7 The concept of information in biology
The use of informational terms is widespread in molecular and developmental biology. The usage dates back to Weismann. In
both protein synthesis and in later development, genes are symbols, in that there is no necessary connection between their
form (sequence) and their effects. The sequence of a gene has been determined by past natural selection, because of the effects
it produces. In biology, the use of informational terms implies intentionality, in that both the form of the signal, and the
response to it, have evolved by selection. Where an engineer sees design, a biologist sees natural selection.
A central idea in contemporary biology is that of information. Developmental biology can be seen as the study of how information in the genome is translated into adult structure, and evolutionary biology of how the information came to be there in the first place. Our excuse for writing a chapter concerning topics as diverse as the origins of genes, of cells, and of language is that all are concerned with the storage and transmission of information.(Szathmáry and Maynard Smith, 1995)
Let us begin with the notions involved in classical information theory … These concepts do not apply to DNA because they presuppose a genuine information system, which is composed of a coder, a transmitter, a receiver, a decoder, and an information channel in between. No such components are apparent in a chemical system (Apter and Wolpert, 1965). To describe chemical processes with the help of linguistic metaphors such as “transcription” and “translation” does not alter the chemical nature of these processes. After all, a chemical process is not a signal that carries a message. Furthermore, even if there were such a thing as information transmission between molecules, transmission would be nearly noiseless (that is, substantially nonrandom), so that the concept of probability, central to the theory of information, does not apply to this kind of alleged information transfer.(Mahner and Bunge, 1997)
It is clear from these quotations that there is something to talk about. I shall be concerned only with the use of information
concepts in genetics, evolution, and development, and not in neurobiology, which I am not competent to discuss.
7.1 The information analogy
The colloquial use of informational terms is all-pervasive in molecular biology. “Transcription,” “translation,” “code,” “redundancy,”
“synonymous,” “messenger,” “editing,” “proofreading,” “library”: these are all technical terms in biology. I am not aware
of any confusions arising because their meanings are not understood. In fact, the similarities between their meanings when
referring to human communication and genetics are surprisingly close. One example must suffice. In “proofreading,” the sequence
of the four bases in a newly synthesized DNA strand is compared with the corresponding sequence of the old strand that acted
as a template for its synthesis. If there is a “mismatch” (that is, if the base in the new strand is not complementary to
that in the old strand according to the pairing rules, A–T and G–C), then it is removed and replaced by the correct base.
The similarity of this process to that in which the letters in a copy are compared – in principle, one by one – with those
in the original, and corrected if they differ, is obvious. It is also relevant that in describing molecular proofreading,
I found it hard to avoid using the words “rule” and “correct.”
Molecular biologists, then, do make use of the information analogy in their daily work. Analogies are used in science in two
ways. Occasionally, there is a formal isomorphism between two different physical systems. Over 50 years ago, I worked as an
aircraft engineer. One thing we wanted to know, in the design stage, was the mode of mechanical vibration of the future airplane.
To find out, we built an electrical analog, in which the masses of different parts of the structure were represented by the
inductances of coils in the circuit, and elasticity by the capacitances of condensers. The vibrations of the circuit then
predicted the vibrations of the aircraft. The justification for this procedure is that the equations describing the electrical
and mechanical vibrations are identical. In effect, we had built a special-purpose analog computer. I remember being annoyed,
later, to discover that I had been talking prose without knowing it.
Cases of exact isomorphism are rather rare. Much commoner is the recognition of a qualitative similarity, useful in giving
insight into an unfamiliar system by comparison with a familiar one. A classic example is Harvey’s recognition that the heart
is a pump: it is unlikely that he would have had this insight had he not been familiar with the engineering use of pumps.
A more controversial example is the fact that both Darwin and Wallace ascribe their idea of evolution by natural selection
to a reading of Malthus’s An Essay on the Principle of Population. A third and more trivial example is that I was led to invent evolutionary game theory by analogy with classical game theory,
which analyzes human behavior: as it happens, the main thing I got out of the analogy was a convenient mathematical notation.
The point is that scientists need to get their ideas from somewhere. Most often, biologists get them by analogy with current
technology, or sometimes with the social sciences. It is therefore natural that during the twentieth century they should have
drawn analogies from machines that transduce information. The first deliberate use of such an analogy, by August Weismann,
occurred towards the end of the last century, and is described below. Of course, as I will demonstrate, if an analogy is only
qualitative, it can mislead as well as illuminate.
However, first I must address the criticisms by Mahner and Bunge quoted at the start of this chapter (Mahner and Bunge, 1997).
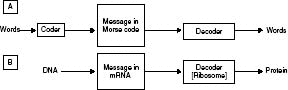
Fig. 7.1 Comparison of transmission of a human message by Morse code (A) and translation of a message coded in DNA into the amino acid
sequence of a protein (B).
