1 Introduction
BioASQ1 is a biomedical document classification, document retrieval, and question answering competition, currently in its seventh year. We provide an overview of our submissions to semantic question answering task (7b, Phase B) of BioASQ 7 (except for ‘ideal answer’ test, in which we did not participate this year). In this task systems are provided with biomedical questions and are required to submit ideal and exact answers to those questions. We have used BioBERT [9] based system, see also Bidirectional Encoder Representations from Transformers (BERT) [4], and we fine tuned it for the biomedical question answering task. Our system scored near the top for factoid questions for all the batches of the challenge. More specifically, in the third test batch set, our system achieved highest ‘MRR’ score for Factoid Question Answering task. Also, for List-type question answering task our system achieved highest recall score in the fourth test batch set. Along with our detailed approach, we present the results for our submissions and also highlight identified downsides for our current approach and ways to improve them in our future experiments.
The QA task is organized in two phases. Phase A deals with retrieval of the relevant document, snippets, concepts, and RDF triples, and phase B deals with exact and ideal answer generations. Exact answer generation is required for factoid, list, and yes/no type question.
BioASQ competition provides the training and testing datasets. The training data consists of questions, golden standard documents, snippets, concepts, and ideal answers (which we did not use in this paper, but we used last year [2]). The test data is split between phase A and phase B. The phase A dataset consists of the questions, unique ids, question types. The phase B dataset consists of the questions, golden standard documents, snippets, unique ids and question types. Exact answers for factoid type questions are evaluated using strict accuracy (consider the top answer), lenient accuracy (consider the top 5 answers), and MRR (Mean Reciprocal Rank) which takes into account the ranks of returned answers. Answers for the list type question are evaluated based on precision, recall, and F-measure.
2 Related Work
2.1 BioASQ
Sharma et al. [16] describe a system with two stage process for factoid and list type question answering. Their system extracts relevant entities and then runs supervised classifier to rank the entities. Wiese et al. [18] propose neural network based model for Factoid and List-type question answering task. The model is based on Fast QA and predicts the answer span in the passage for a given question. The model is trained on SQuAD data set and fine tuned on the BioASQ data. Dimitriadis et al. [5] proposed two stage process for Factoid question answering task. Their system uses general purpose tools such as Metamap, BeCas to identify candidate sentences. These candidate sentences are represented in the form of features, and are then ranked by the binary classifier. Classifier is trained on candidate sentences extracted from relevant questions, snippets and correct answers from BioASQ challenge. For factoid question answering task highest ‘MRR’ achieved in the 6th edition of BioASQ competition is ‘0.4325’. Our system is a neural network model based on contextual word embeddings [4] and achieved a ‘MRR’ score ‘0.6103’ in one of the test batches for Factoid Question Answering task.
