NATIVE AMERICAN
|
Core mutations of Native American mDNA clusters |
|
|
CLUSTER |
MUTATIONS |
|
A1 |
223 390 319 362 |
|
A2 |
111 223 290 319 362 |
|
B |
189 217 |
|
C |
223 298 325 327 |
|
D |
223 325 362 |
|
X |
223 278 |
|
Calibrated origin dates and clan mother names for Native American mDNA clusters |
||
|
CLUSTER |
MOTHER |
AGE (YRS) |
|
A |
Aiyana |
15,800 |
|
B |
Ina |
18,700 |
|
C |
Chochmingwu |
19,600 |
|
D |
Djigonese |
16,900 |
|
X |
Xenia |
15,800 |
EUROPEAN
|
Calibrated origin dates, clan mother names, and frequencies for native European mDNA clusters |
|||
|
CLUSTER |
FREQUENCY (%) |
MOTHER |
AGE (YRS) |
|
U5 |
5.7 |
Ursula |
47,000 |
|
HV |
5.4 |
HV |
34,000 |
|
X (I) |
1.7 |
Xenia |
26,000 |
|
U4 |
3.0 |
Ulrike |
20,000 |
|
H |
37.7 |
Helena |
14,000 |
|
T |
2.2 |
Tara |
13,000 |
|
K |
4.6 |
Katrine |
12,500 |
|
T2 |
2.9 |
Tara |
12,000 |
|
J |
6.1 |
Jasmine |
8,500 |
|
T1 |
2.2 |
Tara |
9,000 |
AFRICAN
|
Clan mothers and ages of native African mDNA clusters |
||
|
NOTATION |
CLAN MOTHER |
AGE (YRS) |
|
Superclan L1 |
||
|
L1A |
Layla |
40,000 |
|
L1B |
Lamia |
30,000 |
|
L1C |
Lalamika |
60,000 |
|
L1D |
Latasha |
50,000 |
|
L1E |
Lalla |
83,000 |
|
L1F |
Labana |
86,000 |
|
L1K |
Lakita |
92,000 |
|
Superclan L2 |
||
|
L2A |
Leisha |
55,000 |
|
L2B |
Lesedi |
32,000 |
|
L2C |
Lingaire |
27,000 |
|
L2D |
Lindewe |
122,000 |
|
Superclan L3 |
||
|
L3A |
Lara |
60,000 |
|
L3B |
Limber |
21,000 |
|
L3D |
Lingaire |
30,000 |
|
L3E |
Lila |
45,000 |
|
L3F |
Lungile |
36,000 |
|
L3G |
Lubaya |
45,000 |
|
