1 Introduction
Language models pre-trained on large-scale text corpora achieve state-of-the-art performance in various natural language processing (NLP) tasks when fine-tuned on a given task [4, 13, 15]. Language models have been shown to be highly effective in question answering (QA), and many current state-of-the-art QA models often rely on pre-trained language models [20]. However, as language models are mostly pre-trained on general domain corpora, they cannot be generalized to biomedical corpora [1, 2, 8, 29]. Hence, similar to using Word2Vec for the biomedical domain [14], a language model pre-trained on biomedical corpora is needed for building effective biomedical QA models.
Recently, Lee et al. [8] have proposed BioBERT which is a pre-trained language model trained on PubMed articles. In three representative biomedical NLP (bioNLP) tasks including biomedical named entity recognition, relation extraction, and question answering, BioBERT outperforms most of the previous state-of-the-art models. In previous works, models were used for a specific bioNLP task [9, 18, 24, 28]. However, the structure of BioBERT allows a single model to be trained on different datasets and used for various tasks with slight modifications in the last layer.
In this paper, we investigate the effectiveness of BioBERT in biomedical question answering and report our results from the 7th BioASQ Challenge [7, 10, 11, 21]. Biomedical question answering has its own unique challenges. First, the size of datasets is often very small (e.g., few thousands of samples in BioASQ) as the creation of biomedical question answering datasets is very expensive. Second, there are various types of questions including factoid, list, and yes/no questions, which increase the complexity of the problem.
We leverage BioBERT to address these issues. To mitigate the small size of datasets, we first fine-tune BioBERT on other large-scale extractive question answering datasets, and then fine-tune it on BioASQ datasets. More specifically, we train BioBERT on SQuAD [17] and SQuAD 2.0 [16] for transfer learning. Also, we modify the last layer of BioBERT so that it can be trained/tested on three different types of BioASQ questions. This significantly reduces the cost of using biomedical question answering systems as the structure of BioBERT does not need to be modified based on the type of question.
The contributions of our paper are three fold: (1) We show that BioBERT pre-trained on general domain question answering corpora such as SQuAD largely improves the performance of biomedical question answering models. Wiese et al. [25] showed that pre-training on SQuAD helps improve performance. We test the performance of BioBERT pre-trained on both SQuAD and SQuAD 2.0. (2) With only simple modifications, BioBERT can be used for various biomedical question types including factoid, list, and yes/no questions. BioBERT achieves the overall best performance on all five test batches of BioASQ 7b Phase B1, and achieves state-of-the-art performance in BioASQ 6b Phase B. (3) We further analyze the role of pre- and post-processing in our system and show that different strategies often lead to different results.
The rest of our paper is organized as follows. First, we introduce our system based on BioBERT. We describe task-specific layers of our system and various pre- and post-processing strategies. We present the results of BioBERT on BioASQ 7b (Phase B), which were obtained using two different transfer learning strategies, and we further test BioBERT on BioASQ 6b on which our system was trained.
2 Methods

Overview of our system
2.1 BioBERT
Word embeddings are crucial for various text mining systems since they represent semantic and syntactic features of words [14, 22]. While traditional models use context-independent word embeddings, recently proposed models use contextualized word representations [4, 13, 15]. Among them, BERT [4], which is built upon multi-layer bidirectional Transformers [23], achieved new state-of-the-art results on various NLP tasks including question answering. BioBERT [8] is the first domain-specific BERT based model pre-trained on PubMed abstracts and full texts. BioBERT outperforms BERT and other state-of-the-art models in bioNLP tasks such as biomedical named entity recognition, relation extraction, and question answering [6, 19].