2.2 A Minimum Background on BERT
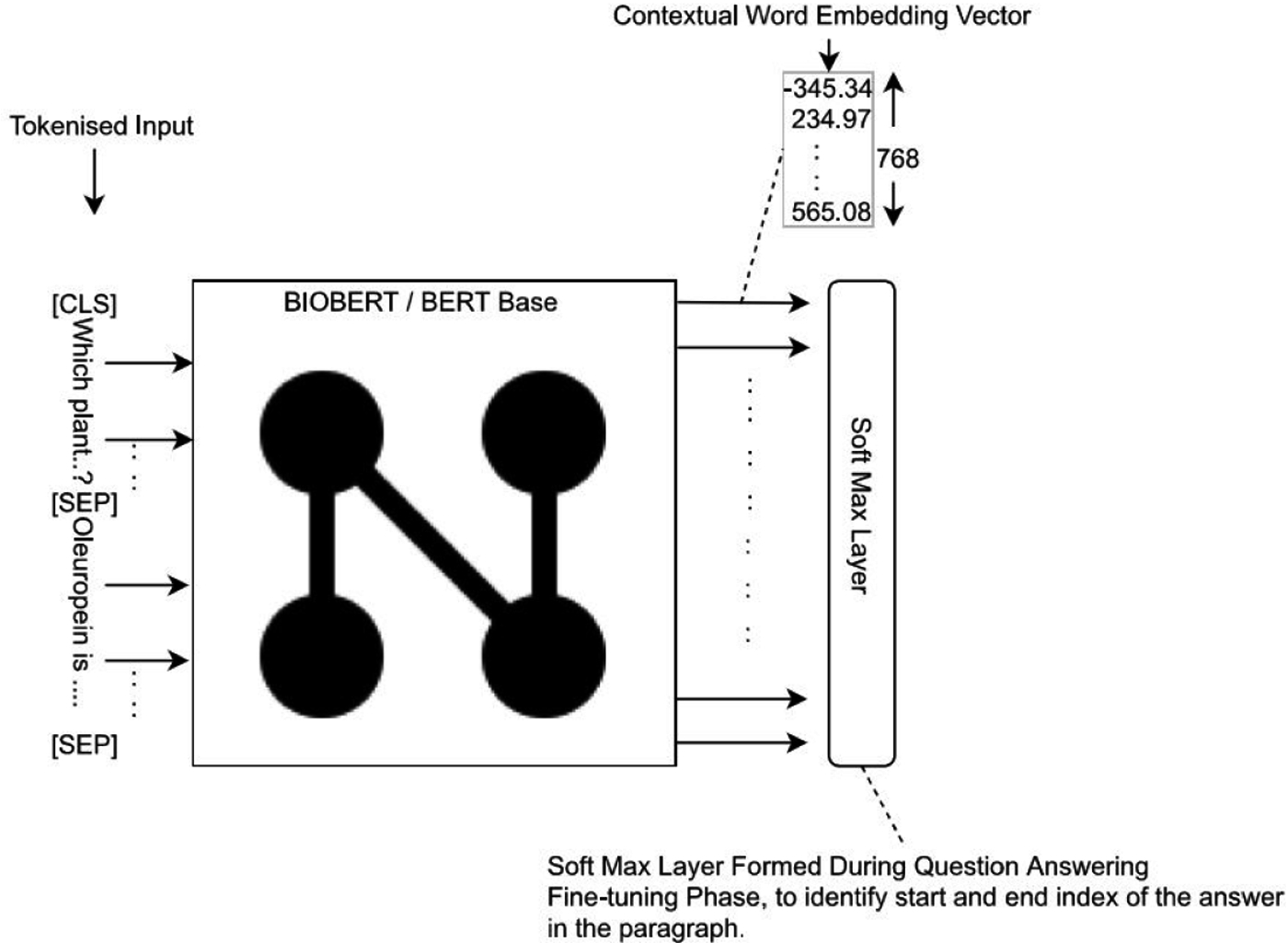
BioBERT fine tuned for question answering task
Comparison of Word Embeddings and Contextual Word Embeddings.
A ‘word embedding’ is a learned representation. It is represented in the form of vector where words that have the same meaning have a similar vector representation. Consider a word embedding model ‘word2vec’ [12] trained on a corpus. Word embeddings generated from the model are context independent that is, word embeddings are returned regardless of where the words appear in a sentence and regardless of e.g. the sentiment of the sentence. However, contextual word embedding models like BERT also takes context of the word into consideration.
2.3 Comparison of BERT and Bio-BERT
‘BERT’ and BioBERT are very similar in terms of architecture. Difference is that ‘BERT’ is pretrained on Wikipedia articles, whereas BioBERT version used in our experiments is pretrained on Wikipedia, PMC and PubMed articles. Therefore BioBERT model is expected to perform well with biomedical text, in terms of generating contextual word embeddings.
BioBERT model used in our experiments is based on BERT-Base Architecture; BERT-Base has 12 transformer Layers where as BERT-Large has 24 transformer layers. Moreover contextual word embedding vector size is 768 for BERT-Base and more for BERT-large. According to [4] Bert-Large, fine-tuned on SQuAD 1.1 question answering data [13] can achieve F1 Score of 90.9 for Question Answering task where as BERT-Base Fine-tuned on the same SQuAD question answering [13] data could achieve F1 score of 88.5. One downside of the current version BioBERT is that word-piece vocabulary2 is the same as that of original BERT Model, as a result word-piece vocabulary does not include biomedical jargon. Lee et al. [9] created BioBERT, using the same pre-trained BERT released by Google, and hence in the word-piece vocabulary (vocab.txt), as a result biomedical jargon is not included in word-piece vocabulary. Modifying word-piece vocabulary (vocab.txt) at this stage would loose original compatibility with ‘BERT’, hence it is left unmodified.
In our future work we would like to build pre-trained ‘BERT’ model from scratch. We would pretrain the model with biomedical corpus (PubMed, ‘PMC’) and Wikipedia. Doing so would give us scope to create word piece vocabulary to include biomedical jargon and there are chances of model performing better with biomedical jargon being included in the word piece vocabulary. We will consider this scenario in the future, or wait for the next version of BioBERT.