Regional distribution within Africa of the most frequent native African mDNA clusters |
|
|
REGION |
TOP 5 MOST FREQUENT CLUSTERS |
|
East |
L1A, L2, L3A, L3F, L3G |
|
Southeast |
L1A, L2A1a, L2A1b, L3E |
|
South |
L1A, L1D, L1K, L3B, L3E |
|
Central |
L1A, L1C, L2A1, L3E |
|
West |
L1B, L2A1, L3B, L3D |
|
North |
L1B, L2A1, L3B, L3D |
|
FROM |
TO |
MUTATIONS |
|
Superclan L1 |
||
|
Root |
L1 |
16230 |
|
Root |
L1D |
16129, 16243 |
|
L1D |
L1D1 |
16294 |
|
L1D |
L1D2 |
16234 |
|
Root |
L1F |
16169, 16327 |
|
Root |
L1A |
16129, 16148, 16172, 16188G, 16278, 16320 |
|
L1A |
L1A1 |
16168 |
|
L1A1 |
L1A1a |
16278 |
|
L1A |
L1A2 |
16129 |
|
Root |
L1K |
16172, 16209, 16214, 16291 |
|
L1K |
L1K1 |
16166C |
|
Root |
L1E |
16129, 16148, 16166 |
|
L1E |
L1E1 |
16111, 16254, 16311 |
|
L1E |
L1E2 |
16355, 16362 |
|
L1 |
L1B/C |
7055R |
|
L1 |
L1B |
16126, 16264, 16270 |
|
L1B |
L1B1 |
16293 |
|
L1B/C |
L1C |
16129, 16294, 16360 |
|
L1C |
L1C1 |
16293 |
|
L1C1 |
L1C1a |
16274 |
|
L1C1a |
L1C1a1 |
16214, 16223, 16234, 16249 |
|
L1C |
L1C2 |
16265C, 16286G |
|
L1C |
L1C3 |
16187, 16215 |
|
L1 |
L2/3 |
16187, 16189, 16311 |
|
Superclan L2 |
||
|
L2/3 |
L2 |
16390 |
|
L2 |
L2C |
13957R |
|
L2C |
L2C1 |
16318 |
|
L2C |
L2C2 |
16264 |
|
L2 |
L2A |
13803R, 16294 |
|
L2A |
L2A1 |
16309 |
|
L2A1 |
L2A1a |
16286 |
|
L2A1 |
L2A1b |
16290 |
|
L2 |
L2D |
3693R, 16399 |
|
L2D |
L2D1 |
16129, 16189, 16223, 16300, 16354 |
|
L2D |
L2D2 |
16111A, 16145, 16239, 16292, 16355 |
|
L2 |
L2B |
4157R, 16129, 16114A, 16213 |
|
L2B |
L2B1 |
16362 |
|
Superclan L3 |
||
|
L2/3 |
L3 |
3592R, 16278 |
|