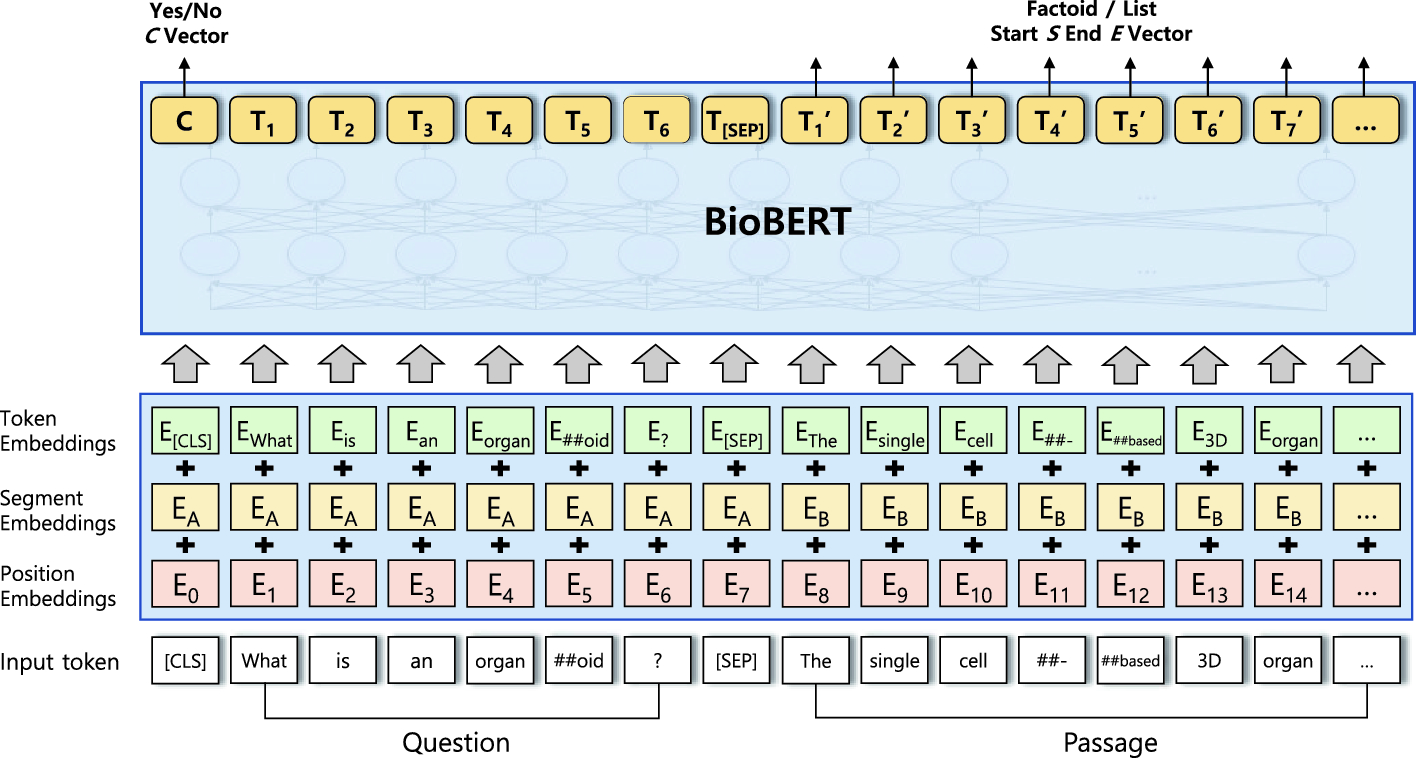
Example of a single sequence (Question-Passage pair) processed by the BioBERT.
2.2 Task-Specific Layer
The BioBERT model for QA is illustrated in Fig. 2. Following the approach of BioBERT [8], a question and its corresponding passage are concatenated to form a single sequence which is marked by different segment embeddings. The task-specific layer for factoid type questions and the layer for list type questions both utilize the output of the passage whereas the layer for yes/no type questions uses the output of the first [CLS] token.
 and the trainable end vector as
and the trainable end vector as  where H denotes the hidden size of BioBERT. The probabilities of the i-th token being the start of the answer token and the j-th token being the end of the answer token can be calculated by the following equations:
where H denotes the hidden size of BioBERT. The probabilities of the i-th token being the start of the answer token and the j-th token being the end of the answer token can be calculated by the following equations:
 denotes l-th token representation from BioBERT and
denotes l-th token representation from BioBERT and  denotes the dot product between two vectors.
denotes the dot product between two vectors. . The parameter learned during training is a sigmoid layer consisting of
. The parameter learned during training is a sigmoid layer consisting of  which is used for binary classification. The probability for the sequence to be “yes” is calculated using the following equation.
which is used for binary classification. The probability for the sequence to be “yes” is calculated using the following equation.
 and
and  , which correspond to the negative log-likelihood for the correct start and end positions, respectively. The ground truth start/end positions are denoted as
, which correspond to the negative log-likelihood for the correct start and end positions, respectively. The ground truth start/end positions are denoted as  for the start token, and
for the start token, and  for the end token. The losses are defined as follows:
for the end token. The losses are defined as follows:

 and the corresponding ground truth was used as the training loss.
and the corresponding ground truth was used as the training loss.