3 Experiments: Factoid Question Answering Task
For Factoid Question Answering task, we fine tuned BioBERT [9] with question answering data and added new features. Figure 1 shows the architecture of BioBERT fine tuned for question answering tasks: Input to BioBERT is word tokenized embeddings for question and the paragraph (Context). As per the ‘BERT’ [4] standards, tokens ‘[CLS]’ and ‘[SEP]’ are appended to the tokenized input as illustrated in the figure. The resulting model has a softmax layer formed for predicting answer span indices in the given paragraph (Context). On test data, the fine tuned model generates n-best predictions for each question. For a question, n-best corresponds that n answers are returned as possible answers in the decreasing order of confidence. Variable n is configurable. In our paper, any further mentions of ‘answer returned by the model’ correspond to the top answer returned by the model.
3.1 Setup
BioASQ provides the training data. This data is based on previous BioASQ competitions. Train data we have considered is aggregate of all train data sets till the 5th version of BioASQ competition. We cleaned the data, that is, question-answering data without answers are removed and left with a total count of 530 question answers. The data is split into train and test data in the ratio of 94 to 6; that is, count of 495 for training and 35 for testing.
The original data format is converted to the BERT/BioBERT format, where BioBERT expects ‘start_index’ of the actual answer. The ‘start_index corresponds to the index of the answer text present in the paragraph/Context. For finding ‘start_index’ we used built-in python function find(). The function returns the lowest index of the actual answer present in the context(paragraph). If the answer is not found ‘−1’ is returned as the index. The efficient way of finding start_index is, if the paragraph (Context) has multiple instances of answer text, then ‘start_index’ of the answer should be that instance of answer text whose context actually matches with what’s been asked in the question.
Example (Question, Answer and Paragraph from [17]):
Question: Which drug should be used as an antidote in benzodiazepine overdose?
Answer: ‘Flumazenil’
Paragraph(context):
“Flumazenil use in benzodiazepine overdose in the UK: a retrospective survey of NPIS data. OBJECTIVE: Benzodiazepine (BZD) overdose (OD) continues to cause significant morbidity and mortality in the UK. Flumazenil is an effective antidote but there is a risk of seizures, particularly in those who have co-ingested tricyclic antidepressants. A study was undertaken to examine the frequency of use, safety and efficacy of flumazenil in the management of BZD OD in the UK. METHODS: A 2-year retrospective cohort study was performed of all enquiries to the UK National Poisons Information Service involving BZD OD. RESULTS: Flumazenil was administered to 80 patients in 4504 BZD-related enquiries, 68 of whom did not have ventilatory failure or had recognised contraindications to flumazenil. Factors associated with flumazenil use were increased age, severe poisoning and ventilatory failure. Co-ingestion of tricyclic antidepressants and chronic obstructive pulmonary disease did not influence flumazenil administration. Seizure frequency in patients not treated with flumazenil was 0.3%”.
Actual answer is ‘Flumazenil’, but there are multiple instances of word ‘Flu-mazenil’. Efficient way to identify the start-index for ‘Flumazenil’ (answer) is to find that particular instance of the word ‘Flumazenil’ which matches the context for the question. In the above example ‘Flumazenil’ highlighted in bold is the actual instance that matches question’s context. Unfortunately, we could not identify readily available tools that can achieve this goal. In our future work, we look forward to handling these scenarios effectively.
Note: The creators of ‘SQuAD’ [13] have handled the task of identifying answer’s start_index effectively. But ‘SQuAD’ data set is much more general and does not include biomedical question answering data.
3.2 Training and Error Analysis
During our training with the BioASQ data, learning rate is set to 3e-5, as mentioned in the BioBERT paper [9]. We started training the model with 495 available train data and 35 test data by setting number of epochs to 50. After training with these hyper-parameters training accuracy (exact match) was 99.3% (overfitting) and testing accuracy is only 4%. In the next iteration we reduced the number of epochs to 25 then training accuracy is reduced to 98.5% and test accuracy moved to 5%. We further reduced number of epochs to 15, and the resulting training accuracy was 70% and test accuracy 15%. In the next iteration set number of epochs to 12 and achieved train accuracy of 57.7% and test accuracy 23.3%. Repeated the experiment with 11 epochs and found training accuracy to be the same 57.7% and the test accuracy 22%. In the next iteration we set number of epochs to 9 and found training accuracy of 48% and test accuracy of 15%. Hence optimum number of epochs is taken as 12 epochs.