L3 |
L3F |
16209, 16311 |
|
L3F |
L3F1 |
16292 |
|
L3 |
L3G |
16293T, 16311, 16355, 16362 |
|
L3 |
L3B/D |
16124 |
|
L3B/D |
L3B |
10084R, 16278, 16362 |
|
L3B |
L3B1 |
16124 |
|
L3B |
L3B2 |
16311 |
|
L3B/D |
L3D |
8616R |
|
L3D |
L3D1 |
16319 |
|
L3D |
L3D2 |
16256 |
|
L3D |
L3D3 |
16189, 16278, 16304, 16311 |
|
L3 |
L3E |
2349R |
|
L3E |
L3E3/4 |
5260R |
|
L3E3/4 |
L3E4 |
16264 |
|
L3E3/4 |
L3E3 |
16265T |
|
L3E |
L3E2 |
16320 |
|
L3E |
L3E2b |
16172, 16189 |
|
L3E |
L3E1 |
16327 |
|
L3E1 |
L3E1a |
16185 |
|
L3E1 |
L3E1b |
16325 |
|
L3A |
M |
10397R |
|
L3A |
N |
10871R |
Mutations in the African mDNA tree.
Numbers are positions of mutations in the mDNA sequence. Positions without a suffix are transitions. Suffixes A, G, C, T indicate transversions to these bases. Suffix del indicates a deletion while R is a restriction enzyme variant.
CLAN DONALD GENEALOGY
|
Identification of individuals on Clan Donald genealogy |
|||
|
CODE |
NAME |
DATES |
BRANCH |
|
A |
Somerled |
c. 1100–1164 |
|
|
B |
Dugall, founder of Clan Dougal of Lorne |
c. 1118–? |
|
|
C |
Donald MacRanald of the Isles |
1190–1269 |
|
|
D |
Alastair Mor MacDonald, founder of Clan Alastair |
d. 1299 |
|
|
E |
John MacDonald, Lord of the Isles |
d. 1386 |
|
|
F |
Ranald Macdonald, 1st of Clanranald and Glengarry |
d. 1386 |
|
|
G |
Donald Gallach MacDonald, 3rd of Sleat |
d. 1506 |
|
|
H |
Ranald Mor MacDonald, 7th of Keppoch |
d. 1547 |
|
|
I |
Alan MacDonald, 4th of Clanranald |
1437–1481 |
|
|
J |
Donald Gruamach MacDonald, 4th of Sleat |
d. 1534 |
|
|
K |
