We can represent distributed computation with two operations: map and reduce. The map operation can be used to transform, filter, or sort data. The reduce operation can be used to group or summarize data. Let’s return to our word count example to see how we can use these two operations for a basic NLP task.
def map(docs):
for doc in docs:
for token in doc:
yield (token, 1)
def reduce(records):
word_counts = {}
for token, count in records:
word_counts[token] = word_counts.get(token, 0) + count
for word, count in word_counts.items():
yield (word, count)
The data is loaded in partitions, with some documents going to each mapper process on the cluster. There can be multiple mappers per machine. Each mapper runs the map function on their documents and saves the results to the disk. After all of the mappers have completed, the data from the mapper stage is shuffled so that, all the records with the same key (word in this case), are in the same partition. This data is now sorted so that within a partition, all the records are ordered by key. Finally, the sorted data is loaded and the reduce step is called for each partition reducer process combining all of the counts. In between stages, the data is saved to the disk.
MapReduce can express most distributed algorithms, but some are difficult or downright awkward in this framework. This is why abstractions to MapReduce were developed rather quickly.
Apache Hadoop is the popular open source implementation of MapReduce along with a distributed file system, Hadoop Distributed File System (HDFS). To write a Hadoop program, you need to select or define an input format, mapper, reducer, and output format. There have been many libraries and frameworks to allow higher levels of implementing a program.
Apache Pig is a framework for expressing MapReduce programs in procedural code. Its procedural nature makes implementing extract, transform, load (ETL) programs very convenient and straightforward. However, other types of programs, model training programs for example, are much more difficult. The language that Apache Pig uses is called Pig Latin. There is some overlap with SQL, so if someone knows SQL well, learning Pig Latin is easy.
Apache Hive is a data warehousing framework originally built on Hadoop. Hive allows users to write SQL that is executed with MapReduce. Now, Hive can run using other distributed frameworks in addition to Hadoop, including Spark.
Apache Spark
Spark is a project started by Matei Zaharia. Spark is a distributed computing framework. There are important differences in how Spark and Hadoop process data. Spark allows users to write arbitrary code against distributed data. Currently, there are official APIs for Spark in Scala, Java, Python, and R. Spark does not save intermediate data to disk. Usually, Spark-based programs will keep data in memory, though this can be changed by configuration to also utilize the disk. This allows for quicker processing but can require more scale out (more machines), or more scale up (machines with more memory).
Let’s look at word_count in Spark.
Architecture of Apache Spark
Spark is organized around a driver that is running the program, a master that manages resources and distributes work, and workers that execute computation. There are a number of possible masters. Spark ships with its own master, which is what is used in standalone and local modes. You can also use Apache YARN or Apache Mesos. In my experience, Apache YARN is the most common choice for enterprise systems.
Let’s take a more detailed look at Spark’s architecture.
Physical Architecture
We start our program on the submitting machine which submits an application. This driver runs the application on the client machine and sends jobs to the spark master to be distributed to the workers. The spark master may not be a completely separate machine. That machine may also be doing work on the cluster and so would be a worker as well. Also, you may be running your program on the spark master, and so it could also be the client machine.
There are two modes that you can use to start a Spark application: cluster mode and client mode. If the machine submitting the application is the same machine that runs the application, you are in client mode, because you are submitting from the client machine. Otherwise, you are in cluster mode. Generally, you use client mode if your machine is inside the cluster and cluster mode if it is not (see Figures 3-1 and 3-2).

Figure 3-1. Physical architecture (client mode)

Figure 3-2. Physical architecture (cluster mode)
You can also run Spark in local mode in which, as the name implies, client machine, spark master, and worker are all the same machine. This is very useful for developing and testing Spark applications. It is also useful if you want to parallelize work on one machine.
Now that we have an idea of the physical architecture, let’s look at the logical architecture.
Logical Architecture
In looking at the logical architecture of Spark, we will treat the client machine, spark master, and worker as if they are different (see Figure 3-3). The driver is a JVM process that will submit work to the spark master. If the program is a Java or Scala program, then it is also the process running the program. If the program is in Python or R, then the driver process is a separate process from that running the program.
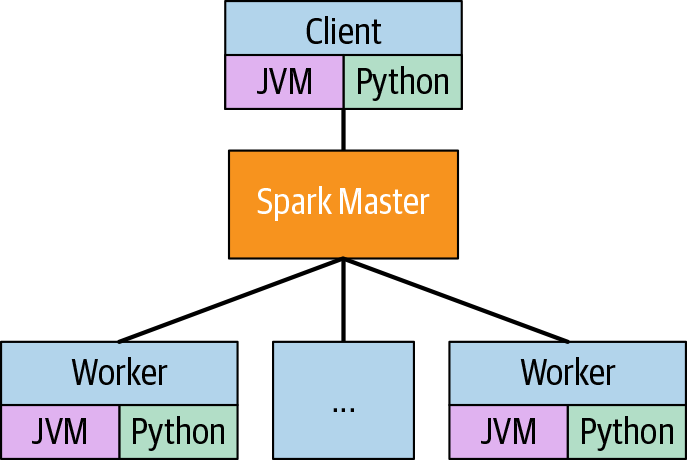
Figure 3-3. Logical architecture
The JVM processes on the workers are called executors. The work to be done is defined on the driver and submitted to the spark master, which orchestrates the executors to do the work. The next step in understanding Spark is understanding how the data is distributed.
Partitioning
In Spark, data is partitioned across the cluster. There are usually more partitions than executors. This allows for each thread on each executor to be utilized. Spark will distribute the data in the RDD evenly across the cluster into the default number of partitions. We can specify the number of partitions, and we can specify a field to partition by. This can be very useful when your algorithm requires some degree of locality—for example, having all the tweets from a single user.
Serialization
Any code that is shipped to the data should refer only to serializable objects. NotSerializableException errors are common and can be nearly inscrutable to those new to Spark. When we map over an RDD, we are creating a Function and sending it to the machines with the data. A function is the code that defines it, and the data needed in the definition. This second part is called the closure of the function. Identifying what objects are needed for a function is a complicated task, and sometimes extraneous objects can be captured. If you are having problems with serializability, there are a couple of possible solutions. The following are questions that can help you find the right solution:
- Are you using your own custom classes? Make sure that they are serializable.
- Are you loading a resource? Perhaps your distributed code should load it lazily so that it is loaded on each executor, instead of being loaded on the driver and shipped to the executors.
- Are Spark objects (
SparkSession,RDDs) being captured in a closure? This can happen when you are defining your function anonymously. If your function is defined anonymously, perhaps you can define it elsewhere.
These tips can help find common errors, but the solution to this problem is something that can be determined only on a case-by-case basis.
Ordering
When working with distributed data, there is not necessarily a guaranteed order to the items in your data. When writing your code, keep in mind that the data exists in partitions across the cluster.
This is not to say that we cannot define an order. We can define an order on the partitions by an index. In this situation, the “first” element of the RDD will be the first element of the first partition. Furthermore, let’s say we want to order an RDD[Int] ascending by value. Using our ordering on partitions, we can shuffle the data such that all elements in partition i are less than all elements in partition i+1. From here, we can sort each partition. Now we have a sorted RDD. This is an expensive operation, however.
Output and logging
When writing functions that are used to transform data, it is often useful to print statements or, preferably, to log statements to look at the state of variables in the function. In a distributed context, this is more complicated because the function is not running on the same machine as the program. Accessing the logs and the stdout generally depends on the configuration of the cluster and which master you are using. In some situations, it may suffice to run your program on a small set of data in local mode.