During our error analysis we found that on test data, model tends to return text in the beginning of the context (paragraph) as the answer. On analysing train data, we found that there are 120 (out of 495) question answering data instances having start_index:0, meaning 120 (about 25%) question answering data has first word(s) in the context(paragraph) as the answer. We removed 70% of those instances in order to make train data more balanced. In the new train data set we are left with 411 question answering data instances. This time we got the highest test accuracy of 26% at 11 epochs. We have submitted our results for BioASQ test batch-2, got strict accuracy of 32% and our system stood in 2nd place. Initially, hyper-parameter- ‘batch size’ is set to 400. Later it is tuned to 32. Although accuracy (exact answer match) remained at 26%, model generated concise and better answers at batch size 32, that is wrong answers are close to the expected answer in good number of cases.
Example.(from [17])
Question: Which mutated gene causes Chediak Higashi Syndrome?
Exact Answer: ‘lysosomal trafficking regulator gene’.
The answer returned by a model trained at 400 batch size is ‘Autosomal-recessive complicated spastic paraplegia with a novel lysosomal trafficking regulator’, and from the one trained at 32 batch size is ‘lysosomal trafficking regulator’.
In further experiments, we have fine tuned the BioBERT model with both ‘SQuAD’ dataset (version 2.0) and BioASQ train data. For training on ‘SQuAD’, hyper parameters- Learning rate and number of epochs are set to ‘3e-3’ and 3 respectively as mentioned in the paper [4]. Test accuracy of the model boosted to 44%. In one more experiment we trained model only on ‘SQuAD’ dataset, this time test accuracy of the model moved to 47%. The reason model did not perform up to the mark when trained with ‘SQuAD’ alongside BioASQ data could be that in formatted BioASQ data, start_index for the answer is not accurate, and affected the overall accuracy.
4 Our Systems and Their Performance on Factoid Questions
We have experimented with several systems and their variations, e.g. created by training with specific additional features (see next subsection). Here is their list and short descriptions. Unfortunately we did not pay attention to naming, and the systems evolved between test batches, so the overall picture can only be understood by looking at the details.
When we started the experiments our objective was to see whether BioBERT and entailment-based techniques can provide value for in the context of biomedical question answering. The answer to both questions was a yes, qualified by many examples clearly showing the limitations of both methods. Therefore we tried to address some of these limitations using feature engineering with mixed results: some clear errors got corrected and new errors got introduced, without overall improvement but convincing us that in future experiments it might be worth trying feature engineering again especially if more training data were available.
We will discuss the performance of these models below and in Sect. 6. But before we do that, let us discuss a feature engineering experiment which eventually produced mixed results, but where we feel it is potentially useful in future experiments.
4.1 LAT Feature Considered and Its Impact (Slightly Negative)
During error analysis we found that for some cases, answer being returned by the model is far away from what it is being asked in the Question.
Example: (from [17])
Question: Hy’s law measures failure of which organ?
Actual Answer: ‘Liver’.
POETS & POETRY: He was a bank clerk in the Yukon before he published “Songs of a Sourdough” in 1907.
The focus is the part of the question that is a reference to the answer. In the example above, the focus is “he”. LATs are terms in the question that indicate what type of entity is being asked for.
(...) In the example, LATs are “he”, “clerk”, and “poet”.
For example in the question “Which plant does oleuropein originate from?” ([17]). The LAT here is ‘plant’. For the BioASQ task we did not need to explicitly distinguish between the focus and the LAT concepts. In this example, the expectation is that answer returned by the model is a plant. Thus it is conceivable that the cosine distance between contextual embedding of word ‘plant’ in the question and contextual embedding for the answer present in the paragraph(context) is comparatively low. As a result model learns to adjust its weights during training phase and returns answers with low cosine distance with the LAT.
We used Stanford CoreNLP [11] library to write rules for extracting lexical answer type present in the question, both ‘parts of speech’(POS) and dependency parsing functionality was used. We incorporated the Lexical Answer Type into one of our systems, UNCC_QA1 in Batch 4. This system underperformed our system FACTOIDS by about 3% in the MRR measure, but corrected errors such as in the example above.