First, is it true that there is no coder, transmitter, receiver, decoder, or information channel? This sentence does draw
attention to some ways in which genetic transcription and translation differ from typical examples of human communication
(Figure 7.1).
In the human example, a message is first coded, and then decoded. In the genetic case, although we think of a message in coded
form in the mRNA being translated at the ribosome into the amino acid sequence of a protein, it is perhaps odd to think of
this as “de”-coding, since it was not “coded” from protein to mRNA in the first place. I do not think this destroys the analogy
between the genetic case and the second part of the human sequence. But it does raise a hard question. If there is “information”
in DNA, copied to RNA, how did it get there? Is there any analogy between the origins of the information in DNA and in Morse
code? Perhaps there is. In human speech, the first “coder” is the person who converts a meaning into a string of phonemes,
later converted to Morse code. In biology, the coder is natural selection. This parallel may seem far-fetched, or even false,
to a non-Darwinist. But it is natural selection that, in the past, produced the sequence of bases out of many possible sequences
that, via the information channel just described, specifies a protein that has a “meaning,” in the sense of functioning in
a way that favors the survival of the organism. Where an engineer sees design, a biologist sees natural selection.
What of the claim that a chemical process is not a signal that carries a message? Why not? If a message can be carried by
a sound wave, an electromagnetic wave, or a fluctuating current in a wire, why not by a set of chemical molecules? A major insight
of information theory is that the same information can be transmitted by different physical carriers. So far, engineers have
not used chemical carriers, essentially because of the difficulty of getting information into and out of a chemical medium.
The living world has solved this problem.
Finally, what of the objection that the concept of probability is central to information theory, but missing in biological
applications? One could as well argue that information cannot be transmitted by the printed word, because typesetting is virtually
noiseless. In information theory, Shannon’s (1948) measure of quantity of information, Σ p log p, is a measure of the capacity of a channel to transmit information, given by the number of different messages that could have been sent. The probabilistic
aspects of Shannon’s theory have been used in neurobiology, but rarely in genetics, because we can get most of what we need
from an assumption of equi-probability. Given a string of n symbols, each of which can be any one of four equally likely alternatives, Shannon’s measure gives 2n bits of information. In the genetic message, there are four alternative bases. If they were equally likely, and if each symbol
was independent of its neighbors, the quantity of information would be two bits per base. In fact, the bases are not equally
likely, and there are correlations between neighbors, so there is some reduction in quantity of information, but it is not
very great, and is usually ignored: a greater reduction results from the redundancy of the code. In brief we do not bother
with Shannon’s measure, because two bits per base is near enough, but we could if we wanted to. As it happens, Gatlin (1972) wrote a whole book applying Shannon’s measure to the genetic message. I am not sure that much came from her approach, but
at least it shows that the concept of probability does apply to the genetic code. There is a formal isomorphism, not merely
a qualitative analogy.
There are difficulties in applying information theory in genetics. They arise principally not in the transmission of informationbut in its meaning. This difficulty is not peculiar to genetics. In the early days, it was customary to assert that the theory
was not concerned with meaning, but only with quantity of information: as Weaver (Shannon and Weaver, 1949) put it: “This word ‘information’ in communication theory relates not so much to what you do say, as to what you could say.”
In biology, the question is: How does genetic information specify form and function?
I now describe five attempts, varyingly successful, to apply concepts of information in biology, ending with the problem of
biological form. Then, in the concluding section, I use the analogy between evolution and engineering design by genetic algorithms
to suggest how ideas drawn from information theory can be applied in biology.
7.2 Weismann and the non-inheritance of acquired characters
Weismann’s assertion that acquired characters are not inherited is one of the decisive moments in the history of evolutionary
biology. Darwin himself believed in “the effects of use and disuse.” What led Weismann to such a counterintuitive notion?
Until I happened, rather by chance, to read The Evolution Theory (Weismann, 1904), I thought that his reasons were, first, that the germ line is segregated early from the soma and, second, that if you cut
the tails off mice, their offspring have normal tails. I thought these were poor reasons. There is no segregation of germ
line and soma in plants, yet they are no more likely than animals to transmit acquired characters; and in any case all the
material and energy for the growth of the germ cells comes via the soma, so what prevents the soma from affecting the germ
cells? As to the mouse tails, this is not the kind of acquired character that one would expect to be transmitted.
I had, of course, done Weismann an injustice. There are two long chapters in The Evolution Theory devoted to the non-inheritance of acquired characters. The one argument not used in these chapters is the segregation of
the germ line: this was important to Weismann for other reasons. His main argument is that there are many traits that are manifestly adaptive, but that could not have evolved
by Lamarckian means, because they could not have arisen as individual adaptations in the first place: an example is the form
of an insect’s cuticle, which is hardened before it is used, and which therefore cannot adapt during an individual lifetime.