Ian Moidartach MacDonald, 8th of Clanranald |
1502–1584 |
|
|
L |
Sir Donald Breac Macdonald, 10th of Sleat |
1605–1678 |
|
|
M |
Ranald Alexander MacDonald, 24th of Clanranald |
Living |
Clanranald |
|
N |
P. M. Macdonald |
Living |
Glenaladale |
|
O |
J. J. Macdonald |
Living |
Bornish |
|
P |
Ranald MacDonell, 23rd of Glengarry |
Living |
Glengarry |
|
Q |
Allan Douglas MacDonald of Vallay |
Living |
Vallay |
|
R |
Sir Iain Godfrey MacDonald, 25th of Sleat |
Living |
Sleat |
|
S |
David Foster Macdonald, 17th of |
Living |
Castle Camus |
|
T |
D. B. Macdonald |
Living |
Sleat |
|
U |
W. F. Macdonald |
Living |
Achnancoichean |
|
V |
L. McDonald |
Living |
Bohuntin |
|
Details of Y chromosome mutations in Clan Donald genealogy (See Fig.2) |
||
|
LINK |
MARKER |
MUTATION |
|
C–E |
458 |
16–15 |
|
E–G |
557 |
15–16 |
|
F–P |
449 |
31–32 |
|
F–P |
456 |
17–16 |
|
F–P |
CDYa |
34–35 |
|
G–T |
GATAH4 |
12–11 |
|
H–U |
448 |
20–21 |
|
H–U |
449 |
30–31 |
|
H–U |
GATAH4 |
12–11 |
|
H–V |
570 |
18–19 |
|
H–V |
390 |
25–24 |
|
I–O |
CDYb |
38–37 |
|
J–L |
448 |
20–19 |
|
J–S |
449 |
31–32 |
|
J–S |
460 |
11–10 |
|
J–S |
557 |
15–17 |
|
K–M |
389a |
14–13 |
|
K–M |
390 |
25–24 |
|
K–M |
449 |
31–32 |
|
K–M |
576 |
17–18 |
|
K–M |
CDYb |
38–37 |
|
K–N |
GATAH4 |
12–10 |
|
K–N |
CDYa |
34–33 |
|
K–N |
449 |
31–30 |
|
K–N |
458 |
15–16 |
|
L–R |
607 |
16–15 |
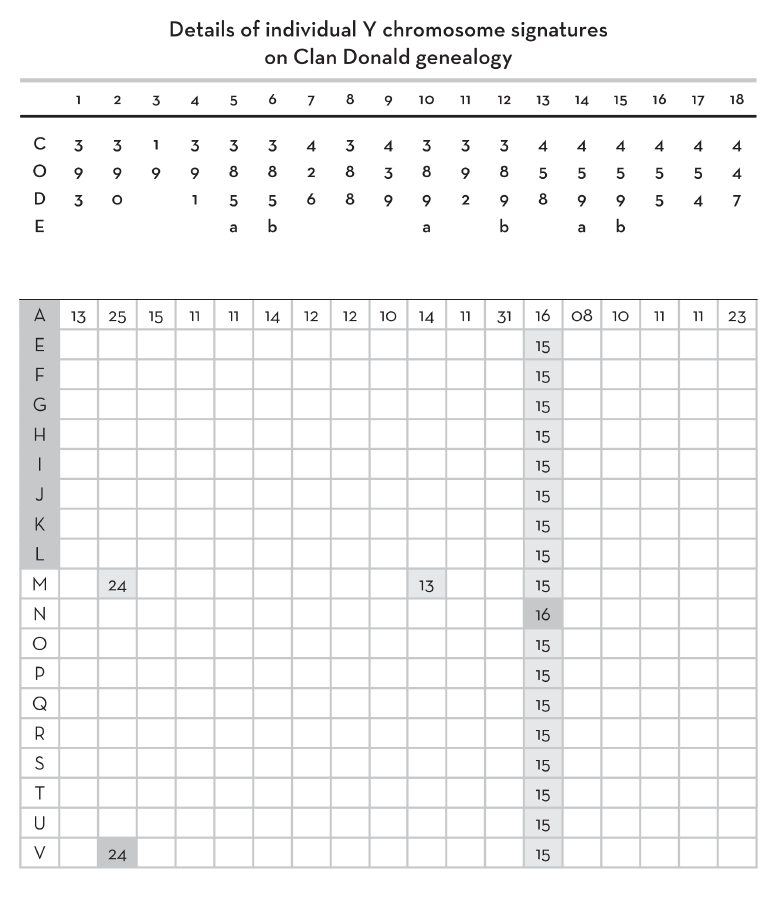
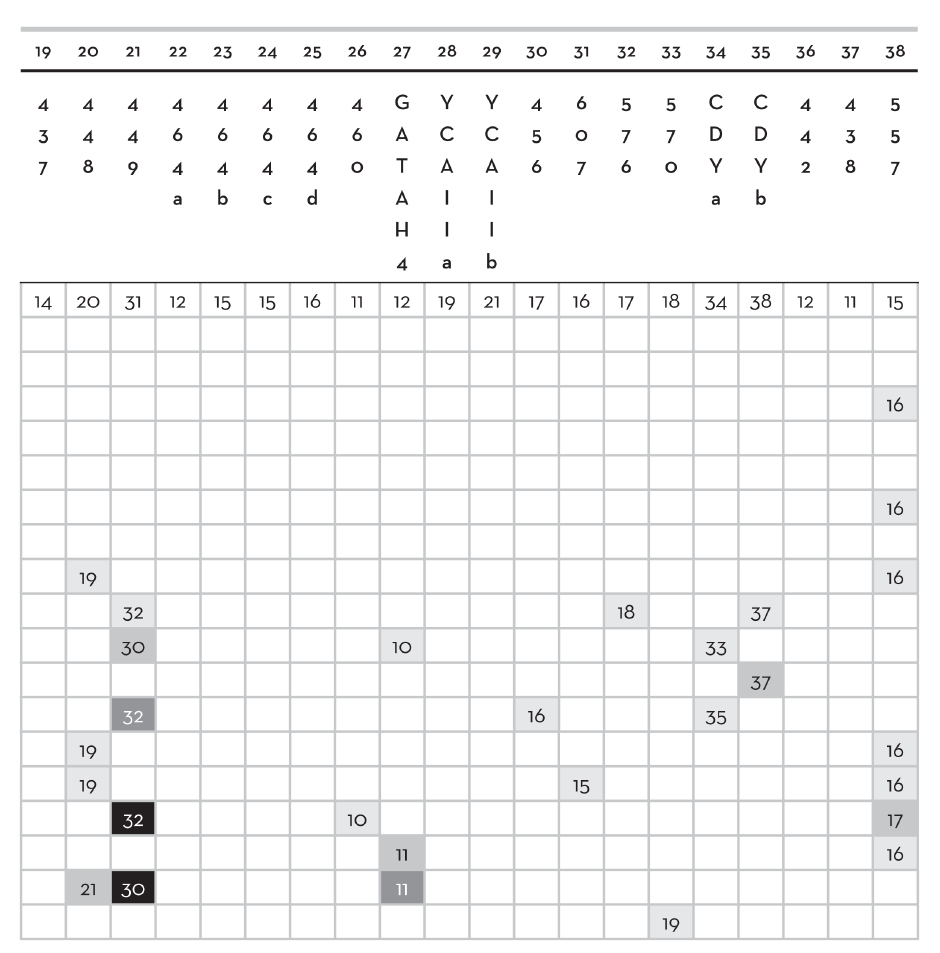
Code letters in column 1 identify deceased (shaded) and living individuals on Fig. 2 (p. 108). Markers 1–38 are identified in row 2.