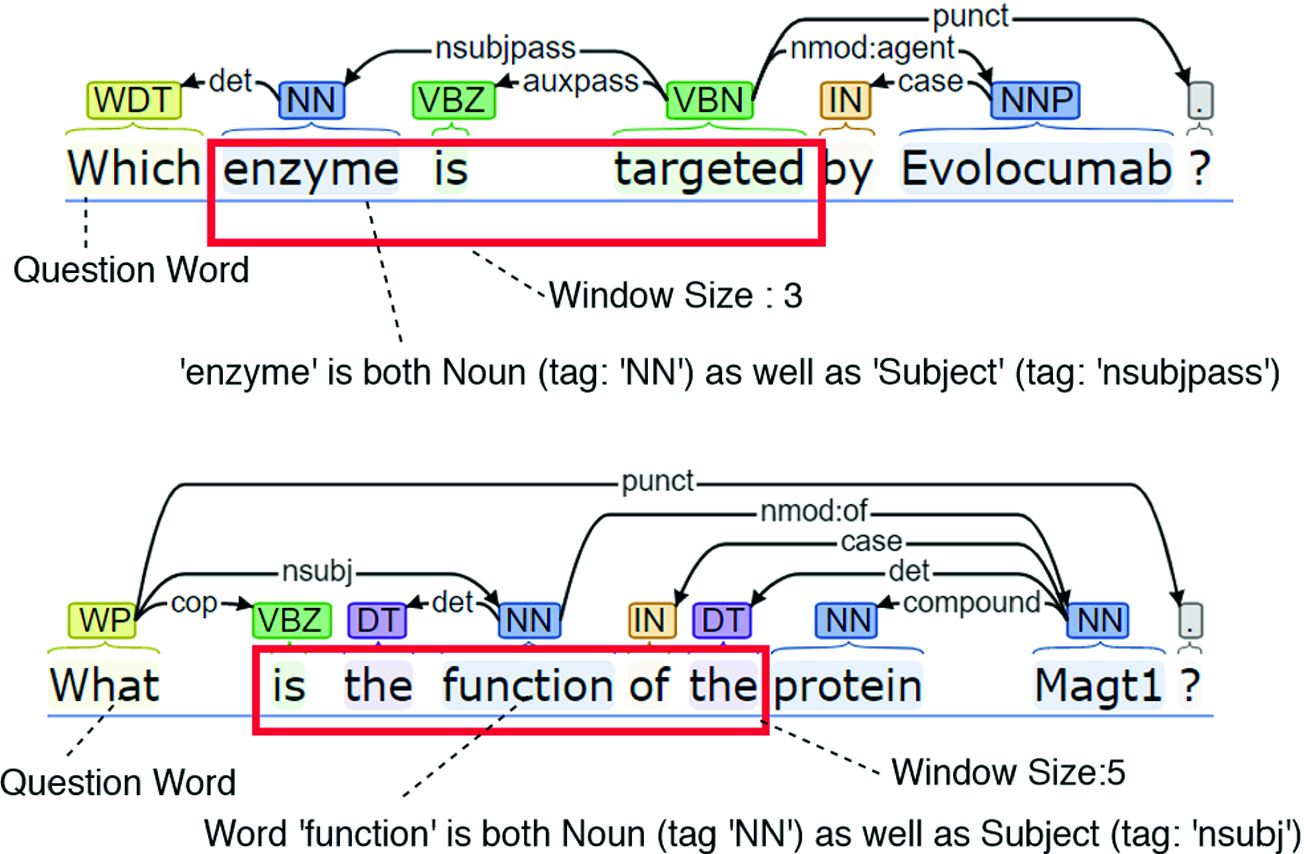
A simple way of finding the lexical answer types, LATs, of factoid questions: using POS tags to find the question word (e.g. ‘which’), and a dependency parse to find the LAT within the window of 3 words. If a noun is not found near the “Wh-” word, we iterate looking for it, as in the second panel.
LAT computation was governed by a few simple rules, e.g. when a question has multiple words that are ‘Subjects’ (and ‘Noun’), a word that is in proximity to the question word is considered as ‘LAT’. These rules are different for each “Wh” word. Perhaps because of using only very simple rules, the accuracy for ‘LAT’ derivation is 75% that is, in the remaining 25% of the cases LAT word is being identified wrong. And similarly the overall performance the system that used LATs was slightly inferior to the system without LATs, but the types of errors changed. We need to improve our ‘LAT’ derivation logic, and then perhaps with the neural network techniques they will yield better results.