It follows that adaptations can evolve without Lamarckian inheritance. But this does not prove that acquired characters are
not inherited. His ultimate reason for thinking that they are not was that he could not conceive of a mechanism whereby it
could happen. Suppose a blacksmith does develop big arm muscles. How could this influence the growth of his sperm cells, in
such a way as to alter the development of an egg fertilized by the sperm, so that the blacksmith’s son develops big muscles?
Explaining why he could not imagine such a mechanism, he wrote that the transmission of an acquired character “is very like
supposing that an English telegram to China is there received in the Chinese language” (in fact, he uses the telegram analogy
twice, in slightly different words). This is remarkable for several reasons. He recognizes that heredity is concerned with
the transmission of information, not just of matter or energy. Second, he draws an analogy with a specific information-transducing
channel, the telegram. Third, although his insight has been of profound importance for biology, his argument is in a sense
fallacious. After all, if a sperm can affect the size of a muscle, why cannot a muscle affect a sperm? In fact, most of the
information-transducing machines we use, such as telephones and tape recorders, transmit both ways; they would not be much
use if they did not. But some resemble the genetic system in that they transmit only one way. A CD player converts patterns
on a disc into sound, but one cannot produce a new disc by singing at the player. I think that the non-inheritance of acquired
characters is a contingent fact, usually but not always true – not a logical necessity. Insofar as it is true, it follows
from the “central dogma” of molecular biology, which asserts that information travels from nucleic acids to proteins, but
not from proteins to nucleic acids.
What, then, of the tails of the mice? Weismann tells us that, when he first spoke of his idea to a zoological meeting in Germany,
people replied, “But this must be wrong: everyone knows that, if the tail of a bitch is docked, her puppies have distorted
tails” – an interesting example of what Haldane once called Aunt Jobiska’s theorem: “It is a fact the whole world knows.”
The mouse experiment was performed to refute this objection.
A failure to see that heredity is concerned with information, and that information transfer is often irreversible, has unfortunate
consequences, as I know to my cost. As a young man, I was a Marxist and a member of the communist party. This is not something
I am proud of, but it is relevant. Philosophically, Marxism is unsympathetic to the notion of a gene that influences development,
but is itself unaffected: it is undialectical. I do not suggest that the only reason for Lysenko’s views was his Marxism –
he had less honorable motives – but I think Marxism must take some of the blame. Certainly, it made me uncomfortable with
Weismann’s views. I spent some 6 months carrying out an experiment to test them. The ability of an adult Drosophila to withstand high temperatures depends on the temperature at which the egg was incubated. Not surprisingly, I found that
the adaptation is not inherited. For me, the exercise was perhaps not a total waste of time.
7.3 The genetic code
The analogy between the genetic code and human-designed codes such as Morse code or the ASCII code is too close to require
justification. But there are some features that are worth noting:
In an article I came across only when this chapter was almost completed, Sarkar (1996) describes in some detail the history of the idea of a “comma-free code” (Crick, Griffith, and Orgel, 1957). I agree with him that this proved to be a red herring, although I have suggested elsewhere (Maynard Smith, 1999) that it was one of the cleverest ideas in the history of science that turned out to be wrong. But it was wrong. It illustrates nicely the fact that analogies in science can be misleading as well as illuminating. But I think that
Sarkar is over-eager to point to the failures of the information analogy and to play down its successes. For example, he does
not explain that the discovery (Crick et al., 1961) of the relationship between DNA and protein – as a triplet code in which the correct “reading frame” is maintained by accurately
counting off in threes, and in which meaning can be destroyed by a “frameshift” mutation – also arose from the coding analogy.
It is intriguing that Francis Crick was one of the authors of both papers. As a second example, Sarkar’s argument that the
code does not enable one to predict amino acid sequences (because of complications such as introns, variations from the universal
code, etc.) is seriously misleading; biologists do it all the time.
(1) The correspondence between a particular triplet and the amino acid it codes for is arbitrary. Although decoding necessarily
depends on chemistry, the decoding machinery (tRNAs, assignment enzymes) could be altered so as to alter the assignments.
Indeed, mutations occur that are lethal because they alter the assignments. In this sense the code is symbolic – a point I
return to later.
(2) The genetic code is unusual in that it codes for its own translating machinery.
(3) The scientists who discovered the nature of the code, and of the translating machinery, had the coding analogy constantly
in mind, as the vocabulary they used to describe their discoveries makes clear. Occasionally, they were misled by the analogy.