Numbers in table body are allele repeat lengths of Y chromosomes that differ from values deduced for Somerled (row A)
|
The location of 146 important genes, showing their order from right to left along each chromosome (Column 1), the body system involved (Column 2), the encoded protein (Column 3), and the international gene symbol (Column 4). |
|||
|
REF |
SYSTEM |
PROTEIN |
SYMBOL |
|
1.1 |
Digestion |
Chymotrysin C |
CTRC |
|
1.2 |
Bones, skin, teeth, and joints |
Bone morphogenetic protein 8a/b |
BMP8A/B |
|
1.3 |
Metabolism |
Leptin receptor |
LEPR |
|
1.4 |
Digestion |
Amylase |
AMY1 and 2 |
|
1.5 |
Digestion |
Cathepsin K and S |
CTSK/S |
|
1.6 |
Muscle |
Tropomyosin 3 |
TPM3 |
|
1.7 |
Brain and nerves |
Laminin gamma 2 |
LAMC2 |
|
1.8 |
Liver and kidney |
Renin |
REN |
|
1.9 |
Muscle |
Actin alpha 1, skeletal muscle |
ACTA1 |
|
1.10 |
Eyes |
Opsin 3 |
OPN3 |
|
2.1 |
Immunity |
Immunoglobulin kappa constant |
IGKC |
|
2.2 |
Bones, skin, teeth, and joints |
Bone morphogenetic protein 10 |
BMP10 |
|
2.3 |
Digestion |
Lactase |
LCT |
|
2.4 |
Brain and nerves |
Sodium channel 9 |
SCN9A |
|
2.5 |
Heart and lung |
Collagen type 3 |
COL1A3 |
|
2.6 |
Eyes |
Crystallin gamma A |
CRYGA |
|
2.7 |
Pigmentation |
Fibronectin |
FN1 |
|
2.8 |
Eyes |
Crystallin beta A2 |
CRYBA2 |
|
2.9 |
Liver and kidney |
Collagen type 4, |
COL4A3/4 |
|
2.10 |
Metabolism |
Glucagon |
GCG |
|
3.1 |
Eyes |
Retinoic acid receptor beta |
RARB |
|
3.2 |
Metabolism |
Parathyroid hormone 1 receptor |
PTH1R |
|
3.3 |
Digestion |
Lactotransferrin |
LTF |
|
3.4 |
Metabolism |
Phosphodiesterase |
PDE12 |
|
3.5 |
Muscle |
Myosin HC 15 |
MYH15 |
|
3.6 |
Eyes |
Rhodopsin |
RHO |
|
4.1 |
Brain and nerves |
Huntingtin |
HTT |
|
4.2 |
Bones, skin, teeth, and joints |
Fibroblast growth factor 3 |
FGFR3 |
|
4.3 |
Bones, skin, teeth, and joints |
Enamalin |
ENAM |
|
4.4 |
Bones, skin, teeth, and joints |
Ameloblastin |
AMBN |
|
4.5 |
Immunity |
Immunoglobulin J |
IGJ |
|
4.6 |
Liver and kidney |
Alcohol dehydrogenase |
ADH1A |
|
4.7 |
Pigmentation |
Hair color 2 (red) |
HCL2 |
|
5.1 |
Blood |
Thrombospondin 4 |
THBS4 |
|
5.2 |
Muscle |
Myosin X |
MYOX |
|
5.3 |
Pigmentation |
Solute carrier, family 45, member A2 |
SLC45A2 |
|
5.4 |
Bones, skin, teeth, and joints |
Growth hormone receptor |
GHR |
|
5.5 |
Bones, skin, teeth, and joints |
Fibroblast growth factor 10 |
FGF10 |
|
5.6 |
Bones, skin, teeth, and joints |
Fibrillin 2 |
FBN2 |
|
5.7 |
Bones, skin, teeth, and joints |
Lysyl oxidase |
LOX |
|
5.8 |
Bones, skin, teeth, and joints |
Fibroblast growth factor 1 |
FGF1 |
|
5.9 |
Metabolism |
Insulin dependent diabetes 10 |
IDDM10 |
|
6.1 |
Immunity |
HLA region |
HGC |
|
6.2 |
Eyes |
Opsin 5 |
OPN5 |
|
6.3 |
Muscle |
Myosin VI |