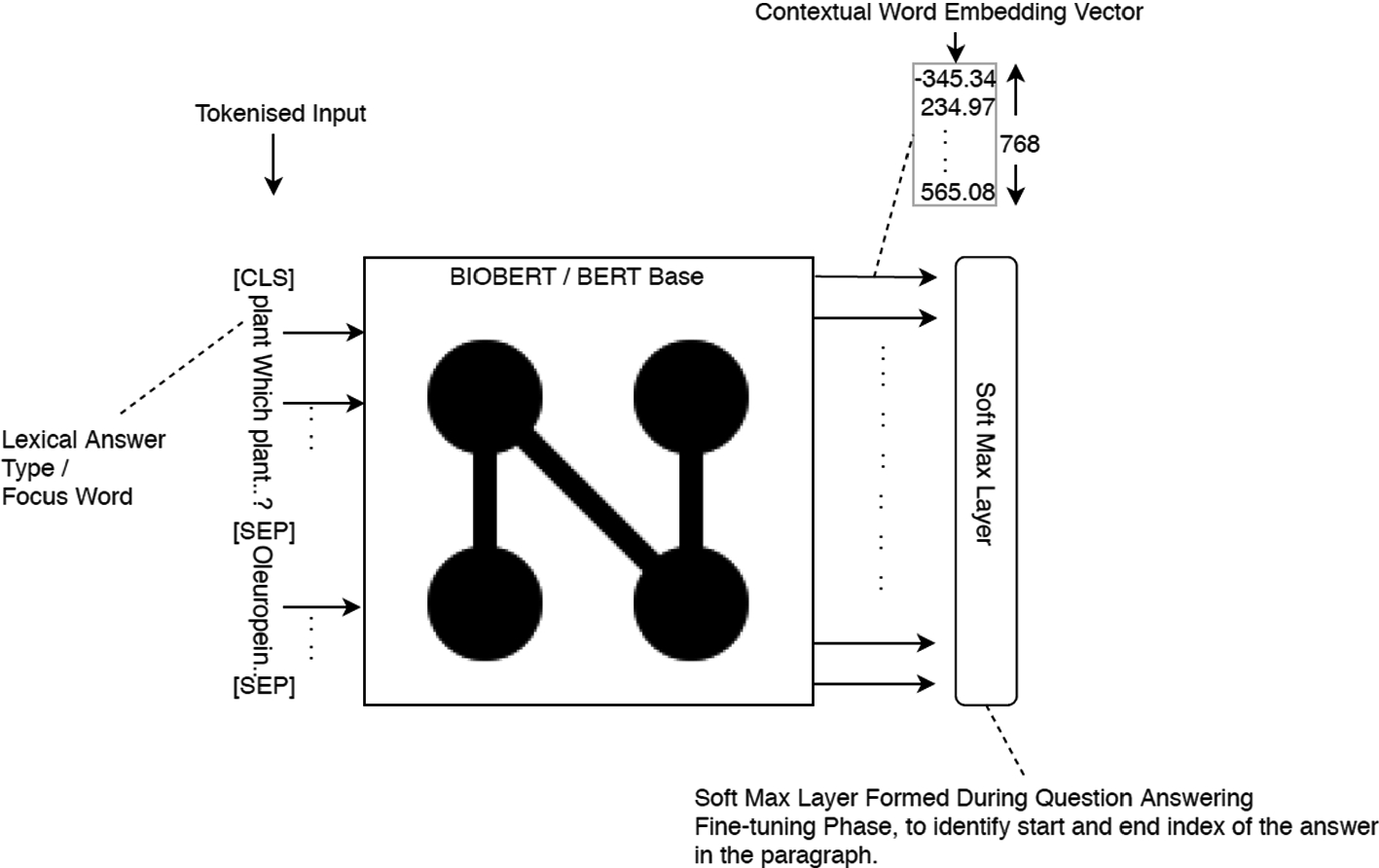
An example of a using BioBERT with additional features: Contextual word embedding for Lexical Answer Type (LAT) given as feature along with the actual contextual embeddings for the words in question and the paragraph. This change produced mixed results and no overall improvement.
4.2 Impact of Training Using BioASQ Data (Slightly Negative)
Training on BioASQ data in our entry in Batch 1 and Batch 2 under the name QA1 showed it might lead to overfitting. This happened both with (Batch 2) and without (Batch 1) hyperparameters tuning: abysmal 18% MRR in Batch 1, and slightly better one, 40% in Batch 2 (although in Batch 2 it was overall the second best result in MRR but 16% lower than the highest score).
