Much of forensic science may be simply applied commonsense, but one does have to collect some scientific data to apply it to. It is this data-collection which is getting more and more technical, even esoteric. We started 35 years ago with test tubes; now we use gas chromatography and electron beams. A modern forensic laboratory is full of mysterious boxes of electronic gadgetry automatically printing out slips of paper and drawing wiggly lines on charts. Most of these mysterious machines were not invented, or at least not available commercially, until after World War II. Now they are spreading like dry rot and keep on proving themselves indispensable everywhere. The forensic scientist trying to keep up with their proliferation is like the Red Queen—he has to run as hard as he can to keep in the same place.
—Dr. Hamish Walls, Expert Witness, 1972
IF DR. WALLS, the former director of Scotland Yard’s Metropolitan Police Laboratory, could see what today’s crime lab looks like and how rapidly new instruments and techniques are being introduced, he would wonder that anyone could keep up. These many new techniques include but are by no means limited to scanning electron microscopy, inductively coupled plasma-optical emission spectroscopy, imaging laser-ablation mass spectroscopy, restriction fragment length polymorphism testing, thin-layer chromatography, and atomic absorption chromatography.
Before becoming a standard component of forensic analysis, each of these technical advances had gone through a slow process of acceptance, first by the scientific community and then by the legal community.
Perhaps no single technological advance has had a greater impact on law enforcement than the discovery of DNA and the subsequent development of DNA profiling. Now the ability to identify suspects and to establish their presence at a crime scene has increased exponentially. But are these technologies as infallible as they are reputed to be?
Let’s begin with a brief primer. DNA is the stuff that people are made from. In fact every living thing comes from a blueprint furnished by the deoxyribonucleic acid molecule. We were a long time in figuring this out.
Many centuries ago we concluded that some traits of appearance and even character are inherited. This is true in the general sense—rabbits give birth only to other rabbits—and in the more specific sense—a child often has the same eye color, hair color, height, and disposition as his parent. Then, in 1866, Gregor Mendel (1822–1884), an Augustinian priest, published the results of a series of experiments with pea plants (Pisum sativum) in which he advanced the idea of dominant and recessive genes. These “factors,” as he called them, control certain traits—wrinkly skin, for example, if you are a pea plant, eye color if you are human.
A couple of years after Mendel’s paper, Friedrich Miescher, a Swiss physician, discovered a substance he called “nuclein” in the nuclei of cells. We now call it nucleic acid, the NA in DNA and RNA.
The discovery of the actual function of Miescher’s nuclein and of DNA is a complex story woven of brilliant science, chance discovery, intense rivalry, and two Nobel Prizes. Linus Pauling was awarded one in 1954 for his work on the chemical bonds and structures of molecules and crystals; in 1953 he proposed that the DNA molecule was shaped like a triple helix—three spiral staircases entwined. He was only one helix off.
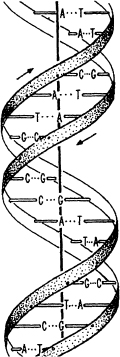
In 1953 James Watson and Francis Crick got it right, aided by an x-ray diffraction image known as Photograph 51 taken by Rosalind Franklin. The actual shape of the DNA molecule, they discovered, was a double helix. They won the Nobel Prize in 1962.
DNA has been described variously as the “blueprint” of life, the “chemical building block,” and the “book of life,” in an attempt to encompass its fundamental importance to life on Earth. Each strand of the DNA double helix is made up of multiples of four organic compounds called “bases,” arranged in a long string: adenine (A), guanine (G), cytosine (C), and thymine (T). In primitive organisms the two strands, effectively mirror images of each other, separate and then rebuild their other halves, thus creating two double helixes of DNA where only one existed before. Thus a cell can divide and become two identical cells.
In sexual reproduction the male and female each contributes half of the genetic material to create a new and unique individual. And since each strand is picked more or less at random, no two beings who are not identical twins have exactly the same genetic makeup.