An example is the belief that the code would be solved as Linear B was deciphered – by discovering the Rosetta stone. What
was needed was a protein of known amino acid sequence, specified by a gene of known base sequence. In fact, the code was not
decoded that way. Instead, it was decoded using a “translating machine” – a piece of cell machinery that, provided with a
piece of RNA of known sequence, would synthesize a peptide with a sequence that could be determined. But despite such false
trails, the information analogy did lead to the solution. If, instead, the problem had been treated as one of the chemistry
of protein–RNA interactions, we might still be waiting for an answer.
(4) It is possible to imagine the evolution of complex, adapted organisms without a genetic code. Godfrey-Smith (2000) imagines a world in which proteins play the same central role that they play in our world, but in which their amino acid sequence is replicated
without coding. In brief, he suggests that proteins could act as templates for themselves, using 20 “connector” molecules,
each with two similar ends, one binding to an amino acid in the template, and another to a similar amino acid in a newly synthesized
strand. In such a system, there would be no “code” connecting one set of molecules to another set of chemically different
molecules. I agree that such a world is conceivable, and that it lacks a code. I will argue below, however, that the notion
of information, and the distinction between genetic and environmental causes in development, would be as relevant in Godfrey-Smith’s
world as it is in the real world.
7.4 Symbol and “gratuity”
Jacques Monod’s (1971) Chance and Necessity did not get a good press from philosophers, particularly in the Anglo-Saxon world. But it contained at least one profound
idea: that of gratuité (translated, not happily, as gratuity). Jacob and Monod (1961) had discovered how a gene can be regulated. In effect, a “repressor”
protein, made by a second “regulatory” gene, binds to the gene and switches it off. The gene can then be switched on by an
“inducer,” usually a small molecule: lactose for this particular gene. What happens is that the inducer binds to the regulatory
protein, and alters its shape, so that the protein no longer binds to the gene and represses it. The point Monod emphasizes
is that the region of the regulatory protein to which the inducer binds is different from the region of the protein that binds
to the gene; the inducer has its effect by altering the shape of the protein. The result is that, in principle, any “inducer”
molecule could switch on, or off, any gene. Of course, all the reactions obey the laws of chemistry, as they must, but there
is no chemical necessity about which inducers regulate which genes. It is this arbitrary nature of molecular biology that
Monod calls “gratuity.”
I think it may be more illuminating to express Monod’s insight by saying that, in molecular biology, inducers and repressors
are “symbolic”: in the terminology of semiotics, there is no necessary connection between their form (chemical composition) and meaning (genes switched on or off). Other features of molecular biology
are symbolic in this sense: for example, CAC codes for histidine, but there is no chemical reason why it should not code for
glycine. (In passing, I have found the semiotic distinction between symbol, icon, and index illuminating also in animal communication
(Maynard Smith and Harper, 1995).)
Sarkar (1996) has an interesting discussion of Monod’s notion of gratuity. He interprets Monod as arguing that “The cybernetic account
of gene regulation is of more explanatory value than a purely physicalist alternative,” but says that this opinion is justified
only if cases of gene regulation other than the lactose operon studied by Monod turn out to be of a similar nature. He concludes
that “Attempts to generalise the operon model to eukaryotic gene regulation have so far shown no trace of success.” I think
it would be hard to find a developmental geneticist who would agree with him. As I explain below, Monod’s ideas are basic
to research in the field.
Linguists would argue that only a symbolic language can convey an indefinitely large number of meanings. I think that it is
the symbolic nature of molecular biology that makes possible an indefinitely large number of biological forms. I return to
the problem of form later, but first I describe a story of how the information analogy led me up a blind alley, but at the
same time prepared me for current discoveries in developmental genetics.
7.5 The quantification of evolution
Around 1960, I conceived the idea that, using information theory, one could quantify evolution simultaneously at three levels:
genetic, selective, and morphological. The genetic aspect is easy: the channel capacity is, approximately, two bits per base.
Things are complicated by the presence of large quantities of repetitive DNA, but this can be allowed for. The selective level
is tricky, but not hopeless. Suppose one asks, “How much selection is needed to program an initially random sequence?” If,
reasonably, the selective removal of half the population is regarded as adding one bit of information, then two bits of selection are needed to program each base. The snag
is that evolution does not start from a random sequence. Instead, an already programmed gene (or set of genes) is duplicated,
and then one copy is altered by selection. However, one can still make a crude estimate of how much selection, measured in
bits, is needed to program an existing genome. Kimura (1961), using Haldane’s (1957) idea of the “cost of selection,” gave a more elegant account of how natural selection accumulates genetic information in
the genome.