Spark jobs
A Spark-based program will have a SparkSession, which is how the driver talks to the master. Before Spark version 2.x, the SparkContext was used for this. There is still a SparkContext, but it is part of the SparkSession now. This SparkSession represents the App. The App submits jobs to the master. The jobs are broken into stages, which are logical groupings of work in the job. The stages are broken into tasks, which represent the work to be done on each partition (see Figure 3-4).

Figure 3-4. Spark jobs
Not every operation on data will start a job. Spark is lazy—in a good way. Execution is done only when a result is needed. This allows the work done to be organized into an execution plan to improve efficiency. There are certain operations, sometimes referred to as actions, that will cause execution immediately. These are operations that return specific values, e.g. aggregate. There are also operations where further execution planning becomes impossible until computed, e.g. zipWithIndex.
This execution plan is often referred to as the Directed Acyclic Graph (DAG). Data in Spark is defined by its DAG, which includes its sources and whatever operations were run to generate it. This allows Spark to remove data from memory as necessary without losing reference to the data. If data that was generated and removed is referred to later, it will be regenerated. This can be time-consuming. Fortunately, we can instruct Spark to keep data if we need to.
Persisting
The basic way of persisting data in Spark is with the persist method. This creates a checkpoint. You can also use the cache method and provide options for configuring how the data will be persisted. The data will still be generated lazily, but once it is generated, it will be kept.
Let’s look at an example:
from operator import concat, itemgetter, methodcaller
import os
from time import sleep
import pyspark
from pyspark import SparkConf
from pyspark.sql import SparkSession
from pyspark.sql import functions as fun
from pyspark.sql.types import *
packages = ','.join([
"com.johnsnowlabs.nlp:spark-nlp_2.11:2.4.5",
])
def has_moon(text):
if 'moon' in text:
sleep(1)
return True
else:
return False
# RDD containing filepath-text pairs
path = os.path.join('data', 'mini_newsgroups', 'sci.space')
text_pairs = spark.sparkContext\
.wholeTextFiles(path)
texts = text_pairs.map(itemgetter(1))
lower_cased = texts.map(methodcaller('lower'))
moon_texts = texts.filter(has_moon).persist()
print('This appears quickly because the previous operations are '
'all lazy')
print(moon_texts.count())
print('This appears slowly since the count method will call '
'has_moon which sleeps')
print(moon_texts.reduce(concat)[:100])
print('This appears quickly because has_moon will not be '
'called due to the data being persisted')
This appears quickly because the previous operations are all lazy 11 This appears slowly since the count method will call has_moon which sleeps Newsgroups: sci.space Path: cantaloupe.srv.cs.cmu.edu!das-news.harvard.edu!noc.near.net! uunet!zaphod This appears quickly because has_moon will not be called due to the data being persisted
Now that we have an idea of how Spark works, let’s go back to our word-count problem.
from collections import Counter
from operator import add
from nltk.tokenize import RegexpTokenizer
# RDD containing filepath-text pairs
texts = spark.sparkContext.wholeTextFiles(path)
print('\n\nfilepath-text pair of first document')
print(texts.first())
tokenizer = RegexpTokenizer(r'\w+', gaps=False)
tokenized_texts = texts.map(
lambda path_text: tokenizer.tokenize(path_text[1]))
print('\n\ntokenized text of first document')
print(tokenized_texts.first())
# This is the equivalent place that the previous implementations
# started
document_token_counts = tokenized_texts.map(Counter)
print('\n\ndocument-level counts of first document')
print(document_token_counts.first().most_common(10))
word_counts = token_counts = document_token_counts.reduce(add)
print('\n\nword counts')
print(word_counts.most_common(10))
filepath-text pair of first document
('file:/.../spark-nlp-book/data/mini_news...')
tokenized text of first document
['Xref', 'cantaloupe', 'srv', 'cs', 'cmu', ..., 'cantaloupe', 'srv']
document-level counts of first document
[('the', 13), ('of', 7), ('temperature', 6), ..., ('nasa', 4)]
word counts
[('the', 1648), ('of', 804), ..., ('for', 305), ('cmu', 288)]
As you see, we use the map and reduce methods here. Spark allows you to implement MapReduce-style programs, but you can also implement in many other ways.
Python and R
Spark is primarily implemented in Scala. The Java API is there to allow more idiomatic Java use of Spark. There is also a Python API (PySpark) and an R API (SparkR). Spark-based programs implemented in Scala or Java run on the same JVM that serves as the driver. Programs implemented in PySpark or SparkR run in Python and R processes, respectively, with the SparkSession ultimately in a different process. This generally does not affect the performance, unless we use functions defined in Python or R.
As can be seen in the previous example, when we are tokenizing, counting, and combining counts we are calling Python code to process our data. This is accomplished by the JVM process serializing and shipping the data to the Python process, which is then deserialized, processed, serialized, and shipped back to the JVM to be deserialized. This adds a lot of extra work to our job. When using PySpark or SparkR it will be faster to use internal Spark functions whenever possible.
Not using custom functions in Python or R seems restrictive when using RDDs, but most likely, your work will be using the DataFrames and DataSets that we discuss in the next section.
Spark SQL and Spark MLlib
Since the release of Spark 2, the primary intended way to work with data within Spark is through the Dataset. The Dataset[T] is an object that allows us to treat our distributed data as tables. The type parameter T is the type used to represent the rows of the table. There is a special kind of Dataset in which the type of the rows is Row, which allows us to have tabular data without defining new classes—this does come at the cost of losing some type safety. The examples we’ll be using will generally be with DataFrames, since they are the best way to work with data in PySpark.
The Dataset and DataFrame are defined in the Spark SQL module, since one of the greatest benefits is the ability to express many operations with SQL. The prebuilt user-defined functions (UDFs) are available in all the APIs. This allows us to do most kinds of processing in the non-JVM languages with the same efficiency as if we were using Scala or Java.
Another module we need to introduce before we begin talking about Spark NLP is MLlib. MLlib is a module for doing machine learning on Spark. Before Spark 2, all the MLlib algorithms were implemented on RDDs. Since then, a new version of MLlib was defined using Datasets and DataFrames. MLlib is similar in design, at a high level, to other machine learning libraries, with a notion of transformers, models, and pipelines.
Before we talk about MLlib, let’s load some data into a DataFrame, since MLlib is built using DataFrames. We will be using the Iris data set, which is often used as an example in data science. It’s small, easy to understand, and can work for clustering and classification examples. It is structured data, so it doesn’t give us any text data to work with. Table-like structure is generally designed around structured data, so this data will help us explore the API before getting into using Spark for text.