In 1984 Alec Jeffreys, a geneticist at the University of Leicester, was comparing DNA patterns of the members of a family, in search of evidence of the genes that caused hereditary diseases. Jeffreys had found that the human DNA molecule contains many places where the AG and TC bonds appear in repeating sequences of the same pattern, sometimes called a “stutter.” A simple stutter would look like this:
The DNA double helix, discovered by Watson and Crick in 1953.
AAGGTC
TTCCAG
Repeated six times, it appears as follows:
AAGGTCAAGGTCAAGGTCAAGGTCAAGGTCAAGGTC TTCCAGTTCCAGTTCCAGTTCCAGTTCCAGTTCCAG
Jeffreys found that these repeating patterns, known as “mini-satellites,” formed sequences of different lengths in different people’s DNA. At the site where a particular repeating pattern occurred, some people might have twenty repeats of the pattern, others fifteen, others thirty-one, and so on. Jeffreys used a “restriction enzyme,” a chemical that cuts the DNA molecule into pieces at the same place in each molecule, to split those pieces (called “restriction fragments”) apart. Then, using a series of complex techniques, he created a radioactive copy of each DNA fragment. This irradiated copy, called a “probe,” left an image, a dark spot called a “band,” on x-ray film. And since every person carries two strands of DNA, one from each parent, each probe produced an image of two bands.
At work one morning, Jeffreys, in what he called a “eureka moment,” suddenly realized that because of the vast number of possible patterns of repeating sequences, no two people were likely to have the same DNA pattern—that the DNA pattern for each human being would be as individual as his or her fingerprints. Jeffreys’s realization would soon revolutionize the science of forensic identification.
In 1985 Jeffreys’s lab was asked to help determine whether a boy whose mother wished to bring him from Ghana to live with her in England was really her child. If he was, he could legally immigrate; if not, he would be deported. The DNA patterning showed that the boy shared all the DNA fragments that Jeffreys measured with either his father or the woman who claimed to be his mother. The test proved “beyond any reasonable doubt” that she was telling the truth; the boy was her son.
Soon afterward, Jeffreys’s new techniques received even more publicity when they were used to establish the paternities of children in several high-profile divorce cases.
Then, in 1987 the police in the village of Narborough, near Leicestershire, turned to Jeffreys for help. A serial rapist-murderer was loose in the area. His first killing had occurred four years earlier, when the body of fifteen-year-old Lynda Mann had been found on the grounds of the Carlton Hayes Psychiatric Hospital near the town. Now a second fifteen-year-old, Dawn Ashworth, had been found raped and strangled in the village of Enderby. The most sophisticated blood tests available at the time had established that both girls had been raped by a secretor—a man whose blood type showed up in his semen—and that the blood type was PGM1 + Group A. This eliminated 90 percent of the men in England, but it was not enough to convict the man they had in custody, the person they believed to be guilty of the crimes.
The suspect, seventeen-year-old Richard Buckland, was a porter at the Carlton Hayes Hospital. The learning-disabled young man allegedly knew details of Dawn Ashworth’s murder, had made several confused admissions about it, and had then retracted them. But he refused to admit or discuss anything about the first murder, that of Lynda Mann, who had been found on the hospital grounds.
Serologic testing of his blood revealed that he had blood type PGM1 + Group A, the same as that of the semen found in the victims’ bodies. With a DNA match, the police felt certain they could clinch the case against him.
Jeffreys’s lab ran DNA profiles of the semen from the victims’ bodies and of the blood of the suspect. The DNA typing showed that both victims had been raped by the same man, but that the rapist’s DNA did not match Buckland’s. The first DNA test ever done in a criminal case thus resulted in the exoneration of the accused man.
The police were now back to square one. They decided to try a sweep of local men to see if a DNA match could be found. From the records of their earlier investigations, they identified a large number of men who would have been between the ages of fourteen and forty at the time of the first murder and who lacked a credible alibi for the time frame of either crime. They “invited” all those men, more than four thousand of them, to provide a sample of their blood for testing. To save the time and expense of performing DNA testing on all four thousand men, they would first isolate those with the PGM1 + Group A serological type, statistically no more than 10 percent of the sample.