MYO6 |
|
6.4 |
Brain and nerves |
Laminin alpha 2/4 |
LAMA2/4 |
|
6.5 |
Brain and nerves |
Opioid receptor mu 1 |
OPRM1 |
|
6.6 |
Blood |
Plasminogen |
PLG |
|
6.7 |
Blood |
Thrombospondin 2 |
THBS2 |
|
7.1 |
Muscle |
Actin beta |
ACTB |
|
7.2 |
Muscle |
Myosin light chain 7 |
MYL7 |
|
7.3 |
Bones, skin, teeth, and joints |
Elastin |
ELN |
|
7.4 |
Blood |
Erythropoietin |
EPO |
|
7.5 |
Bones, skin, teeth, and joints |
Collagen type 1, alpha 2 |
COL1A2 |
|
7.6 |
Brain and nerves |
Acetylcholinesterase |
ACHE |
|
7.7 |
Eyes |
Opsin 1 cone short wave |
OPN1SW |
|
7.8 |
Brain and nerves |
Olfactory receptor 2A.25 |
OR2A25 |
|
8.1 |
Heart and lung |
Surfactant protein C |
SFTPC |
|
8.2 |
Bones, skin, teeth, and joints |
Fibroblast growth factor receptor 1 |
FGFR1 |
|
8.3 |
Blood |
Plasminogen activator (tissue) |
HBB |
|
8.4 |
Brain and nerves |
Opioid receptor kappa 1 |
OPRK1 |
|
8.5 |
Blood |
Angiopoietin 1 |
ANGPT1 |
|
8.6 |
Brain and nerves |
Otoconin |
OC90 |
|
9.1 |
Immunity |
Inteferon alpha |
IFNA |
|
9.2 |
Muscle |
Tropomyosin 2 |
TPM2 |
|
9.3 |
Digestion |
Beta glucosidase (bile acid) |
GBA2 |
|
9.4 |
Bones, skin, teeth, and joints |
Osteoclast stimulating factor 1 |
OSTF1 |
|
9.5 |
Bones, skin, teeth, and joints |
Osteoglycin |
OGN |
|
9.6 |
Blood |
Hemogen |
HEMGN |
|
9.7 |
Blood |
ABO blood group |
ABO |
|
10.1 |
Eyes |
Optineurin |
OPTN |
|
10.2 |
Muscle |
Myosin IIIA |
MYO3A |
|
10.3 |
Eyes |
Opsin 4 |
OPN4 |
|
10.4 |
Muscle |
Actin alpha 2, smooth muscle |
ACTA2 |
|
10.5 |
Digestion |
Trypsin |
TYSND1 |
|
10.6 |
Heart and lung |
Adrenergic alpha 2A/beta receptor |
ADRA2A/B1 |
|
10.7 |
Metabolism |
Cytochrome P450, family 2C, polypeptide |
CYP2C9 |
|
10.8 |
Heart and lung |
Pancreatic lipase |
PNLIP |
|
11.1 |
Blood |
Beta globin |
HBB |
|
11.2 |
Metabolism |
Insulin |
INS |
|
11.3 |
Brain and nerves |
Otogelin |
OTOG |
|
11.4 |
Digestion |
Pepsin |
PGA3 |
|
11.5 |
Digestion |
Cathepsin C |
CTSC |
|
11.6 |
Pigmentation |
Tyrosinase |
TYR |
|
11.7 |
Eyes |
Crystallin alpha B |
CRYAB |
|
11.8 |
Immunity |
Thymocyte cell surface antigen |
THY1 |
|
12.1 |
Immunity |
Alpha 2 macroglobulin |
A2M |
|
12.2 |
Digestion |
Islet amyloid polypeptide |
IAPP |
|
12.3 |
Pigmentation |
Keratin 1-5 |
KRT1-5 |
|
12.4 |
Bones, skin, teeth, and joints |
Collagen type 2 |
COL2A1 |
|
12.5 |
Digestion |
Peptidase B |
PEPB |
|
12.6 |
Pigmentation |
KIT ligand |
KITLG |
|
12.7 |
Immunity |
Thympoietin |
TMPO |
|
12.8 |
Digestion |
Phopholipase A2 |
PLA2G1B |
|
13.1 |
Brain and nerves |
Olfactomedin 4 |
OLFM4 |
|
13.2 |
Brain and nerves |
Olfactory receptor E |
OR7E |
|
13.3 |
Muscle |
Myosin XVI |
MYO16 |
|
13.4 |
Liver and kidney |
Collagen type 4, alpha 1 and 2 |
COL4A1/2 |
|
14.1 |
Heart and lung |
