Here, we are taking a breast cancer dataset wherein we have classified according to whether the cancer is benign/malignant.
The following is for importing all the required libraries:
import pandas as pd
import numpy as np
from sklearn import svm, datasets
from sklearn.svm import SVC
import matplotlib.pyplot as plt
from sklearn.model_selection import train_test_split
from sklearn.model_selection import GridSearchCV
from sklearn.metrics import classification_report
from sklearn.utils import shuffle
%matplotlib inline
Now, let's load the breast cancer dataset:
BC_Data = datasets.load_breast_cancer()
The following allows us to check the details of the dataset:
print(BC_Data.DESCR)
This if for splitting the dataset into train and test:
X_train, X_test, y_train, y_test = train_test_split(BC_Data.data, BC_Data.target, random_state=0)
This is for setting the model with the linear kernel and finding out the accuracy:
C= 1.0
svm= SVC(kernel="linear",C=C)
svm.fit(X_train, y_train)
print('Accuracy-train dataset: {:.3f}'.format(svm.score(X_train,y_train)))
print('Accuracy- test dataset: {:.3f}'.format(svm.score(X_test,y_test)))
We get the accuracy output as shown:
Accuracy-train dataset: 0.967
Accuracy- test dataset: 0.958
Setting the model with the Gaussian/RBF kernel and accuracy is done like this:
svm= SVC(kernel="rbf",C=C)
svm.fit(X_train, y_train)
print('Accuracy-train dataset: {:.3f}'.format(svm.score(X_train,y_train)))
print('Accuracy- test dataset: {:.3f}'.format(svm.score(X_test,y_test)))
The output can be seen as follows:
Accuracy-train dataset: 1.000
Accuracy- test dataset: 0.629
It's quite apparent that the model is overfitted. So, we will go for normalization:
min_train = X_train.min(axis=0)
range_train = (X_train - min_train).max(axis=0)
X_train_scaled = (X_train - min_train)/range_train
X_test_scaled = (X_test - min_train)/range_train
This code is for setting up the model again:
svm= SVC(kernel="rbf",C=C)
svm.fit(X_train_scaled, y_train)
print('Accuracy-train dataset: {:.3f}'.format(svm.score(X_train_scaled,y_train)))
print('Accuracy test dataset: {:.3f}'.format(svm.score(X_test_scaled,y_test)))
The following shows the output:
Accuracy-train dataset: 0.948
Accuracy test dataset: 0.951
Now, the overfitting issue cannot be seen any more. Let's move on to having an optimal result:
parameters = [{'kernel': ['rbf'],
'gamma': [1e-4, 1e-3, 0.01, 0.1, 0.2, 0.5],
'C': [1, 10, 100, 1000]},
{'kernel': ['linear'], 'C': [1, 10, 100, 1000]}]
clf = GridSearchCV(SVC(decision_function_shape='ovr'), parameters, cv=5)
clf.fit(X_train, y_train)
print("Best parameters set found on development set:")
print()
print(clf.best_params_)
print()
print("Grid scores on training set:")
print()
means = clf.cv_results_['mean_test_score']
stds = clf.cv_results_['std_test_score']
for mean, std, params in zip(means, stds, clf.cv_results_['params']):
print("%0.3f (+/-%0.03f) for %r"
% (mean, std * 2, params))
print()
With the help of grid search, we get the optimal combination for gamma, kernel, and C as shown:
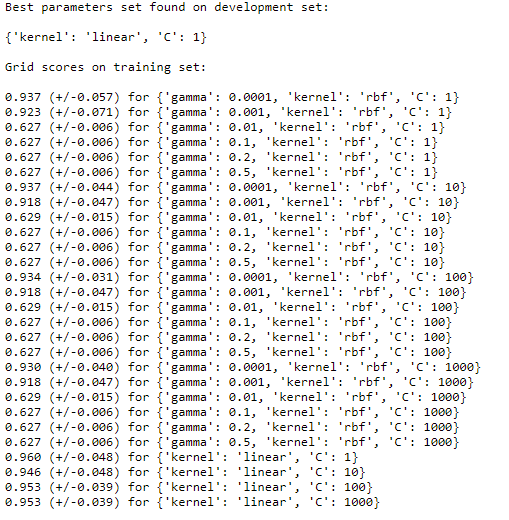
With the help of this, we can see and find out which combination of parameters is giving us the better result.
Here, the best combination turns out to be a linear kernel with a C value of 1.