But Jeffreys did not have a chance to do the work. One of the group, a man named Colin Pitchfork, had asked a co-worker to take the blood test for him. Pitchfork’s deception was discovered when his co-worker told a friend, and the friend in turn told the police. When the police confronted Pitchfork at his home, he confessed to the two murders. His DNA matched that found in the victims. He was convicted of the two rapes and murders and sentenced to life in prison.
After the first few successes, Jeffreys’s lab was inundated with requests for DNA typing. Soon the flood of requests was much greater than the lab could handle, and in almost no time several commercial laboratories stepped into the gap by performing DNA typing for criminal and paternity cases. By 1989 the FBI had become convinced of the value of the new technique and opened its own DNA laboratory.
The commercial laboratories developed tests that were simpler to standardize and interpret, using four or five probes (irradiated fragments), each of which bound to only one part of the DNA molecule. For analyzing DNA in sexual assault cases, where the material collected is often a mixture of sperm and vaginal cells, they developed ways of separating the male and female DNA and analyzing them separately.
Even with only four probes, the commercial labs claimed that the DNA patterns produced were so individual that they were shared by only one in millions of people. In a criminal case this could be strong, even overwhelming, evidence of guilt.
Because DNA typing used such complex technology and claimed to produce incontrovertible identifications, it was carefully scrutinized by the scientific community. Unlike most forensic techniques, DNA identification emerged directly from methods used by scientists in medicine, biochemistry, and genetics. Scientists from these fields were able to point out the shortcomings of the testing methods used by the commercial laboratories as well as the fallacies in the astounding statistics they reported. In spite of its reputation for infallibility, there are indeed weaknesses in DNA testing.
One double strand of human DNA contains about three billion base pairs. Yet forensic DNA typing counts DNA fragments from only a few regions of this enormous molecule. In that sense, DNA typing is unlike fingerprint identification. Where fingerprint examiners can compare the entire print of a suspect to a large fraction of an unknown print, DNA technicians compare only a few scattered pieces of DNA. So far, the sequencing of the entire DNA molecule of every sample is too expensive for forensic use. So all the estimates of the frequency of DNA in the population are actually calculations of the frequency of occurrence of a very limited number of bands.
While it is probably true that no two people on earth share an entire DNA sequence, the likelihood of there being no two people with the same small DNA fragments is not known.
Another question was whether the testing methods originally used on pristine laboratory samples would work on the kinds of samples found in crime scenes—samples in which the DNA might be decomposed or contaminated with DNA from the place where they were found, from the person doing the collecting, or even from the lab itself. Could a lab get a reliable result from a bloodstain found on a street, a dirty carpet, or a pair of jeans? Contamination from material around the DNA sample can prevent the restriction enzymes from breaking up the DNA completely and render the test result unreliable.
Contamination can produce extra bands on the x-ray film. So can leaving the x-ray film on the sample for a longer-than-recommended time. And so can the presence of more than one person’s DNA in the sample. How does a lab decide which bands belong to the DNA being tested and which to something else? If there are bands on a DNA profile that don’t fit the suspect’s DNA, does this mean that the extra bands came from an extraneous source or from another suspect? When DNA decomposes, it breaks up into ever smaller fragments. So, when you break up an old piece of DNA using restriction enzymes, how can you tell whether the resulting pattern is from DNA fragments produced by those enzymes or is the result of decomposition? If a DNA sample is old or small in size, or if a person inherits the same repeating gene sequence from both parents, bands may be missing from the profile. What then should the lab conclude? Can it still be called a match?
No two tests, even under the best of circumstances, produce totally identical results. DNA doesn’t always move the same distance up the gel in response to the electric current. For example, a sample with a lot of DNA will move more slowly than one with less. Can you be sure that two DNA samples came from the same person just because the bands moved the same distance on the gel? If they didn’t move the same distance, how far apart can two bands be and still be said to match?