Myosin heavy chain 6 and 7, cardiac |
MYH6/7 |
|
14.2 |
Metabolism |
Estrogen receptor 2 |
ESR2 |
|
14.3 |
Bones, skin, teeth, and joints |
Bone morphogenetic protein 4 |
BMP4 |
|
14.4 |
Blood |
Spectrin beta |
SPTB |
|
14.5 |
Digestion |
Chymotrypsin |
CTRB1 |
|
14.6 |
Brain and nerves |
Creatine kinase (brain) |
CKB |
|
14.7 |
Immunity |
Immunoglobulin H alpha 1 |
IGHA1 et al. |
|
15.1 |
Muscle |
Actin alpha 1, cardiac muscle |
ACTC1 |
|
15.2 |
Pigmentation |
Oculocutaneous albinism II |
OCA2 |
|
15.3 |
Brain and nerves |
Cholinergic receptor nicotinic alpha 7 |
CHRNA7 |
|
15.4 |
Pigmentation |
Solute carrier, family 24 member 5 |
SLC24A5 |
|
15.5 |
Blood |
Thrombospondin 1 |
THBS1 |
|
15.6 |
Muscle |
Myosin 1E |
MYO1E |
|
15.7 |
Pigmentation |
Hair color 3 (brown) |
HCL3 |
|
15.8 |
Bones, skin, teeth, and joints |
Chondroitin sulfate proteoglycan 4 |
CSPG4 |
|
15.9 |
Bones, skin, teeth, and joints |
Aggrecan |
ACAN |
|
16.1 |
Blood |
Alpha globin |
HBA1 and 2 |
|
16.2 |
Digestion |
Myosin HC 11, smooth muscle |
MYH11 |
|
16.3 |
Blood |
Alpha hemoglobin stabilizing protein |
AHSP |
|
16.4 |
Muscle |
Myosin light chain kinase 3 |
MYLK3 |
|
16.5 |
Blood |
Haptoglobin |
HP |
|
16.6 |
Digestion |
Chymotrypsinogen B1/2 |
CTRB1/2 |
|
16.7 |
Pigmentation |
Melanocortin 2 receptor |
MC2R |
|
17.1 |
Muscle |
Myosin, heavy chain 1, skeletal adult |
MYH1-4 |
|
17.2 |
Eyes |
Crystallin beta A1 |
CRYBA1 |
|
17.3 |
Bones, skin, teeth, and joints |
Collagen type 1, alpha 1 |
COL1A1 |
|
17.4 |
Digestion |
Gastrin |
GAST |
|
17.5 |
Heart and lung |
Angiotensin 1 converting enzyme |
ACE |
|
17.6 |
Metabolism |
Glucagon receptor |
GCGR |
|
17.7 |
Muscle |
Actin gamma |
ACTG1 |
|
18.1 |
Pigmentation |
Melanocortin 2 receptor |
MC2R |
|
18.2 |
Metabolism |
Insulin-dependent diabetes 6 |
IDDM6 |
|
18.3 |
Digestion |
Gastrin-releasing peptide |
GRP |
|
18.4 |
Brain and nerves |
Myelin basic protein |
MBP |
|
19.1 |
Metabolism |
Insulin receptor |
INSR |
|
19.2 |
Pigmentation |
Hair color |
HCL1 |
|
19.3 |
Brain and nerves |
Myelin-associated glycoprotein |
MAG |
|
19.4 |
Digestion |
Gastric inhibitory polypeptide receptor |
GIPR |
|
20.1 |
Liver and kidney |
Arginine vasopressin |
AVP |
|
20.2 |
Pigmentation |
Agouti signaling peptide |
ASIP |
|
20.3 |
Metabolism |
Topoisomerase 1 |
TOP1 |
|
20.4 |
Bones, skin, teeth, and joints |
Bone morphogenetic protein 7 |
BMP7 |
|
20.5 |
Bones, skin, teeth, and joints |
Collagen, type IX, alpha 3 |
COL9A3 |
|
21.1 |
Bones, skin, teeth, and joints |
Chondrolectin |
CHODL |
|
21.2 |
Eyes |
Crystallin alpha A |
CRYAA |
|
22.1 |
Immunity |
Immunoglobulin lambda constant |
IGKC |
|
22.2 |
Eyes |
Crystallin beta A4 |
CRYBA4 |
|
22.3 |
Brain and nerves |
Synaptogyrin 1 |
SYNGR1 |