The iris.data file does not have a header, so we have to tell Spark what the columns are when they are loaded. Let’s construct a schema. The schema is the definition of the columns and their types in the DataFrame. The most common task is to build a model to predict what class an iris flower is (I. virginica, I. setosa, or I. versicolor) based on its sepals and petals.
from pyspark.sql.types import *
schema = StructType([
StructField('sepal_length', DoubleType(), nullable=False),
StructField('sepal_width', DoubleType(), nullable=False),
StructField('petal_length', DoubleType(), nullable=False),
StructField('petal_width', DoubleType(), nullable=False),
StructField('class', StringType(), nullable=False)
])
Now that we have created the schema, we can load our CSV. Table 3-3 shows the summary of the data.
iris = spark.read.csv('./data/iris/iris.data', schema=schema)
iris.describe().toPandas()
| summary | sepal_length | sepal_width | petal_length | petal_width | class | |
|---|---|---|---|---|---|---|
| 0 | count | 150 | 150 | 150 | 150 | 150 |
| 1 | mean | 5.843 | 3.054 | 3.759 | 1.199 | None |
| 2 | stddev | 0.828 | 0.434 | 1.764 | 0.763 | None |
| 3 | min | 4.3 | 2.0 | 1.0 | 0.1 | Iris-setosa |
| 4 | max | 7.9 | 4.4 | 6.9 | 2.5 | Iris-virginica |
Let’s start by looking at the classes (species of Iris) in our data (see Table 3-4).
iris.select('class').distinct().toPandas()
iris.where('class = "Iris-setosa"').drop('class').describe().toPandas()
| summary | sepal_length | sepal_width | petal_length | petal_width | |
|---|---|---|---|---|---|
| 0 | count | 50 | 50 | 50 | 50 |
| 1 | mean | 5.006 | 3.418 | 1.464 | 0.244 |
| 2 | stddev | 0.352 | 0.381 | 0.174 | 0.107 |
| 3 | min | 4.3 | 2.3 | 1.0 | 0.1 |
| 4 | max | 5.8 | 4.4 | 1.9 | 0.6 |
We can register a DataFrame, which will allow us to interact with it purely through SQL. We will be registering our DataFrame as a temporary table. This means that the table will exist only for the lifetime of our App, and it will be available only through our App’s SparkSession (see Table 3-6).
iris.registerTempTable('iris')
spark.sql('''
SELECT *
FROM iris
LIMIT 5
''').toPandas()
| sepal_length | sepal_width | petal_length | petal_width | class | |
|---|---|---|---|---|---|
| 0 | 5.1 | 3.5 | 1.4 | 0.2 | Iris-setosa |
| 1 | 4.9 | 3.0 | 1.4 | 0.2 | Iris-setosa |
| 2 | 4.7 | 3.2 | 1.3 | 0.2 | Iris-setosa |
| 3 | 4.6 | 3.1 | 1.5 | 0.2 | Iris-setosa |
| 4 | 5.0 | 3.6 | 1.4 | 0.2 | Iris-setosa |
Let’s look at some of the fields grouped by their class in Table 3-7.
spark.sql('''
SELECT
class,
min(sepal_length), avg(sepal_length), max(sepal_length),
min(sepal_width), avg(sepal_width), max(sepal_width),
min(petal_length), avg(petal_length), max(petal_length),
min(petal_width), avg(petal_width), max(petal_width)
FROM iris
GROUP BY class
''').toPandas()
| class | Iris-virginica | Iris-setosa | Iris-versicolor |
|---|---|---|---|
| min(sepal_length) | 4.900 | 4.300 | 4.900 |
| avg(sepal_length) | 6.588 | 5.006 | 5.936 |
| max(sepal_length) | 7.900 | 5.800 | 7.000 |
| min(sepal_width) | 2.200 | 2.300 | 2.000 |
| avg(sepal_width) | 2.974 | 3.418 | 2.770 |
| max(sepal_width) | 3.800 | 4.400 | 3.400 |
| min(petal_length) | 4.500 | 1.000 | 3.000 |
| avg(petal_length) | 5.552 | 1.464 | 4.260 |
| max(petal_length) | 6.900 | 1.900 | 5.100 |
| min(petal_length) | 1.400 | 0.100 | 1.000 |
| avg(petal_length) | 2.026 | 0.244 | 1.326 |
| max(petal_length) | 2.500 | 0.600 | 1.800 |
Transformers
A Transformer is a piece of logic that transforms the data without needing to learn or fit anything from the data. A good way to understand transformers is that they represent functions that we wish to map over our data. All stages of a pipeline have parameters so that we can make sure that the transformation is being applied to the right fields and with the desired configuration. Let’s look at a few examples.
SQLTransformer
The SQLTransformer has only one parameter—statement—which is the SQL statement that will be executed against our DataFrame. Let’s use an SQLTransformer to do the group-by we performed previously. Table 3-8 shows the result.
from pyspark.ml.feature import SQLTransformer
statement = '''
SELECT
class,
min(sepal_length), avg(sepal_length), max(sepal_length),
min(sepal_width), avg(sepal_width), max(sepal_width),
min(petal_length), avg(petal_length), max(petal_length),
min(petal_width), avg(petal_width), max(petal_width)
FROM iris
GROUP BY class
'''
sql_transformer = SQLTransformer(statement=statement)
sql_transformer.transform(iris).toPandas()
| class | Iris-virginica | Iris-setosa | Iris-versicolor |
|---|---|---|---|
| min(sepal_length) | 4.900 | 4.300 | 4.900 |
| avg(sepal_length) | 6.588 | 5.006 | 5.936 |
| max(sepal_length) | 7.900 | 5.800 | 7.000 |
| ... |
We get the same output as when we ran the SQL command.
SQLTransformer is useful when you have preprocessing or restructuring that you need to perform on your data before other steps in the pipeline. Now let’s look at a transformer that works on one field and returns the original data with a new field.
Binarizer
The Binarizer is a Transformer that applies a threshold to a numeric field, turning it into 0s (when below the threshold) and 1s (when above the threshold). It takes three parameters:
inputCol- The column to be binarized
outputCol- The column containing the binarized values
threshold- The threshold we will apply
Table 3-9 shows the results.
from pyspark.ml.feature import Binarizer
binarizer = Binarizer(
inputCol='sepal_length',
outputCol='sepal_length_above_5',
threshold=5.0
)
binarizer.transform(iris).limit(5).toPandas()
| sepal_length | ... | class | sepal_length_above_5 | |
|---|---|---|---|---|
| 0 | 5.1 | ... | Iris-setosa | 1.0 |
| 1 | 4.9 | ... | Iris-setosa | 0.0 |
| 2 | 4.7 | ... | Iris-setosa | 0.0 |
| 3 | 4.6 | ... | Iris-setosa | 0.0 |
| 4 | 5.0 | ... | Iris-setosa | 0.0 |
Unlike the SQLTransformer, the Binarizer returns a modified version of the input DataFrame. Almost all Transformers behave this way.
The Binarizer is used when you want to convert a real valued property into a class. For example, if we want to mark social media posts as “viral” and “not-viral” we could use a Binarizer on the views property.
VectorAssembler
Another import Transformer is the VectorAssembler. It takes a list of numeric and vector-valued columns and constructs a single vector. This is useful because all MLlib’s machine learning algorithms expect a single vector-valued input column for features. The VectorAssembler takes two parameters:
inputCols- The list of columns to be assembled
outputCol- The column containing the new vectors
from pyspark.ml.feature import VectorAssembler
assembler = VectorAssembler(
inputCols=[
'sepal_length', 'sepal_width',
'petal_length', 'petal_width'
],
outputCol='features'
)
Let’s persist this data (see Table 3-10).
iris_w_vecs = assembler.transform(iris).persist()
iris_w_vecs.limit(5).toPandas()
| sepal_length | sepal_width | petal_length | petal_width | class | features | |
|---|---|---|---|---|---|---|
| 0 | 5.1 | 3.5 | 1.4 | 0.2 | Iris-setosa | [5.1, 3.5, 1.4, 0.2] |
| 1 | 4.9 | 3.0 | 1.4 | 0.2 | Iris-setosa | [4.9, 3.0, 1.4, 0.2] |
| 2 | 4.7 | 3.2 | 1.3 | 0.2 | Iris-setosa | [4.7, 3.2, 1.3, 0.2] |
| 3 | 4.6 | 3.1 | 1.5 | 0.2 | Iris-setosa | [4.6, 3.1, 1.5, 0.2] |
| 4 | 5.0 | 3.6 | 1.4 | 0.2 | Iris-setosa | [5.0, 3.6, 1.4, 0.2] |
Now we have our features as vectors. This is what the machine learning Estimators in MLlib need to work with.