2.3 Pre-processing
To solve the BioASQ 7b Phase B dataset as extractive question answering, the challenge datasets containing factoid and list type questions were converted into the format of the SQuAD datasets [16, 17]. For yes/no type questions, we used 0/1 labels for each question-passage pair.
The dataset in the SQuAD format consists of passages and their respective question-answer sets. A passage is an article which contains answers or clues for answers and is denoted as the context in the dataset. The length of a passage varies from a sentence to a paragraph. An exact answer may or may not exist in the passage, depending on the task. According to the rules of the BioASQ Challenge, all the factoid and list type questions should be answerable with the given passages [21]. An exact answer and its starting position are provided in the answers field. We used various sources including snippets and PubMed abstracts, as passages. Multiple passages attached to a single question were divided to form question-passage pairs, which increased the number of question-passage pairs. The predicted answers of the question-passage pairs which share the same question are later combined in the post-processing layer.
Yes/no type questions are in the same format as the questions in the SQuAD dataset. However, binary answers are given to yes/no type questions, rather than answers selected based on their location in passages. Instead of providing an exact answer and its starting position in the answers field, we marked yes/no type questions using the strings “yes” or “no” and the Boolean values “false” and “true” in the is_impossible field. Since the distribution of yes/no answers in the training set is usually skewed, we undersampled the training data to balance the number of “yes” and “no” answers.
We used the following strategies for developing the datasets: Snippet as-is Strategy, Full Abstract Strategy, and Appended Snippet Strategy.
Snippet as-is Strategy Using snippets in their original form is a basic method for filling passages. The starting positions of exact answers indicate the positional offsets of exact matching words. If a single snippet has more than one exact matching answer word, we form multiple question-passage pairs for the snippet.
Full Abstract Strategy In the Full Abstract Strategy, we use an entire abstract, including the title of an article, as a passage. Full abstracts are retrieved from PubMed using their provided PMIDs. The snippets field of the original dataset is used to find the location of the correct answer. First, we look for the given snippet (e.g., a sentence in a typical case) from the retrieved abstract. Then, we search for the offset of the first exact matching words in the snippet, and add it to the offset of the snippet in the paragraph. In this way, we can find a plausible location of the answer within the paragraph.
Appended Snippet Strategy The Appended Snippet Strategy is a compromise between using snippets as-is and full abstracts. We first search a given snippet from an abstract and concatenate
 sentences before and after the given snippet, forming
sentences before and after the given snippet, forming  sentences into a passage (k denotes the number of sentences in a snippet, which is usually 1).
sentences into a passage (k denotes the number of sentences in a snippet, which is usually 1).
2.4 Post-processing
Since our pre-processing step involves dividing multiple passages with a same single question into multiple question-passage pairs, a single question can have multiple predicted answers. The probabilities of predicted answers for question-passage pairs sharing the same question, were merged to form a single list of predicted answers and their probabilities for a question. The answer candidate with the highest probability is considered as the final answer for a given factoid type question. For list type questions, probability thresholding was the default method for providing answers. Answer candidates with a probability higher than the threshold were included in the answer list. However, a considerable number (28.6% of BioASQ 6b list type questions) of list type questions contain the number of required answers. From the training example “Please list 6 symptoms of Scarlet fever,” we can extract the number 6 from the given question. We extracted the number provided in the question and used it to limit the length of the answer list for the question. For questions that contain the number of answers, the extracted number of answers were yielded.
For factoid and list type questions, we also filtered incomplete answers. Answers with non-paired parenthesis were removed from the list of possible answers. Pairs of round brackets and commas at the beginning and end of answers were removed.
3 Experimental Setup
3.1 Dataset
For factoid and list type questions, exact answers are included in the given snippets, which is consistent with the extractive QA setting of the SQuAD [17] dataset. Only binary answers are provided for yes/no questions. For each question, regardless of the question type, multiple snippets or documents are provided as corresponding passages.
The statistics of the BioASQ datasets are listed in Table 1. A list type question can have one or more than one answer; question-context pairs are made for every answer of a list type question. In our pre-processing step, 3,722 question-context pairs were made from 779 factoid questions in the BioASQ 7b training set. For yes/no questions, we undersampled the training data to balance the number of “yes" and “no" answers.
Statistics of the BioASQ training set.