The hard step is to quantify morphology, but before tackling that question, I want to suggest that the quantification of genetic
and selective information in the same units has one, perhaps trivial, use. Occasionally someone, often a mathematician, will
announce that there has not been time since the origin of the Earth for natural selection to produce the astonishing diversity
and complexity we see. The odd thing about these assertions is that, although they sound quantitative, they never tell us
by how much the time would have to be increased: twice as much, or a million times, or what? The only way I know to give a
quantitative answer is to point out that if one estimates, however roughly, the quantity of information in the genome, and
the quantity that could have been programmed by selection in 5000 MY, there has been plenty of time. If, remembering that
for most of the time our ancestors were microbes, we allow an average of 20 generations a year, there has been time for selection
to program the genome ten times over. But this assumes that the genome contains enough information to specify the form of
the adult. This is a reasonable assumption, because it is hard to see where else the information is coming from.
How much information is needed to specify the form of the adult? Clearly, one does not have to specify the nature and position
of every atom in the body, because not everything is specified. This suggested that one asks how much information is required
to specify those features shared by two individuals of the same genotype – for example, monovular twins. For simplicity, imagine
a pair of two-dimensional organisms (it is easy to extend the argument to three dimensions). Form an image of each as a matrix of black
and white dots (in effect, pixels: again, one can extend the argument to more than two kinds of pixel). Start with minute
pixels: then identical twins will differ. Gradually enlarge the pixels, until the images of identical twins are the same.
Then the information required equals the number of pixels in the image.
It is only necessary to describe the method to see what is wrong with it. Imagine three black-and-white pictures: the first
a pattern of random dots, the second the Mona Lisa, and the third a black circle on a white ground. The first would indeed require a quantity of information equal to the number
of pixels. The Mona Lisa could be described in fewer bits, because of the correlations between neighboring dots, but would still require a lot of
information. The circle could be specified by saying, if (x–a)2 + (y–b)2 < r2, then black, else white (where ab is the center of the circle, and r its radius). One might argue that this is irrelevant, because genes do not know about coordinate geometry, but this would
be a mistake. Most simple forms – and a circle is an example – can be generated by simple physical processes, so that all
the genome need do is to specify a few physical parameters: for example, reaction rates can be fixed by specifying enzymes.
The fallacy of the “pixel” line of approach is that the genome is not a description of the adult form but a set of instructions
on how to make it: it is a recipe, not a blueprint.
7.6 Is the genome a developmental program?
There is, I think, no serious objection to speaking of a genetic code, or to asserting that a gene codes for the sequence
of amino acids in a protein. Certainly, a gene requires the translating machinery of a cell – ribosomes, tRNAs, etc. – but
this does not invalidate the analogy: a computer program needs a computer before it can do anything. For an evolutionary biologist,
the point is that the translating machinery can remain constant in a lineage (although it needs an unchanging genetic program to specify it), yet changes in the genetic program can lead to changes in proteins.
One objection could be that a gene specifies only the amino acid sequence of a protein, but not its three-dimensional folded
shape. In most cases, given appropriate physical and chemical conditions, the linear string of amino acids will fold itself
up. Folding is a complex dynamic process: it is not yet possible to predict the three-dimensional structure from the sequence.
But the laws of chemistry and physics do not have to be coded for by the genes: they are given and constant. In evolution,
changes in genes can cause changes in proteins, while the laws of chemistry remain unchanged.
However, an organism is more than a bag of specific proteins. Development requires that different proteins be made at different
times, in different places. A revolution is now taking place in our understanding of this process. The picture that is emerging
is one of a complex hierarchy of genes regulating the activity of other genes. Today, the notion of genes sending signals
to other genes is as central as the notion of a genetic code was 40 years ago.
First, an experiment (Halder, Callaerts, and Gehring, 1995). There is a gene, eyeless (also known as ptx3), in the mouse. Mutations in this gene (in homozygotes) cause the mouse to develop without eyes, suggesting that the unmutated
form of the gene plays some role in eye development. The normal mouse gene has been transferred to the fruit fly, Drosophila, and activated at various sites in the developing fly (Halder, Callaerts, and Gehring, 1995). If it is activated in a developing leg, then an eye develops at the site: not, of course, a mouse eye, but a compound fly
eye. This suggests that the gene is sending a signal, “make an eye here”; more precisely, it is locally switching on other
genes concerned with eye development.
Why should a mouse gene work in a fly? Presumably, the common ancestor of mouse and fly, some 500 million years ago, had the
ancestor of the gene: this is confirmed by the presence in Drosophila of a gene with a base sequence very similar to the mouse eyeless gene. What was the gene doing in that remote ancestor? We do not know, but a plausible guess is that the ancestor had a pair of sense organs on its head – perhaps one or a small cluster of
light-sensitive cells – and that the differentiation of these cells, from undifferentiated epidermal cells, was triggered
by the ancestral gene.