Estimators and Models
Estimators allow us to create transformations that are informed by our data. Classification models (e.g., decision trees) and regression models (e.g., linear regressions) are prominent examples, but some preprocessing algorithms are like this as well. For example, preprocessing that needs to know the whole vocabulary first will be Estimators. The Estimator is fit with a DataFrame and returns a Model, which is a kind of Transformer. The Models created from classifier and regression Estimators are PredictionModels.
This is a similar design to scikit-learn, with the exception that in scikit-learn when we call fit we mutate the estimator instead of creating a new object. There are pros and cons to this, as there always are when debating mutability. Idiomatic Scala strongly prefers immutability.
Let’s look at some examples of Estimators and Models.
MinMaxScaler
The MinMaxScaler allows us to scale our data to be between 0 and 1. It takes four parameters:
inputCol- The column to be scaled
outputCol- The column containing the scaled values
max- The new maximum value (optional, default = 1)
min- The new minimum value (optional, default = 0)
The results are shown in Table 3-11.
from pyspark.ml.feature import MinMaxScaler
scaler = MinMaxScaler(
inputCol='features',
outputCol='petal_length_scaled'
)
scaler_model = scaler.fit(iris_w_vecs)
scaler_model.transform(iris_w_vecs).limit(5).toPandas()
Notice that the petal_length_scaled column now has values between 0 and 1. This can help some training algorithms, specifically those that have difficulty combining features of different scales.
StringIndexer
Let’s build a model! We will try and predict the class from the other features, and we will use a decision tree. First, though, we must convert our target into index values.
The StringIndexer Estimator will turn our class values into indices. We want to do this to simplify some of the downstream processing. It is simpler to implement most training algorithms with the assumption that the target is a number. The StringIndexer takes four parameters:
inputCol- The column to be indexed
outputCol- The column containing the indexed values
handleInvalid- The policy for how the model should handle values not seen by the estimator (optional, default = error)
stringOrderType- How to order the values to make the indexing deterministic (optional, default = frequencyDesc)
We will also want an IndexToString Transformer. This will let us map our predictions, which will be indices, back to string values. IndexToString takes three parameters:
inputCol- The column to be mapped
outputCol- The column containing the mapped values
labels- The mapping from index to value, usually generated by
StringIndexer
from pyspark.ml.feature import StringIndexer, IndexToString
indexer = StringIndexer(inputCol='class', outputCol='class_ix')
indexer_model = indexer.fit(iris_w_vecs)
index2string = IndexToString(
inputCol=indexer_model.getOrDefault('outputCol'),
outputCol='pred_class',
labels=indexer_model.labels
)
iris_indexed = indexer_model.transform(iris_w_vecs)
Now we are ready to train our DecisionTreeClassifier. This Estimator has many parameters, so I recommend you become familiar with the APIs. They are all well documented in the PySpark API documentation. Table 3-12 shows our results.
from pyspark.ml.classification import DecisionTreeClassifier
dt_clfr = DecisionTreeClassifier(
featuresCol='features',
labelCol='class_ix',
maxDepth=5,
impurity='gini',
seed=123
)
dt_clfr_model = dt_clfr.fit(iris_indexed)
iris_w_pred = dt_clfr_model.transform(iris_indexed)
iris_w_pred.limit(5).toPandas()
| ... | class | features | class_ix | rawPrediction | probability | prediction | |
|---|---|---|---|---|---|---|---|
| 0 | ... | Iris-setosa | [5.1, 3.5, 1.4, 0.2] | 0.0 | [50.0, 0.0, 0.0] | [1.0, 0.0, 0.0] | 0.0 |
| 1 | ... | Iris-setosa | [4.9, 3.0, 1.4, 0.2] | 0.0 | [50.0, 0.0, 0.0] | [1.0, 0.0, 0.0] | 0.0 |
| 2 | ... | Iris-setosa | [4.7, 3.2, 1.3, 0.2] | 0.0 | [50.0, 0.0, 0.0] | [1.0, 0.0, 0.0] | 0.0 |
| 3 | ... | Iris-setosa | [4.6, 3.1, 1.5, 0.2] | 0.0 | [50.0, 0.0, 0.0] | [1.0, 0.0, 0.0] | 0.0 |
| 4 | ... | Iris-setosa | [5.0, 3.6, 1.4, 0.2] | 0.0 | [50.0, 0.0, 0.0] | [1.0, 0.0, 0.0] | 0.0 |
Now we need to map the predicted classes back to their string form using our IndexToString (see Table 3-13).
iris_w_pred_class = index2string.transform(iris_w_pred)
iris_w_pred_class.limit(5).toPandas()
| ... | class | features | class_ix | rawPrediction | probability | prediction | pred_class | |
|---|---|---|---|---|---|---|---|---|
| 0 | ... | Iris-setosa | [5.1, 3.5, 1.4, 0.2] | 0.0 | [50.0, 0.0, 0.0] | [1.0, 0.0, 0.0] | 0.0 | Iris-setosa |
| 1 | ... | Iris-setosa | [4.9, 3.0, 1.4, 0.2] | 0.0 | [50.0, 0.0, 0.0] | [1.0, 0.0, 0.0] | 0.0 | Iris-setosa |
| 2 | ... | Iris-setosa | [4.7, 3.2, 1.3, 0.2] | 0.0 | [50.0, 0.0, 0.0] | [1.0, 0.0, 0.0] | 0.0 | Iris-setosa |
| 3 | ... | Iris-setosa | [4.6, 3.1, 1.5, 0.2] | 0.0 | [50.0, 0.0, 0.0] | [1.0, 0.0, 0.0] | 0.0 | Iris-setosa |
| 4 | ... | Iris-setosa | [5.0, 3.6, 1.4, 0.2] | 0.0 | [50.0, 0.0, 0.0] | [1.0, 0.0, 0.0] | 0.0 | Iris-setosa |
How well did our model fit the data? Let’s see how many predictions match the true class.
Evaluators
The evaluation options in MLlib are still limited compared to libraries like scikit-learn, but they can be useful if you are looking to create an easy-to-run training pipeline that calculates metrics.
In our example, we are trying to solve a multiclass prediction problem, so we will use the MulticlassClassificationEvaluator.
from pyspark.ml.evaluation import MulticlassClassificationEvaluator
evaluator = MulticlassClassificationEvaluator(
labelCol='class_ix',
metricName='accuracy'
)
evaluator.evaluate(iris_w_pred_class)
1.0
This seems too good. What if we are overfit? Perhaps we should try using cross-validation to evaluate our models. Before we do that, let’s organize stages into a pipeline.