Police and laboratories can and do make mistakes. Samples are mixed up or are contaminated with DNA from other suspects, either during collection or in the lab. Sometimes these mistakes are discovered; at other times, particularly when the entire sample is used up in one test, they are not.
If a crime scene sample contains a mixture of DNA from the blood of two or three people, testing can determine whether a sample contains male or female DNA or a mixture of both. And in samples from sexual assaults, sperm can be physically separated from vaginal cells before testing. But in a mixture of DNA where the individual contributions cannot be separated, it is often impossible to distinguish which bands in a sample came from which contributor. If some of the bands in a mixed sample match the suspect in a case but others do not, can you really say that the matching bands all came from the suspect rather than from some other source?
Even when a DNA pattern is clear, calculating the frequency of its occurrence in the world population is not a straightforward operation. Some forensic labs say that if you determine how frequently each band occurs in the general population, you can simply multiply these occurrences to arrive at a probability that one person will have those bands in those particular places. For example, when throwing dice you have a one in six chance of throwing a six on one of the dice, and the same odds on the other. When you multiply the probabilities (6 × 6 = 36), you find that you have a 1 in 36 chance of throwing two 6s on one throw.
According to population geneticists, the frequency of genetically determined traits such as hair, skin, eye color, blood type, and certain diseases varies significantly between different racial and ethnic groups, meaning that the DNA that codes for these variations is also group-dependent. Genes are not always inherited independently from one another. People with light hair and light skin are much more likely to have blue eyes than people with dark hair and dark skin. It would not be accurate to say, therefore, that if one in ten people has blond hair and one in ten has blue eyes, you can multiply the frequency of occurrence of those two genes and get an accurate figure for the frequency of blond-haired, blue-eyed people in the population. And people within an ethnic group tend to marry people in the same group, meaning that it is likely that the genes of a person’s parents would have many similarities. Simply multiplying the bands based on these factors would therefore give a fallacious result.
Much research needed to be done before it could be confidently stated that the likelihood of two people sharing the same DNA profile on a commercial laboratory test was as small as the laboratories claimed it was. DNA testing was litigated in the courts for years, both over the accuracy of the tests and over the statistics used to interpret them. Continuing research was conducted to determine the effects of contamination on the tests as well as on the distribution within different racial and ethnic groups of the DNA fragments used in forensic testing. The National Research Council, a prestigious organization of scientists, published reports in 1992 and 1996 on forensic DNA typing. The reports addressed the questions raised about the accuracy of DNA and whether these had been adequately answered by research. Eventually courts around the world decided that enough was known about how the tests worked and how frequently the patterns occurred to allow DNA test results to be routinely considered as evidence.
Meanwhile the tests themselves were changing. Simpler and more powerful techniques were developed. The most authoritative test, called the RFLP test (Restriction Fragment Length Polymorphism), had limitations that made it less than ideal for forensic purposes. It required a large sample of DNA and took six to eight weeks to complete. Other tests were available that worked more rapidly and on smaller samples, but they did not yield the same stunning statistical outcomes. The other tests might produce a match probability in the range of one in a thousand or two, but not one in a million or even a hundred million as in RFLP testing.
To create tests that would give better statistical results on smaller samples, scientists began working with a set of smaller repeating DNA fragments called short tandem repeats, or STRs. These smaller fragments were amplified using a then-new technique called polymerase chain reaction, or PCR.
PCR uses enzymes to break a length of DNA at each end and split the AT and CG bonds in that length to make two complementary strands. When this is gently heated in a bath of A, T, C, and G molecules, the loose molecules bind to the split strands to create two whole pieces of DNA. The enzyme splits them apart again, and more molecules bind to them to create more strands of DNA, and so on until the quantity of DNA has grown to hundreds or thousands of times the original amount. PCR allows labs to obtain profiles from very small samples. Some analysts claim results can be obtained from a sample with as few as nine DNA molecules.