This raises questions about the nature of the signals that are passing. I argued above that the inducers and repressors of
gene activity are symbolic, in the sense that there is no necessary chemical connection between the nature of an inducer and
its effects. In Jacob and Monod’s original experiments, genes metabolizing the sugar lactose were switched on by the presence
of lactose in the medium. This is obviously adaptive; there would be no point in switching on the genes if there was nothing
for them to do. But if it was selectively advantageous for these genes to be switched on by a different sugar – say maltose
– then changes in the regulatory genes that brought this about would no doubt have evolved.
Yet the experiment described above suggests that the gene responsible for initiating eye development has been conserved for
500 million years. If genes are symbolic, why should this be so? Words are symbols, and are not conserved. The words used
to describe a given object change, so why has not the gene used to elicit an eye changed? The question is made more acute
by the fact that signaling genes do sometimes acquire new meanings. In evolution, it often happens that a regulatory gene
is duplicated: one copy retains its original function, and the other changes slightly, and acquires a new function. I think
that the extreme conservatism of many signaling genes can be explained as follows. Regulatory genes are often arranged hierarchically:
gene A controls genes B, C, D … and each of B, C, and D controls yet other genes. Adaptive evolutionary changes are likely
to be gradual, and this rules out changes in the initial gene in a regulatory hierarchy. The gene eyeless, specifying where an eye is to develop, is likely to be such an initial gene, and so has been conserved. But the point I
want to make here is that it is hard even to think about the problem if one does not think of genes sending signals, and if
one does not recognize that the signals are symbolic.
To date, then, there is talk of genes “signaling” to other genes, of the genome “programming” development, and so on. Informational
terminology is invading developmental biology, as it earlier invaded molecular biology. In the next section I try to justify
this usage.
7.7 Evolution theory and the concept of information in biology
I start with a concept of information that has the virtue of clarity, but that would rule out the current usage of the concept
in biology. Dretske (1981) argues as follows. If some variable, A, is correlated with a second variable, B, then we can say that B carries information
about A; for example, if the occurrence of rain (A) is correlated with a particular type of cloud (B), then the type of cloud
tells us whether it will rain. Such correlations depend on the laws of physics, and on local conditions, which Dretske calls
“channel conditions.”
With this definition, there is no difficulty in saying that a gene carries information about adult form; an individual with
the gene for achondroplasia will have short arms and legs. But we can equally well say that a baby’s environment carries information
about its growth; if it is malnourished, it will be underweight. Colloquially, this is fine; a child’s environment does indeed
predict its future. But biologists draw a distinction between two types of causal chain – genetic and environmental, or “nature”
and “nurture” – for a number of reasons. Differences due to nature are likely to be inherited, whereas those due to nurture
are not; evolutionary changes are changes in nature, not nurture; traits that adapt an organism to its environment are likely
to be due to nature. For these reasons, the nature–nurture distinction has become fundamental in biology. Of course, the distinction
could be drawn without using the concept of information, or applying it specifically to genetic causes. However, as the examples
discussed above demonstrate, informational language has been used to characterize genetic, as opposed to environmental, causes.
I want now to try to justify this usage.
I will argue that the distinction can be justified only if the concept of information is used in biology only for causes that
have the property of intentionality (Dennett, 1987). In biology, the statement that A carries information about B implies that A has the form it does because it carries that
information. A DNA molecule has a particular sequence because it specifies a particular protein, but a cloud is not black
because it predicts rain. This element of intentionality comes from natural selection.
I start with an engineering analogy. An engineer interested in genetic algorithms wants to devise a program to play a competitive
game. For simplicity, he chooses Fox and Geese, a game played on a draughts board in which four “geese” try to corner a “fox.”
(As it happens, I played with the “evolution” of a program to play this game as long ago as the 1940s. Without a computer,
I could not tackle more difficult games, but Fox and Geese proved easily soluble.) The engineer first invents a number of
“rules” for the geese (for example, keep in line, do not leave gaps, keep opposite the fox). Each rule has one or more parameters
(for example, for the gap rule, specifying the position of any gaps). He then arranges for a bit string to specify these parameters,
and the weightings to be given to the different rules when selecting the next move. He then performs a typical genetic algorithm
experiment, starting with a population of random strings, allowing each to play against an efficient fox, selecting the most
successful, and generating a new population of strings, with random mutation. For a simple game like Fox and Geese, he will
finish up with a program that wins against any fox strategy; things are a bit harder for chess. This procedure is illustrated
in panel A of Figure 7.2.