Pipelines
Pipelines are a special kind of Estimator that takes a list of Transformers and Estimators and allows us to use them as a single Estimator (see Table 3-14).
from pyspark.ml import Pipeline
pipeline = Pipeline(
stages=[assembler, indexer, dt_clfr, index2string]
)
pipeline_model = pipeline.fit(iris)
pipeline_model.transform(iris).limit(5).toPandas()
| ... | class | features | class_ix | rawPrediction | probability | prediction | pred_class | |
|---|---|---|---|---|---|---|---|---|
| 0 | ... | Iris-setosa | [5.1, 3.5, 1.4, 0.2] | 0.0 | [50.0, 0.0, 0.0] | [1.0, 0.0, 0.0] | 0.0 | Iris-setosa |
| 1 | ... | Iris-setosa | [4.9, 3.0, 1.4, 0.2] | 0.0 | [50.0, 0.0, 0.0] | [1.0, 0.0, 0.0] | 0.0 | Iris-setosa |
| 2 | ... | Iris-setosa | [4.7, 3.2, 1.3, 0.2] | 0.0 | [50.0, 0.0, 0.0] | [1.0, 0.0, 0.0] | 0.0 | Iris-setosa |
| 3 | ... | Iris-setosa | [4.6, 3.1, 1.5, 0.2] | 0.0 | [50.0, 0.0, 0.0] | [1.0, 0.0, 0.0] | 0.0 | Iris-setosa |
| 4 | ... | Iris-setosa | [5.0, 3.6, 1.4, 0.2] | 0.0 | [50.0, 0.0, 0.0] | [1.0, 0.0, 0.0] | 0.0 | Iris-setosa |
Cross validation
Now that we have a Pipeline and an Evaluator we can create a CrossValidator. The CrossValidator itself is also an Estimator. When we call fit, it will fit our pipeline to each fold of data, and calculate the metric determined by our Evaluator. CrossValidator takes five parameters:
estimator- The
Estimatorto be tuned estimatorParamMaps- The hyperparameter values to try in a hyperparameter grid search
evaluator- The
Evaluatorthat calculates the metric numFolds- The number of folds to split the data into
seed- A seed for making the splits reproducible
We will make a trivial hyperparameter grid here, since we are only interested in estimating how well our model does on data it has not seen.
from pyspark.ml.tuning import CrossValidator, ParamGridBuilder
param_grid = ParamGridBuilder().\
addGrid(dt_clfr.maxDepth, [5]).\
build()
cv = CrossValidator(
estimator=pipeline,
estimatorParamMaps=param_grid,
evaluator=evaluator,
numFolds=3,
seed=123
)
cv_model = cv.fit(iris)
Now, we can see how the model does when trained on two-thirds and evaluated on one-third. The avgMetrics in cv_model contains the average value of the designated metric across folds for each point in the hyperparameter grid tested. In our case, there is only one point in the grid.
cv_model.avgMetrics
[0.9588996659642801]
Keep in mind that 95% accuracy is much more believable than 100%.
There are many other Transformers, Estimators, and Models. We will look into more as we continue, but for now, there is one more thing we need to discuss—saving our pipelines.
Serialization of models
MLlib allows us to save Pipelines so that we can use them later. We can also save individual Transformers and Models, but we will often want to keep all the stages of a Pipeline together. Generally speaking, we use separate programs for building models and using models.
pipeline_model.write().overwrite().save('pipeline.model')
! ls pipeline.model/*
pipeline.model/metadata: part-00000 _SUCCESS pipeline.model/stages: 0_VectorAssembler_45458c77ca2617edd7f6 1_StringIndexer_44d29a3426fb6b26b2c9 2_DecisionTreeClassifier_4473a4feb3ff2cf54b73 3_IndexToString_4157a15742628a489a18
NLP Libraries
There are two kinds of NLP libraries, generally speaking: functionality libraries and annotation libraries.
Functionality Libraries
A functionality library is a collection of functions built for specific NLP tasks and techniques. Often, the functions are built without assuming that other functions will be used first. This means that functions like part-of-speech (POS) tagging will also perform tokenization. These libraries are good for research because it is often much easier to implement novel functions. On the other hand, because there is no unifying design, the performance of these libraries is generally much worse than that of annotation libraries.
The Natural Language Tool Kit (NLTK) is a great functionality library. It was originally created by Edward Loper. The landmark NLP book Natural Language Processing with Python (O’Reilly) was written by Steven Bird, Ewan Klein, and Edward Loper. I strongly recommend that book to anyone learning NLP. There a many useful and interesting modules in NLTK. It is, and will likely remain, the best NLP library for teaching NLP. The functions are not necessarily implemented with runtime performance or other productionization concerns in mind. If you are working on a research project and using a data set manageable on a single machine, you should consider NLTK.
Annotation Libraries
Annotation libraries are libraries in which all the functionality is built around a document-annotation model. There are three objects to keep in mind with annotation libraries: document, annotation, and annotator. The idea behind annotation libraries is to augment the incoming data with the results of our NLP functions.
- Document
- The document is the representation of the piece of text we wish to process. Naturally, the document must contain the text. Additionally, we often want to have an identifier associated with each document so that we can store our augmented data as structured data. This identifier will often be a title if the texts we are processing have titles.
- Annotation
- The annotation is the representation of the output of our NLP functions. For the annotation we need to have a type so that later processing knows how to interpret the annotations. Annotations also need to store their location within the document. For example, let’s say the word “pacing” occurs 134 characters into the document. It will have 134 as the start, and 140 as the end. The lemma annotation for “pacing” will have the location. Some annotation libraries also have a concept of document-level annotation that does not have a location. There will be additional fields, depending on the type. Simple annotations like tokens generally don’t have extra fields. Stem annotations usually have the stem that was extracted for the range of the text.
- Annotator
- The annotator is the object that contains the logic for using the NLP function. The annotator will often require configuration or external data sets. Additionally, there are model-based annotators. One of the benefits of an annotation library is that annotators can take advantage of the work done by previous annotators. This naturally creates a notion of a pipeline of annotators.
- spaCy
spaCy is an “industrial strength” NLP library. I will give a brief description, but I encourage you to go and read their fantastic documentation. spaCy combines the document model just described with a model for the language being processed (English, Spanish, etc.), which has allowed spaCy to support multiple languages in a way that is easy for developers to use. Much of its functionality is implemented in Python to get the speed of native code. If you are working in an environment that is using only Python, and you are unlikely to run distributed processes, then spaCy is a great choice.
NLP in Other Libraries
There are some non-NLP libraries that have some NLP functionality. It is often in machine learning libraries to support machine learning on text data.
- scikit-learn
- A Python machine learning library that has functionality for extracting features from text. This functionality is generally of the bag-of-words kind of processing. The way these processes are built allows them to easily take advantage of more NLP-focused libraries.
- Lucene
- A Java document search framework that has some text-processing functionality necessary for building a search engine. We will use Lucene later on when we talk about information retrieval.
- Gensim
- A topic-modeling library (and it performs other distributional semantics techniques). Like spaCy, it is partially implemented in Cython, and like scikit-learn, it allows plug-and-play text processing in its API.
Spark NLP
The Spark NLP library was originally designed in early 2017 as an annotation library native to Spark to take full advantage of Spark SQL and MLlib modules. The inspiration came from trying to use Spark to distribute other NLP libraries, which were generally not implemented with concurrency or distributed computing in mind.
Annotation Library
Spark NLP has the same concepts as any other annotation library but differs in how it stores annotations. Most annotation libraries store the annotations in the document object, but Spark NLP creates columns for the different types of annotations.
The annotators are implemented as Transformers, Estimators, and Models. Let’s take a look at some examples.