In Batch 3 (only), our UNCC_QA3 system was fine tuned on BioASQ and SQuAD 2.0 [13], and for data preprocessing Context paragraph is generated from relevant snippets provided in the test data. This system underperformed, by about 2% in MRR, our other entry UNCC_QA1, which was also an overall category winner for this batch. The latter was also trained on SQuAD, but not on BioASQ. We suspect that the reason could be the simplistic nature of the find() function described in Sect. 3.1. So, this could be an area where a better algorithm for finding the best occurrence of an entity could improve performance.
4.3 Impact of Using Context from URLs (Negative)
In some experiments, for context in testing, we used documents for which URL pointers are provided in BioASQ. However, our system UNCC_QA3 underperformed our other system tested only on the provided snippets.
In Batch 5 the underperformance was about 6% of MRR, compared to our best system UNCC_QA1, and by 9% to the top performer (Fig. 3).
5 Performance on Yes/No and List Questions
Our work focused on Factoid questions. But we also have done experiments on List-type and Yes/No questions.
5.1 Entailment Improves Yes/No Accuracy
We started by answering always YES (in batch 2 and 3) to get the baseline performance. For batch 4 we used entailment. Our algorithm was very simple: Given a question we iterate through the candidate sentences and try to find any candidate sentence is contradicting the question (with confidence over 50%), if so ‘No’ is returned as answer, else ‘Yes’ is returned. In batch 4 this strategy produced better than the BioASQ baseline performance, and compared to our other systems, the use of entailment increased the performance by about 13% (macro F1 score). We used ‘AllenNlp’ [7] entailment library to find entailment of the candidate sentences with question.
5.2 For List-Type the URLs Have Negative Impact
Overall, we followed the similar strategy that’s been followed for Factoid Question Answering task. We started our experiment with batch 2, where we submitted 20 best answers (with context from snippets). Starting with batch 3, we performed post processing: once models generate answer predictions (n-best predictions), we do post-processing on the predicted answers. In test batch 4, our system (called FACTOIDS) achieved highest recall score of ‘0.7033’ but low precision of 0.1119, leaving open the question of how could we have better balanced the two measures.

6 Summary of Our Results
Factoid Questions. In Batch 3 we obtained the highest score. Also the relative distance between our best system and the top performing system shrunk between Batch 4 and 5.
System | Strict accuracy | Lenient accuracy | MRR |
|---|---|---|---|
Batch 1 | |||
QA1 | 0.1538 | 0.2308 | 0.1761 |
Top Competitor | 0.4103 | 0.5385 | 0.4637 |
Batch 2 | |||
QA1 | 0.36 | 0.48 | 0.4033 |
Top Competitor | 0.52 | 0.64 | 0.5667 |
Batch 3 | |||
UNCC_QA1 | 0.4483 | 0.5862 | 0.5115 |
UNCC QA2 | 0.4138 | 0.5862 | 0.4856 |
UNCC_QA3 | 0.4138 | 0.5862 | 0.4943 |
Top Competitor | 0.36 | 0.48 | 0.5023 |
Batch 4 | |||
FACTOIDS | 0.5294 | 0.7353 | 0.6103 |
UNCC QA1 | 0.4706 | 0.7353 | 0.5833 |
Top Competitor | 0.5882 | 0.8235 | 0.6912 |
Batch 5 | |||
UNCC_QA1 | 0.2857 | 0.4286 | 0.3305 |
UNCC_QA3 | 0.2286 | 0.3143 | 0.2643 |
QA1 | 0.2286 | 0.3714 | 0.2938 |
Top Competitor | 0.2857 | 0.5143 | 0.3638 |
6.1 Factoid Questions
Systems Used in Batch 5 Experiments
System description for ‘UNCC_QA1’: The system was finetuned on the SQuAD 2.0. For data preprocessing Context/paragraph was generated from relevant snippets provided in the test data.
System description for ‘QA1’: ‘LAT’ feature was added and finetuned with SQuAD 2.0. For data preprocessing Context/paragraph was generated from relevant snippets provided in the test data.
System Description for ‘UNCC_QA3’: Fine tuning process is same as it is done for the system ‘UNCC_QA1’ in test batch-5. Difference is during data preprocessing, Context/paragraph is generated from the relevant documents for which URLS are included in the test data.