If, instead of using a genetic algorithm approach, the engineer had simply written an appropriate program, no one, I think,
would object to saying that the program carried information, or at least instructions, embodying his intentions. By analogy,
I want to say that, in the process illustrated in panel A of Figure 7.2, there is information in the bit string, which has been programmed by selection,
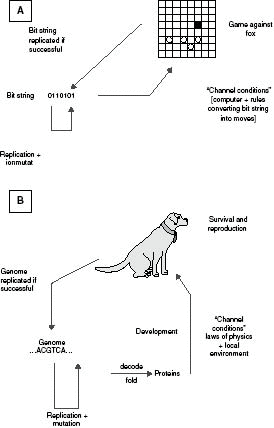
Fig. 7.2 Comparison of selection of a “genetic algorithm” to play a game of Fox and Geese (A) and biological evolution (B).
and not by the engineer. This usage is justified by the fact that, presented with a bit string and the moves that it generated,
it would be impossible to tell whether it had been designed by the engineer directly, or by selection between genetic algorithms.
Biological evolution is illustrated in panel B of Figure 7.2. It differs from panel A in two ways. First, a coding stage is present. Second, selection based on success in the game is
replaced by survival and reproduction (“fitness”) in a specific environment. I do not think the latter difference is important.
I think that the analogy between panels A and B justifies biologists in saying: that DNA contains information that has been
programmed by natural selection; that this information codes for the amino acid sequence of proteins; that, in a sense that
is much less well understood, the DNA and proteins carry instructions, or a program, for the development of the organism;
that natural selection of organisms alters the information in the genome; and finally, that genomic information is “meaningful”
in that it generates an organism able to survive in the environment in which selection has acted.
The weakness of these models, both engineering and biological, is that they do not tell us where the “rules” come from. In
the engineering case, the success of the procedure depends on the ingenuity with which the rules were chosen. In the biological
case, the rules depend on the laws of physics and chemistry; organisms do not have to invent, or evolve, rules to tell a string
of amino acids how to fold up. But there are higher-level rules, depending on the following facts: that cells divide repeatedly;
that every cell contains a complete genome; that cells can signal to their neighbors; that genes can be switched on or off
by other genes; and that states of gene switching can be transmitted through cell division to daughter cells. Research in
developmental biology is concerned with identifying regulatory genes, and with identifying the higher-level rules with parameters
that the genes control.
It should now be clear why biologists wish to distinguish between genetic and environmental causes. The environment is represented
in panel B of Figure 7.2 by the “channel conditions.” The laws of physics do not change, but the local environment might do. Fluctuations in the environment
are a source of noise in the system, not of information. Sometimes, organisms do adapt to changes in the environment during their lifetime, without genetic evolution. For example, pigment develops in the skin of humans exposed
to strong sunlight, protecting against ultraviolet rays. Such adaptive responses require that the genome has evolved under
natural selection to cope with a varying environment. What is inherited is not the dark pigment itself, but the genetic mechanism
causing it to appear in response to sunlight.
This has been a natural history of the concept of information in biology, rather than a philosophical analysis. The concept
played a central role in the growth of molecular genetics. The image of development that is emerging is one of a complex hierarchy
of regulatory genes, and of a signaling system that is essentially symbolic. Such a system depends on genetic information,
but the way in which that information is responsible for biological form is so different from the way in which a computer
program works that the analogy between them has not, I think, been particularly helpful – although it is a lot nearer the
truth than the idea that complex dynamic systems will generate biological forms “for free.” A less familiar idea that has
been central both to molecular biology and to development is Monod’s notion of “gratuity,” which I think is most clearly expressed
by saying that molecular signals in biology are symbolic.
Given the central role that ideas drawn from a study of human communication have played, and continue to play, in biology,
it is strange that so little attention has been paid to them by philosophers of biology. I think it is a topic that would
reward serious study.
7.8 Conclusions
In colloquial speech, the word “information” is used in two different contexts. It may be used without semantic implications;
for example, we may say that the form of a cloud provides information about whether it will rain. In such cases, no one would
think that the cloud had the shape it did because it provided information. By contrast, a weather forecast contains information
about whether it will rain, and it has the form it does because it conveys that information. The difference can be expressed by saying that the forecast has intentionality (Dennett, 1987), whereas the cloud does not. The notion of information as it is used in biology is of the former kind; it implies intentionality.
It is for this reason that we speak of genes carrying information during development, and of environmental fluctuations not
doing so.
A gene can be said to carry information, but what of a protein coded for by that gene? I think one must distinguish between
two cases. A protein might have a function directly determined by its structure – for example, it may be a specific enzyme,
or a contractile fiber. Alternatively, it might have a regulatory function, switching other genes on or off. Such regulatory
functions are arbitrary, or symbolic. They depend on specific receptor DNA sequences, which have themselves evolved by natural
selection. The activity of an enzyme depends on the laws of chemistry and on the chemical environment (for example, the presence
of a suitable substrate), but there is no structure that can be thought of as an evolved “receiver” of a “message” from the
enzyme. By contrast, the effect of a regulatory protein does depend on an evolved receiver of the information it carries:
the eyeless gene signals “make an eye here,” but only because the genes concerned with making an eye have an appropriate receptor sequence.