Stages
One of the design principles of Spark NLP is easy interoperability with the existing algorithms in MLlib. Because there is no notion of documents or annotations in MLlib there are transformers for turning text columns into documents and converting annotations into vanilla Spark SQL data types. The usual usage pattern is as follows:
- Load data with Spark SQL.
- Create document column.
- Process with Spark NLP.
- Convert annotations of interest into Spark SQL data types.
- Run additional MLlib stages.
We have already looked at how to load data with Spark SQL and how to use MLlib stages in the standard Spark library, so we will look at the middle three stages now. First, we will look at the DocumentAssembler (stage 2).
Transformers
To explore these five stages we will again use the mini_newsgroups data set (see Table 3-15).
from sparknlp import DocumentAssembler, Finisher
# RDD containing filepath-text pairs
texts = spark.sparkContext.wholeTextFiles(path)
schema = StructType([
StructField('path', StringType()),
StructField('text', StringType()),
])
texts = spark.createDataFrame(texts, schema=schema)
texts.limit(5).toPandas()
| path | text | |
|---|---|---|
| 0 | file:/.../spark-nlp-book/data/... | Xref: cantaloupe.srv.cs.cmu.edu sci.astro:3522... |
| 1 | file:/.../spark-nlp-book/data/... | Newsgroups: sci.space\nPath: cantaloupe.srv.cs... |
| 2 | file:/.../spark-nlp-book/data/... | Xref: cantaloupe.srv.cs.cmu.edu sci.space:6146... |
| 3 | file:/.../spark-nlp-book/data/... | Path: cantaloupe.srv.cs.cmu.edu!rochester!udel... |
| 4 | file:/.../spark-nlp-book/data/... | Newsgroups: sci.space\nPath: cantaloupe.srv.cs... |
DocumentAssembler
The DocumentAssembler takes five parameters (see Table 3-16):
inputCol- The column containing the text of the document
outputCol- The name of the column containing the newly constructed document
idCol- The name of the column containing the identifier (optional)
metadataCol- The name of a
Map-type column that represents document metadata (optional) trimAndClearNewLines->- Determines whether to remove new line characters and trim strings (optional, default = True)
document_assembler = DocumentAssembler()\
.setInputCol('text')\
.setOutputCol('document')\
.setIdCol('path')
docs = document_assembler.transform(texts)
docs.limit(5).toPandas()
| path | text | document | |
|---|---|---|---|
| 0 | file:/.../spark-nlp-book/data/... | Xref: cantaloupe.srv.cs.cmu.edu sci.astro:3522... | [(document, 0, 1834, Xref: cantaloupe.srv.cs.c... |
| 1 | file:/.../spark-nlp-book/data/... | Newsgroups: sci.space\nPath: cantaloupe.srv.cs... | [(document, 0, 1804, Newsgroups: sci.space Pat... |
| 2 | file:/.../spark-nlp-book/data/... | Xref: cantaloupe.srv.cs.cmu.edu sci.space:6146... | [(document, 0, 1259, Xref: cantaloupe.srv.cs.c... |
| 3 | file:/.../spark-nlp-book/data/... | Path: cantaloupe.srv.cs.cmu.edu!rochester!udel... | [(document, 0, 8751, Path: cantaloupe.srv.cs.c... |
| 4 | file:/.../spark-nlp-book/data/... | Newsgroups: sci.space\nPath: cantaloupe.srv.cs... | [(document, 0, 1514, Newsgroups: sci.space Pat... |
docs.first()['document'][0].asDict()
{'annotatorType': 'document',
'begin': 0,
'end': 1834,
'result': 'Xref: cantaloupe.srv.cs.cmu.edu sci.astro:...',
'metadata': {
'id': 'file:/.../spark-nlp-book/data/mini_newsg...'
}
}
SentenceDetector
The SentenceDetector uses a rule-based algorithm inspired by Kevin Dias’s Ruby implementation. It takes the following parameters (see Table 3-17):
inputCols- A list of columns to sentence-tokenize.
outputCol- The name of the new sentence column.
useAbbrevations- Determines whether to apply abbreviations at sentence detection.
useCustomBoundsOnly- Determines whether to only utilize custom bounds for sentence detection.
explodeSentences- Determines whether to explode each sentence into a different row, for better parallelization. Defaults to false.
customBounds- Characters used to explicitly mark sentence bounds.
from sparknlp.annotator import SentenceDetector
sent_detector = SentenceDetector()\
.setInputCols(['document'])\
.setOutputCol('sentences')
sentences = sent_detector.transform(docs) sentences.limit(5).toPandas()
| path | text | document | sentences | |
|---|---|---|---|---|
| 0 | file:/.../spark-nlp-book/data/... | ... | [(document, 0, 1834, Xref: cantaloupe.srv.cs.c... | [(document, 0, 709, Xref: cantaloupe.srv.cs.cm... |
| 1 | file:/.../spark-nlp-book/data/... | ... | [(document, 0, 1804, Newsgroups: sci.space Pat... | [(document, 0, 288, Newsgroups: sci.space Path... |
| 2 | file:/.../spark-nlp-book/data/... | ... | [(document, 0, 1259, Xref: cantaloupe.srv.cs.c... | [(document, 0, 312, Xref: cantaloupe.srv.cs.cm... |
| 3 | file:/.../spark-nlp-book/data/... | ... | [(document, 0, 8751, Path: cantaloupe.srv.cs.c... | [(document, 0, 453, Path: cantaloupe.srv.cs.cm... |
| 4 | file:/.../spark-nlp-book/data/... | ... | [(document, 0, 1514, Newsgroups: sci.space Pat... | [(document, 0, 915, Newsgroups: sci.space Path... |
Tokenizer
A Tokenizer is a fundamental Annotator. Almost all text-based data processing begins with some form of tokenization. Most classical NLP algorithms expect tokens as the basic input. Many deep learning algorithms are being developed that take characters as basic input. Most NLP applications still use tokenization. The Spark NLP Tokenizer is a little more sophisticated than just a regular expression-based tokenizer. It has a number of parameters. The following are some of the basic ones (see Table 3-18 for the results):
inputCols- A list of columns to tokenize.
outputCol- The name of the new token column.
targetPattern- Basic regex rule to identify a candidate for tokenization. Defaults to \S+ which means anything not a space (optional).
prefixPattern- Regular expression (regex) to identify subtokens that come in the beginning of the token. Regex has to start with \A and must contain groups (). Each group will become a separate token within the prefix. Defaults to nonletter characters—for example, quotes or parentheses (optional).
suffixPattern- Regex to identify subtokens that are in the end of the token. Regex has to end with \z and must contain groups (). Each group will become a separate token within the prefix. Defaults to nonletter characters—for example, quotes or parentheses (optional).
from sparknlp.annotator import Tokenizer
tokenizer = Tokenizer()\
.setInputCols(['sentences'])\
.setOutputCol('tokens')\
.fit(sentences)
tokens = tokenizer.transform(sentences) tokens.limit(5).toPandas()
| path | text | document | sentences | tokens | |
|---|---|---|---|---|---|
| 0 | file:/.../spark-nlp-book/data/... | ... | ... | [(document, 0, 709, Xref: cantaloupe.srv.cs.cm... | [(token, 0, 3, Xref, {’sentence’: ’1'}), (toke... |
| 1 | file:/.../spark-nlp-book/data/... | ... | ... | [(document, 0, 288, Newsgroups: sci.space Path... | [(token, 0, 9, Newsgroups, {’sentence’: ’1'}),... |
| 2 | file:/.../spark-nlp-book/data/... | ... | ... | [(document, 0, 312, Xref: cantaloupe.srv.cs.cm... | [(token, 0, 3, Xref, {’sentence’: ’1'}), (toke... |
| 3 | file:/.../spark-nlp-book/data/... | ... | ... | [(document, 0, 453, Path: cantaloupe.srv.cs.cm... | [(token, 0, 3, Path, {’sentence’: ’1'}), (toke... |
| 4 | file:/.../spark-nlp-book/data/... | ... | ... | [(document, 0, 915, Newsgroups: sci.space Path... | [(token, 0, 9, Newsgroups, {’sentence’: ’1'}),... |
There are some Annotators that require additional resources. Some require reference data, like the following example, the lemmatizer.