With PCR available to amplify collected DNA, testable amounts of DNA can be taken from cigarette butts, licked envelopes, clothing, soda-can rims, and even fingerprints. Testing methods have been developed that allow for the testing of up to fourteen areas on the DNA molecule. Instead of band patterns, these new methods yield a computer printout that shows a pattern of color-coded peaks and valleys. Each peak represents an STR sequence on the DNA molecule, one each from the mother and father of the person from whom the sample came. Profiles in which the likelihood of a match is one in tens of billions are common. And the test can be completed in two days. STR has become the name commonly used for the test method itself.
Current STR testing methods are highly automated. First a test sample is amplified using PCR. As part of the amplification process, primer sequences containing a fluorescent dye are added to the mix and bind to the DNA fragments. To help distinguish the primers from one another, four different colors of dye are used. The amplified DNA sample is then loaded into a specialized machine that completes the test. In a process called capillary electrophoresis, the machine uses an electric current to “pull” the DNA sample through a polymer solution in an extremely narrow “capillary” tube. The smaller DNA fragments move more quickly through the tube, the larger ones more slowly.
One section of the capillary tube is constructed of clear plastic, and the machine aims a laser light at that place. As each dyed primer moving through the tube reaches the clear section, it fluoresces under the light. Sensors measure the brightness and color of the fluorescence and transfer the information to a computer, where specialized software translates the light patterns into a printout of peaks of various colors.
Whatever concerns the scientific community had about the science behind STR testing were quickly brushed aside, and today STR profiling is universally accepted as courtroom evidence. Nevertheless some problems with the tests remain. The amplification of such small amounts of DNA can also amplify contaminants, creating extra peaks in the printout; mutations may result in patterns that contain extra peaks; peaks can be produced by the test process itself and sometimes for no known reason. Not everyone agrees on how low a peak must be before it should not be classified a peak; and small quantities of DNA as well as old DNA may yield incomplete profiles with very small peaks or none at all. DNA lab scientists say they can tell most false peaks from real ones by their shapes. But the difference is not always obvious.
When a profile is not an obvious match and requires interpretation to distinguish real peaks from false ones, it is all too easy for the analyst to see what he expects or wants to see in the profile. On occasion a DNA lab analyst has reviewed a printout of DNA evidence containing some peaks consistent with a suspect’s DNA, some peaks in areas inconsistent with the suspect, and no peaks at some points where the suspect’s DNA would have shown them. The analyst has called it a match, rationalizing that the inconsistent peaks were caused by contamination or accidents, and the missing peaks dropped out of the sample.
Because of the extremely tiny quantities of DNA involved, contamination is an even larger problem with STR testing than with the older tests. Several cases have been reported in which a DNA result has identified the wrong person because of contamination in the laboratory. Often the wrongly identified “suspect” is one of the lab workers. In one instance in the Mid-west, the testing of semen from a decades-old rape and murder case resulted in a match with a known sex offender in the area, one whose DNA was being processed in the same government lab for inclusion in an offender database. But for the fact that the identified suspect was only four years old and living in another city at the time the crime was committed, the error might never have been discovered. An adult suspect might have ended up serving prison time for an offense he did not commit.
In a widely reported case in Australia, DNA found on the body of a murdered toddler was matched to a mentally disabled woman living hundreds of miles away. Her family insisted that she had never left her hometown. It turned out that the woman had been sexually assaulted and that the samples from her case had been analyzed on the same day as the samples from the case of the murdered child.
Mixtures are still a problem. Even with the new techniques, bands from multiple sources of DNA cannot generally be distinguished from one another in a printout. New test methods are being developed, however, that will supposedly make it possible to tell which of several people left the DNA in a mix of profiles.
DNA typing of animals and even plants has also been used to solve crimes. Animal DNA has been used to convict poachers of endangered animals, birds, and fish as well as smugglers of prohibited animal products. DNA typing of dog and cat hair has been used to link animal hair found at a crime scene with the suspect’s pets. In at least one case, DNA typing of plant material found in the defendant’s truck was used to show that the truck had been in the area of the crime.