In the same way, the effect of a gene depends on the cell’s translating machinery: ribosomes, tRNAs, and assignment enzymes.
For these reasons, I want to say that genes and regulatory proteins carry information, but enzymes do not.
A very similar conclusion about the concept of information in biology has been reached by Sterelny and Griffiths (1999). In particular, they write: “Intentional information seems like a better candidate for the sense in which genes carry developmental
information and nothing else does.” Justifying this view, they add, “A distinctive test of intentional or semantic information
is that talk of error or misrepresentation makes sense.” In biology, misrepresentation is possible because there is both an
evolved structure carrying the information, and an evolved structure that receives it.
In human communication, the form of a message depends on an intelligent human agent; forecasts are written by humans (or by
computers that were programmed by humans), and are intended to alter the behavior of people who read them. There are intelligent
senders and receivers. How, then, can a genome be said to have intentionality? I have argued that the genome is as it is because
of millions of years of selection, favoring those genomes that cause the development of organisms to be able to survive in
a given environment. As a result, the genome has the base sequence it does because it generates an adapted organism. It is
in this sense that genomes have intentionality. Intelligent design and natural selection produce similar results. One justification
for this view is that programs designed by humans to produce a result are similar to, and may be indistinguishable from, programs
generated by mindless selection.
References
Apter, M. J., and Wolpert, L. (1965). Cybernetics and development I. Information theory. Journal of Theoretical Biology, 8: 244–257.
Crick, F. H. C., Griffith, J. S., and Orgel, L. E. (1957). Codes without commas. Proceedings of the National Academy of Sciences of the United States of America, 43: 416–421.
Crick, F. H. C., Barnett, L., Brenner, S., and Watts-Tobin, R. J. (1961). General nature of the genetic code. Nature, 192: 1227–1232.
Dennett, D. (1987). The Intentional Stance. Cambridge, MA: MIT Press.
Dretske, F. (1981). Knowledge and the Flow of Information. Cambridge, MA: MIT Press.
Gatlin, L. L. (1972). Information Theory and the Living System. New York: Columbia University Press.
Godfrey-Smith, P. (2000). On the theoretical role of “genetic coding.”. Philosophy of Science, 67: 26–44.
Haldane, J. B. S. (1957). The cost of natural selection. Journal of Genetics, 55: 511–524.
Halder, G., Callaerts, P., and Gehring, W. J. (1995). Induction of ectopic eyes by targeted expression of the eyeless gene in Drosophila. Science, 267: 1758–1791.
Jacob, F., and Monod, J. (1961). On the regulation of gene activity. Cold Spring Harbor Symposia on Quantitative Biology, 26: 193–211.
Kimura, M. (1961). Natural selection as a process of accumulating genetic information in adaptive evolution. Genetical Research, Cambridge, 2: 127–140.
Mahner, M., and Bunge, M. (1997). Foundations of Biophilosophy. Berlin, Heidelberg, New York: Springer-Verlag.
Maynard Smith, J. (1999). Too good to be true. Nature, 400: 223.
Maynard Smith, J., and Harper, D. G. C. (1995). Animal signals: Models and terminology. Journal of Theoretical Biology, 177: 305–311.
Monod, J. (1971). Chance and Necessity. New York: Knopf.
Sarkar, S. (1996). Biological information: A skeptical look at some central dogmas of molecular biology. In The Philosophy and History of Molecular Biology, ed. S. Sarkar. Dordrecht: Kluwer Academic Publishers, 157–231.
Shannon, C. E. (1948). A mathematical theory of communication. Bell System Technical Journal, 27: 279–423, 623–656.
Shannon, C. E., and Weaver, W. (1949). The Mathematical Theory of Communication. Urbana, IL: University of Illinois Press.
Sterelny, K., and Griffiths, P. E. (1999). Sex and Death: An Introduction to the Philosophy of Biology. Chicago, IL: University of Chicago Press.
Szathmáry, E., and Maynard Smith, J. (1995). The major evolutionary transitions. Nature, 374: 227–232.
Weismann, A. (1904). The Evolution Theory, trans. J. A. and M. R. Thomson. London: Edward Arnold.
Information and the Nature of Reality: From Physics to Metaphysics, eds. Paul Davies and Niels Henrik Gregersen. Published by Cambridge University Press © P. Davies and N. Gregersen 2010.