Lemmatizer
The lemmatizer finds the lemmas for the tokens. Lemmas are the entry words in dictionaries. For example, “cats” lemmatizes to “cat,” and “oxen” lemmatizes to “ox.” Loading the lemmatizer requires a dictionary and the following three parameters:
inputCols- A list of columns to tokenize
outputCol- The name of the new token column
dictionary- The resource to be loaded as the lemma dictionary
from sparknlp.annotator import Lemmatizer
lemmatizer = Lemmatizer() \
.setInputCols(["tokens"]) \
.setOutputCol("lemma") \
.setDictionary('en_lemmas.txt', '\t', ',')\
.fit(tokens)
lemmas = lemmatizer.transform(tokens) lemmas.limit(5).toPandas()
Table 3-19 shows the results.
| path | text | document | sentences | tokens | lemma | |
|---|---|---|---|---|---|---|
| 0 | file:/.../spark-nlp-book/data/... | ... | ... | ... | [(token, 0, 3, Xref, {’sentence’: ’1'}), (toke... | [(token, 0, 3, Xref, {’sentence’: ’1'}), (toke... |
| 1 | file:/.../spark-nlp-book/data/... | ... | ... | ... | [(token, 0, 9, Newsgroups, {’sentence’: ’1'}),... | [(token, 0, 9, Newsgroups, {’sentence’: ’1'}),... |
| 2 | file:/.../spark-nlp-book/data/... | ... | ... | ... | [(token, 0, 3, Xref, {’sentence’: ’1'}), (toke... | [(token, 0, 3, Xref, {’sentence’: ’1'}), (toke... |
| 3 | file:/.../spark-nlp-book/data/... | ... | ... | ... | [(token, 0, 3, Path, {’sentence’: ’1'}), (toke... | [(token, 0, 3, Path, {’sentence’: ’1'}), (toke... |
| 4 | file:/.../spark-nlp-book/data/... | ... | ... | ... | [(token, 0, 9, Newsgroups, {’sentence’: ’1'}),... | [(token, 0, 9, Newsgroups, {’sentence’: ’1'}),... |
POS tagger
There are also Annotators that require models as resources. For example, the POS tagger uses a perceptron model, so it is called PerceptronApproach. The PerceptronApproach has five parameters:
inputCols- A list of columns to tag
outputCol- The name of the new tag column
posCol- Column of
Arrayof POS tags that match tokens corpus- POS tags delimited corpus; needs “delimiter” in options
nIterations- Number of iterations in training, converges to better accuracy
We will load a pretrained model here (see Table 3-20) and look into training our own model in Chapter 8.
from sparknlp.annotator import PerceptronModel
pos_tagger = PerceptronModel.pretrained() \
.setInputCols(["tokens", "sentences"]) \
.setOutputCol("pos")
postags = pos_tagger.transform(lemmas) postags.limit(5).toPandas()
| path | ... | sentences | tokens | pos | |
|---|---|---|---|---|---|
| 0 | file:/.../spark-nlp-book/data/... | ... | [(document, 0, 709, Xref: cantaloupe.srv.cs.cm... | [(token, 0, 3, Xref, {’sentence’: ’1'}), (toke... | [(pos, 0, 3, NNP, {'word’: ‘Xref'}), (pos, 4, ... |
| 1 | file:/.../spark-nlp-book/data/... | ... | [(document, 0, 288, Newsgroups: sci.space Path... | [(token, 0, 9, Newsgroups, {’sentence’: ’1'}),... | [(pos, 0, 9, NNP, {'word’: ‘Newsgroups'}), (po... |
| 2 | file:/.../spark-nlp-book/data/... | ... | [(document, 0, 312, Xref: cantaloupe.srv.cs.cm... | [(token, 0, 3, Xref, {’sentence’: ’1'}), (toke... | [(pos, 0, 3, NNP, {'word’: ‘Xref'}), (pos, 4, ... |
| 3 | file:/.../spark-nlp-book/data/... | ... | [(document, 0, 453, Path: cantaloupe.srv.cs.cm... | [(token, 0, 3, Path, {’sentence’: ’1'}), (toke... | [(pos, 0, 3, NNP, {'word’: ‘Path'}), (pos, 4, ... |
| 4 | file:/.../spark-nlp-book/data/... | ... | [(document, 0, 915, Newsgroups: sci.space Path... | [(token, 0, 9, Newsgroups, {’sentence’: ’1'}),... | [(pos, 0, 9, NNP, {'word’: ‘Newsgroups'}), (po... |
Pretrained Pipelines
We saw earlier how we can organize multiple MLlib stages into a Pipeline. Using Pipelines is especially useful in NLP tasks because there are often many stages between loading the raw text and extracting structured data.
Spark NLP has pretrained pipelines that can be used to process text. This doesn’t mean that you do not need to tune pipelines for application. But it is often convenient to begin experimenting with a prebuilt NLP pipeline and find what needs tuning.
Explain document ML pipeline
The BasicPipeline does sentence splitting, tokenization, lemmatization, stemming, and POS tagging. If you want to get a quick look at some text data, this is a great pipeline to use (see Table 3-21).
from sparknlp.pretrained import PretrainedPipeline
pipeline = PretrainedPipeline('explain_document_ml', lang='en')
pipeline.transform(texts).limit(5).toPandas()
| ... | sentence | token | spell | lemma | stem | pos | |
|---|---|---|---|---|---|---|---|
| 0 | ... | ... | ... | [(token, 0, 9, Newsgroups, {'confidence’: ’0.0... | ... | [(token, 0, 9, newsgroup, {'confidence’: ’0.0’... | [(pos, 0, 9, NNP, {'word’: ‘Newsgroups'}, [], ... |
| 1 | ... | ... | ... | [(token, 0, 3, Path, {'confidence’: ’1.0'}, []... | ... | [(token, 0, 3, path, {'confidence’: ’1.0'}, []... | [(pos, 0, 3, NNP, {'word’: ‘Path'}, [], []), (... |
| 2 | ... | ... | ... | [(token, 0, 9, Newsgroups, {'confidence’: ’0.0... | ... | [(token, 0, 9, newsgroup, {'confidence’: ’0.0’... | [(pos, 0, 9, NNP, {'word’: ‘Newsgroups'}, [], ... |
| 3 | ... | ... | ... | [(token, 0, 3, xref, {'confidence’: ’0.3333333... | ... | [(token, 0, 3, pref, {'confidence’: ’0.3333333... | [(pos, 0, 3, NN, {'word’: ‘pref'}, [], []), (p... |
| 4 | ... | ... | ... | [(token, 0, 3, tref, {'confidence’: ’0.3333333... | ... | [(token, 0, 3, xref, {'confidence’: ’0.3333333... | [(pos, 0, 3, NN, {'word’: ‘pref'}, [], []), (p... |
You can also use the annotate function to process a document without Spark:
text = texts.first()['text']
annotations = pipeline.annotate(text)
list(zip(
annotations['token'],
annotations['stems'],
annotations['lemmas']
))[100:120]
[('much', 'much', 'much'),
('argument', 'argum', 'argument'),
('and', 'and', 'and'),
('few', 'few', 'few'),
('facts', 'fact', 'fact'),
('being', 'be', 'be'),
('offered', 'offer', 'offer'),
('.', '.', '.'),
('The', 'the', 'The'),
('summaries', 'summari', 'summary'),
('below', 'below', 'below'),
('attempt', 'attempt', 'attempt'),
('to', 'to', 'to'),
('represent', 'repres', 'represent'),
('the', 'the', 'the'),
('position', 'posit', 'position'),
('on', 'on', 'on'),
('which', 'which', 'which'),
('much', 'much', 'much'),
('of', 'of', 'of')]
There are many other pipelines, and there is additional information available.