The success of RFLP and STR testing has further encouraged forensic scientists to look at other forms of DNA typing being used in biology and medicine.
Mitochondria are often called the energy factories of cells. There are as many as two thousand microbe-sized mitochondria in the cells of every animal, plant, and fungus. They reside outside the nucleus of the cell, have their own separate membranes, and convert sugar and oxygen into a compound called adenosine triphosphate (ATP). ATP provides the energy that allows the cells and the organisms they comprise to do what they do. Mitochondria are passed from mother to child, and, except for random mutations, remain unchanged through the maternal line for many generations.
The DNA found in mitochondria is unique to the mitochondria and bears no relation to the cell’s own DNA. The mitochondrial DNA genome was first sequenced in 1981.
Our bodies contain a great deal more mitochondrial DNA than nuclear DNA. Mitochondrial DNA testing has become popular with archaeologists and anthropologists because testable amounts of mitochondrial DNA may continue to exist in ancient bones and teeth long after the nuclear DNA has disintegrated. Scientists have been able to obtain sequences of mitochondrial DNA from samples that are thousands of years old. Mitochondrial DNA testing can also be done on a material such as rootless hair, which contains no nuclear DNA.
In forensics, mitochondrial DNA typing may help make an identification when a specimen is too old or degraded to permit even STR typing.
Mitochondrial DNA typing is more difficult to perform than STR typing, and the results are less discriminating. Because we receive our mitochondrial DNA only from our mothers, any person’s mitochondrial DNA is the same as that of his or her mother and of all his or her maternal relations. So many people can have the same mitochondrial DNA, in fact, that it is not as conclusive as STR typing result for identifying the source of a sample.
Mitochondrial DNA testing has been used in the aftermath of wars and oppressive political regimes to identify bodies in mass graves. In 1993 it was used to help solve the mystery of the disappearance of the Russian royal family during the Bolshevik Revolution. The Romanovs—Tsar Nicholas II, his wife Alexandra, and their five children—were imprisoned and killed by the Bolsheviks in 1918. But the location of their remains was not revealed until 1991, after the fall of the Soviet Union. Their bodies were found in an unmarked grave in a forest near Yekaterinburg.
Sulfuric acid had been poured on the bodies after the murders, and the bones had been moved from their original site and reburied. Besides the incomplete skeletons, little was left with which to identify the bodies. To confirm that the bones found at the site were in fact those of the Romanovs, researchers used both STR and mitochondrial DNA testing. STR testing of tiny amounts of nuclear DNA recovered from the bones revealed that they had come from a family—a father, mother, and three daughters—as well as from five unrelated persons. According to records, the additional remains were probably those of four servants and the family doctor, killed along with the tsar and his family.
Mitochondrial DNA was recovered from the bones, sequenced, and compared to that of known maternal relatives of Nicholas and Alexandra, a group of European aristocrats that included England’s Prince Philip. The mitochondrial DNA in the bones believed to be the tsar’s were mostly a match with Nicholas’s relatives, but they did show a rare mutation that was not present in any of the living relations. To confirm the match, researchers recovered mitochondrial DNA from the body of Nicholas’s brother; testing showed the same mutation.
More bone fragments found near the burial site in 2007 were also tested and appear to be those of the remaining two children.
Mitochondrial DNA testing was also done on a tissue sample from Anna Anderson, a woman who, until her death in 1984, claimed to be the youngest Romanov daughter, Anastasia. The tests confirmed that she was not a member of the Romanov family.
Mitochondrial DNA testing was also used to identify the Vietnam War–era soldier whose remains were placed in the tomb of the unknown soldiers in Washington, D.C. The remains were believed to be those of air force lieutenant Michael J. Blassie, but this identification was not confirmed until 1998 when mitochondrial DNA was extracted from his bones, tested, and compared with that of Lieutenant Blassie’s mother.