6.2 List Questions
List questions
System | Mean precision | Recall | F-measure |
|---|---|---|---|
Batch 2 | |||
QA1 | 0.0471 | 0.2898 | 0.0786 |
Top Competitor | 0.5826 | 0.4839 | 0.4732 |
Batch 3 | |||
UNCC_QA1 | 0.0780 | 0.4711 | 0.1297 |
Top Competitor | 0.4267 | 0.3058 | 0.3298 |
Batch 4 | |||
FACTOIDS | 0.1119 | 0.7033 | 0.1893 |
UNCC QA1 | 0.1087 | 0.6968 | 0.1846 |
UNCC_QA3 | 0.1087 | 0.6968 | 0.1846 |
Top Competitor | 0.4841 | 0.5051 | 0.4604 |
Batch 5 | |||
UNCC_QA1 | 0.2051 | 0.5127 | 0.2862 |
Top Competitor | 0.5653 | 0.4131 | 0.4619 |
6.3 Yes/No Questions
Yes/No questions
System | Accuracy | F1 Yes | F1 No | Macro F1 |
|---|---|---|---|---|
Batch 1 | ||||
QA1 | 0.7931 | 0.8846 | – | 0.4423 |
Top Competitor | 0.8276 | 0.8980 | 0.4444 | 0.6712 |
Batch 2 | ||||
QA1 | 0.5667 | 0.7234 | – | 0.3617 |
Top Competitor | 0.8333 | 0.8387 | 0.8276 | 0.8331 |
Batch 3 | ||||
QA1 | 0.7826 | 0.8780 | – | 0.4390 |
UNCC_QA3 | 0.7826 | 0.8780 | – | 0.4390 |
Top Competitor | 0.8696 | 0.9231 | 0.5714 | 0.7473 |
Batch 4 | ||||
UNCC_QA1 | 0.6087 | 0.7097 | 0.4000 | 0.5548 |
FACTOIDS | 0.7391 | 0.8500 | – | 0.4250 |
UNCC_QA3 | 0.7391 | 0.8500 | – | 0.4250 |
Top Competitor | 0.8696 | 0.9143 | 0.7273 | 0.8208 |
Batch 5 | ||||
UNCC QA2 | 0.5429 | 0.7037 | – | 0.3519 |
Top Competitor | 0.8286 | 0.8500 | 0.8000 | 0.8250 |
7 Discussion, Future Experiments, and Conclusions
Summary. In contrast to 2018, when we submitted [2] to BioASQ a system based on extractive summarization (and scored very high in the ideal answer category), this year we mainly targeted factoid question answering task and focused on experimenting with BioBERT. After these experiments we see the promise of BioBERT in QA tasks, but we also see its limitations. The latter we tried to address with mixed results using feature engineering. Overall these experiments allowed us to secure a best and a second best score in different test batches. Along with Factoid-type question, we also tried ‘Yes/No’ and ‘List’-type questions, and did reasonably well with our very simple approach.
For Yes/No the moral worth remembering is that reasoning has a potential to influence results, as evidenced by our adding the AllenNLP entailment [7] system increased its performance.
All our data and software is available at Github, in the previously referenced URL (end of Sect. 2).
Future Experiments. In the current model, we have a shallow neural network with a softmax layer for predicting answer span. Shallow networks however are not good at generalizations. In our future experiments we would like to create dense question answering neural network with a softmax layer for predicting answer span. The main idea is to get contextual word embedding for the words present in the question and paragraph (Context) and feed the contextual word embeddings retrieved from the last layer of BioBERT to the dense question answering network. The mentioned dense layered question answering neural network need to be tuned for finding right hyper parameters.
In one more experiment, we would like to add a better version of ‘LAT’ contextual word embedding as a feature, along with the actual contextual word embeddings for question text, and Context and feed them as input to the dense question answering neural network. By this experiment, we would like to find if ‘LAT’ feature is improving overall answer prediction accuracy. Adding ‘LAT’ feature this way instead of feeding this word piece embedding directly to the BioBERT (as we did in our above experiments) would not downgrade the quality of contextual word embeddings generated form ‘BioBERT’. Quality contextual word embeddings would lead to efficient transfer learning and chances are that it would improve the model’s answer prediction accuracy.
We also see potential for incorporating domain specific inference into the task e.g. using the MedNLI dataset [15]. For all types of experiments it might be worth exploring clinical BERT embeddings [1], explicitly incorporating domain knowledge (e.g. [10]) and possibly deeper discourse representations (e.g. [14]).