Now let’s talk about how we will perform step 4, converting the annotations into native Spark SQL types using the Finisher.
Finisher
The annotations are useful for composing NLP steps, but we generally want to take some specific information out to process. The Finisher handles most of these use cases. If you want to get a list of tokens (or stems, or what have you) to use in downstream MLlib stages, the Finisher can do this (see Table 3-22). Let’s look at the parameters:
inputCols- Name of input annotation cols
outputCols- Name of finisher output cols
valueSplitSymbol- Character separating annotations
annotationSplitSymbol- Character separating annotations
cleanAnnotations- Determines whether to remove annotation columns
includeMetadata- Annotation metadata format
outputAsArray- Finisher generates an
Arraywith the results instead of string
finisher = Finisher()\
.setInputCols(['tokens', 'lemma'])\
.setOutputCols(['tokens', 'lemmata'])\
.setCleanAnnotations(True)\
.setOutputAsArray(True)
custom_pipeline = Pipeline(stages=[
document_assembler,
sent_detector,
tokenizer,
lemmatizer,
finisher
]).fit(texts)
custom_pipeline.transform(texts).limit(5).toPandas()
| path | text | tokens | lemmata | |
|---|---|---|---|---|
| 0 | ... | ... | [Newsgroups, :, sci.space, Path, :, cantaloupe... | [Newsgroups, :, sci.space, Path, :, cantaloupe... |
| 1 | ... | ... | [Path, :, cantaloupe.srv.cs.cmu.edu!rochester!... | [Path, :, cantaloupe.srv.cs.cmu.edu!rochester!... |
| 2 | ... | ... | [Newsgroups, :, sci.space, Path, :, cantaloupe... | [Newsgroups, :, sci.space, Path, :, cantaloupe... |
| 3 | ... | ... | [Xref, :, cantaloupe.srv.cs.cmu.edu, sci.space... | [Xref, :, cantaloupe.srv.cs.cmu.edu, sci.space... |
| 4 | ... | ... | [Xref, :, cantaloupe.srv.cs.cmu.edu, sci.astro... | [Xref, :, cantaloupe.srv.cs.cmu.edu, sci.astro... |
Now we will use the StopWordsRemover transformer from Spark MLlib. The results are shown in Table 3-23.
from pyspark.ml.feature import StopWordsRemover
stopwords = StopWordsRemover.loadDefaultStopWords('english')
larger_pipeline = Pipeline(stages=[
custom_pipeline,
StopWordsRemover(
inputCol='lemmata',
outputCol='terms',
stopWords=stopwords)
]).fit(texts)
larger_pipeline.transform(texts).limit(5).toPandas()
| ... | lemmata | terms | |
|---|---|---|---|
| 0 | ... | [Newsgroups, :, sci.space, Path, :, cantaloupe... | [Newsgroups, :, sci.space, Path, :, cantaloupe... |
| 1 | ... | [Path, :, cantaloupe.srv.cs.cmu.edu!rochester!... | [Path, :, cantaloupe.srv.cs.cmu.edu!rochester!... |
| 2 | ... | [Newsgroups, :, sci.space, Path, :, cantaloupe... | [Newsgroups, :, sci.space, Path, :, cantaloupe... |
| 3 | ... | [Xref, :, cantaloupe.srv.cs.cmu.edu, sci.space... | [Xref, :, cantaloupe.srv.cs.cmu.edu, sci.space... |
| 4 | ... | [Xref, :, cantaloupe.srv.cs.cmu.edu, sci.astro... | [Xref, :, cantaloupe.srv.cs.cmu.edu, sci.astro... |
Now that we have reviewed Spark and Spark NLP, we are almost ready to start building an NLP application. There is an extra benefit to learning an annotation library—it helps you understand how to structure a pipeline for NLP. This knowledge will be applicable even if you are using other technologies.
The only topic left for us to cover is deep learning, which we cover in the next chapter.
Exercises: Build a Topic Model
One of the easiest things you can do to begin exploring your data set is to create a topic model. To do this we need to transform the text into numerical vectors. We will go into this more in the next part of the book. For now, let’s build a pipeline for processing text.
First, we need to split the texts into sentences. Second, we need to tokenize. Next, we need to normalize our words with a lemmatizer and the normalizer. After this, we need to finish our pipeline and remove stop words. (So far, everything except the Normalizer has been demonstrated in this chapter.) After that, we will pass the information into our topic modeling pipeline.
Check the online documentation for help with the normalizer.
# document_assembler = ??? # sent_detector = ??? # tokenizer = ??? # lemmatizer = ??? # normalizer = ??? # finisher = ??? # sparknlp_pipeline = ???
# stopwords = ??? # stopword_remover = ??? # use outputCol='terms' #text_processing_pipeline = ??? # first stage is sparknlp_pipeline
# from pyspark.ml.feature import CountVectorizer, IDF # from pyspark.ml.clustering import LDA # tf = CountVectorizer(inputCol='terms', outputCol='tf') # idf = IDF(inputCol='tf', outputCol='tfidf') # lda = LDA(k=10, seed=123, featuresCol='tfidf') # pipeline = Pipeline(stages=[ # text_processing_pipeline, # tf, # idf, # lda # ])
# model = pipeline.fit(texts)
# tf_model = model.stages[-3] # lda_model = model.stages[-1]
# topics = lda_model.describeTopics().collect()
# for k, topic in enumerate(topics):
# print('Topic', k)
# for ix, wt in zip(topic['termIndices'], topic['termWeights']):
# print(ix, tf_model.vocabulary[ix], wt)
# print('#' * 50)
Congratulations! You have built your first full Spark pipeline with Spark NLP.
Resources
- Spark API: this is a great resource to have handy. With each new release there can be new additions, so I rely on the API documentation to keep up to date.
- Natural Language Processing with Python: this is the book written explaining NLP with NLTK. It is available for free online.
- spaCy: this library is doing a lot of cool things and has a strong community.
- Spark NLP: the website of the Spark NLP library. Here you can find documentation, tutorials, and videos about the library.
- Foundations of Statistical Natural Language Processing by Christopher D. Manning and Hinrich Schütze (MIT Press)
- This is considered a classic for natural language processing. It is an older book, so you won’t find information on deep learning. If you want to have a strong foundation in NLP this is a great book to have.
- Spark: The Definitive Guide by Matei Zaharia and Bill Chambers (O’Reilly)