A recent addition to forensic DNA typing, Y-STR testing, was made possible by the sequencing of the Y chromosome of the human genome. The X and Y chromosomes determine the sex of a human being: women’s DNA has two X chromosomes while men’s has an X and a Y. Since only men have Y chromosomes, DNA typing using short tandem repeats unique to the Y chromosome (Y-STR) can detect and identify male DNA in a mixture. In samples in which the DNA of more than one male is present, it can also help distinguish one profile from another.
Standard STR typing can also detect the presence of male DNA using a sequence on the amelogenin gene that has been shown to be different in men and women. But detecting male DNA with this marker can be difficult when very little of it is present in a sample.
The usefulness of Y-STR typing is limited because, like mitochondrial DNA, it is transmitted by only one parent. Since women do not have a Y chromosome, a man’s Y chromosome must come from his father. And, unless it mutates, this DNA will be identical to that of his father, grandfather, paternal uncles, sons, and so forth—all his paternal relatives.
Y-STR typing has been used in genealogy to determine relatedness. Perhaps the most famous example of Y-STR typing was its use in determining if Thomas Jefferson fathered the children of Sally Hemings, one of his slaves.
Stories have circulated from as long ago as Jefferson’s own lifetime that Sally Hemings was Jefferson’s mistress and that some or all of her six or seven children were his. In the 1990s researchers interested in seeing whether the story could be confirmed by DNA typing sought out relatives who could be tested. They were able to find living descendants, in an unbroken male line, of two of Hemings’s sons. On the other hand, there are no living descendants in the male line of either Thomas Jefferson or his only brother. But researchers found some male descendants of Jefferson’s paternal uncle.
Y-STR testing on the living male descendants of both Jefferson and Hemings showed that one of Sally Hemings’s great-great-great grandsons, the descendant of her youngest son, had a Y-STR profile identical to that of Jefferson’s uncle—and, by implication, identical to that of Jefferson himself. The descendants of another of Hemings’s sons had a profile that differed at several points from the Jefferson family profile, showing that he was not a descendant of Jefferson’s.
The results in this case demonstrate both the power and the limitations of Y-STR testing. While the Y-STR profile match is conclusive evidence that one of Sally Hemings’s great-great-great grandsons is directly related to Thomas Jefferson, it does not prove conclusively that Jefferson was his great-great-great grandfather. The father of Sally Hemings’s son might have been Jefferson, but it might also have been one of Jefferson’s relatives, a number of whom spent time at Monticello. The Y-STR typing provided evidence that bolsters the stories about Jefferson’s relationship with Sally Hemings, but not enough to prove conclusively that they are true.
Once DNA’s power to identify people was recognized, it did not take long to decide that a large-scale database of DNA profiles, like the AFIS database of fingerprints, could be a major resource for law enforcement in identifying suspects in criminal cases. Most states now have laws that allow the police to collect saliva and blood samples from arrestees or convicted criminal offenders. These DNA profiles are then uploaded into databanks. The FBI also keeps a national database of offender profiles. The standard method for creating database profiles uses thirteen particular STR markers plus the amelogenin marker for the gender of the subject.
The FBI’s DNA database, NDIS, now contains about three million profiles. Various state databases altogether contain many more. As the number of stored profiles has increased, so has the number of cases in which a suspect has been located by comparing DNA found at a crime scene to that of known criminals in the offender databases. “Cold hits,” as these matches are called, have resulted in convictions in many previously unsolved cases, some of them many years old. Because of the success of DNA databases in identifying perpetrators of crimes, some authorities have proposed expanding them to include the profiles of all Americans.
More recently, DNA databases have been used to locate suspects through their close relatives. In a case in Great Britain, a killer whose DNA was not in the national database was located when blood he left on the murder weapon turned out to be a close, but not quite complete, match to another profile in the database. Police turned the focus of their investigation to that man and eventually found that the killer was his brother.
Privacy advocates have raised alarms about the use of DNA databanks to harass innocent people who happen to be relatives of a criminal suspect. And scientists have pointed out that in the huge offender databases now in existence, even people who are not related may have profiles that share many similarities. So the likelihood that profile-searching will actually bring police closer to a perpetrator may not be great enough to justify the expense of investigating close matches or the risk of disrupting the lives of the innocent.
As research scientists develop faster, better, and more sensitive methods of detecting, sequencing, and comparing DNA, forensic laboratories continue to look for ways to adapt those methods in order to improve the quality of their own testing. Around the world forensic DNA test methods are constantly being sought that would allow forensic scientists to discriminate among profiles in a mixture and obtain test results from ever smaller amounts of DNA.
Among the forensic sciences, DNA typing is unique in its close relationship to the biological sciences and in the continuing interest of scientists in the forensic uses of DNA typing techniques. The courts have been willing to listen when the scientific community weighs in on the validity of a particular use of DNA typing. This interest has made DNA typing one of the most reliable of the forensic sciences.
On December 23, 1972, Diana Sue Sylvester, a twenty-two-year-old nurse, was raped and murdered in her San Francisco apartment. The police had one suspect, based on a neighbor’s identification, but the identification was shaky, and no other evidence was ever developed against the man.
On February 21, 2008, thirty-five years later, a wheelchair-bound seventy-one-year-old handyman named John Puckett was convicted of her murder (the statute of limitations had run out on the rape charge). He had been convicted of rape previously, but there was no known connection between him and Diana Sue Sylvester. The only evidence against him was a DNA “cold hit” on the California CODIS system.
CODIS, the Combined DNA Index System, was set up by the FBI in 1994. Originally intended to keep DNA records of all sex offenders, the index has been broadened to include the DNA profiles of all persons arrested on federal charges, the profiles of missing persons, and DNA samples gathered at crime scenes but as yet unidentified. Most states now have CODIS systems of their own.
During Puckett’s trial the prosecution held that although the DNA “hit” was the only evidence against the defendant, it was enough to convict him. They argued that the odds of a coincidental match between Puckett’s DNA and the sample found at the crime scene were one in 1.1 million. Based on this argument, Puckett was convicted and sentenced to seven years to life in prison.
The judge, however, had refused to allow the jurors in the case to hear that there was considerable controversy over that one in 1.1 million figure, and that some statisticians believed the figure should be closer to one in three.
There are two ways of looking at the problem presented by Puckett’s cold hit. Statistically, the results can be vastly different. The odds of finding a particular match in a database are drastically different from the odds of finding any old match. The “birthday paradox” explains what I mean.
What are the odds of anyone in the room having the same birthday as yours? The size of the group doesn’t matter. It could be a small gathering or could include everyone at Grand Central Station at 8:30 in the morning. The odds are in fact one in 365. And the odds of having a better-than-even chance of finding a match are one in 183—half of 365 for a 50 percent chance.
But now let’s look at the problem differently and make it nonexclusive. What are the chances that any two people will have the same birthday? How many people must be in a room before the odds are 50–50 that if everyone calls out their birth date, two of them will match? The math is a bit complex, involving combinational statistics, but the number works out to 23. That means that if you take random groups of 23 people and check their birthdays, half the time you will get a match. Just changing it from a specific match—someone else having your birthday—to a nonspecific match—any two people having the same birthday—changes the number of people in the group from 183 to 23.
And this is what searching for cold hits in CODIS does—it checks everyone in the system against everyone else in the system. And if you run the same math on CODIS as you do on the birthday paradox, you will find that the odds of a cold hit are one in three. Yes, but what are the odds that the person you find will be a sex offender? The odds of that are 100 percent since that is the nature of the database you are running the test on.
The acceptance of DNA evidence in the courtroom and the belief in its accuracy is now so complete that complacency has begun to set in. Rather than giving any statistics at all, the criminalist on the stand need only say, “The DNA evidence says it’s him, so it’s him.” He may not be telling the court and the jury about technical difficulties—the unexplained allele dropout at position five, or the mysterious peak somewhere else on the chart. It may not be the actual DNA evidence that the court hears but rather the criminalist’s interpretation of the DNA evidence. The jury needs to know this.