Question type | BioASQ version | # of questions in original datasets | # of pre-processed question-passage pairs |
|---|---|---|---|
Factoid | 6b | 618 | 3,121 |
7b | 779 | 3,722 | |
List | 6b | 485 | 6,896 |
7b | 556 | 7,716 | |
Yes/No | 6b | 612 | 5,921 |
7b | 745 | 6,676 |
3.2 Training
Our system is composed of BioBERT, task-specific layers, and a post-processing layer. The parameters of BioBERT and a task-specific layer are trainable. Our training procedure starts with pre-training the system on the SQuAD dataset. The trainable parameters for factoid and list type questions were pre-trained on the SQuAD 1.1 dataset, and the parameters for yes/no type questions were pre-trained on the SQuAD 2.0 dataset. The pre-trained system is then fine-tuned on each task.
We tuned the hyperparameters on the BioASQ 4/5/6b training and test sets. We used a probability threshold of 0.42 as one of the hyperparameters for list type questions. The probability threshold was decided using the tuning procedure.
4 Results and Discussion
In this section, we first report our results for the BioASQ 7b (Phase B) Challenge, which are shown in Table 2. Please note that the results and ranks were obtained from the leaderboard of BioASQ 7b [3]. Then we evaluate our system and other competing systems on the validation set (BioASQ 6b). The results are presented in Table 3. Finally, we investigate the performance gain due to the sub-structures of the system (Tables 5 and 6). Mean reciprocal rank (MRR) and mean average F-measure ( ) were used as official evaluation metrics to measure the performance on factoid and list type questions from BioASQ, respectively. We reported strict accuracy (SAcc), lenient accuracy (LAcc) and MRR for factoid questions and mean average precision, mean average recall, and mean average F1 score for list questions4. Since the label distribution was skewed, macro average F1 score was used as an evaluation metric for yes/no questions.
) were used as official evaluation metrics to measure the performance on factoid and list type questions from BioASQ, respectively. We reported strict accuracy (SAcc), lenient accuracy (LAcc) and MRR for factoid questions and mean average precision, mean average recall, and mean average F1 score for list questions4. Since the label distribution was skewed, macro average F1 score was used as an evaluation metric for yes/no questions.
4.1 Results on BioASQ 7b
Batch results of the BioASQ 7b Challenge. We report the rank of the systems in parentheses.
Batch | Yes/no | Factoid | List | # of systems | |||
|---|---|---|---|---|---|---|---|
Participating system | Mac F1 | Participating system | MRR | Participating system | F1 | ||
1 | (1) Ours | 67.12 | (1) Ours | 46.37 | (3) Ours | 30.51 | 17 |
(2) auth-qa-1 | 53.97 | (2) BJUTNLPGroup | 34.83 | (1) Lab Zhu, Fudan Univer | 32.76 | ||
(3) BioASQ_Baseline | 47.27 | (3) auth-qa-1 | 27.78 | (4) auth-qa-1 | 25.94 | ||
2 | (1) Ours | 83.31 | (1) Ours | 56.67 | (1) Ours | 47.32 | 21 |
(2) auth-qa-1 | 62.96 | (3) QA1 | 40.33 | (3) LabZhu, FDU | 25.79 | ||
(4) BioASQ_Baseline | 42.58 | (4) transfer-learning | 32.67 | (5) auth-qa-1 | 23.21 | ||
3 | (5) Ours | 46.23 | (6) Ours | 47.24 | (1) Ours | 32.98 | 24 |
(1) unipi-quokka-QA-2 | 74.73 | (1) QA1/UNCC_QA_1 | 51.15 | (2) auth-qa-1 | 25.13 | ||
(3) auth-qa-2 | 51.65 | (3) google-gold-input | 50.23 | (4) BioASQ_Baseline | 22.75 | ||
4 | (2) Ours | 79.28 | (1) Ours | 69.12 | (1) Ours | 46.04 | 36 |
(1) unipi-quokka-QA-1 | 82.08 | (4) FACTOIDS/UNCC | 61.03 | (2) google-gold-input-nq | 43.64 | ||
(8) bioasq_experiments | 58.01 | (9) google-gold-input | 54.95 | (9) LabZhu, FDU | 32.14 | ||
5 | (1) Ours | 82.50 | (1) Ours | 36.38 | (1) Ours | 46.19 | 40 |
(2) unipi-quokka-QA-5 | 79.39 | (3) BJUTNLPGroup | 33.81 | (6) google-gold-input-nq | 28.89 | ||
(6) google-gold-input-ab | 69.41 | (4) UNCC_QA_1 | 33.05 | (7) UNCC_* | 28.62 | ||
4.2 Validating on the BioASQ 6b Dataset
Performance comparison between existing systems and our system on the BioASQ 6b dataset (from last year). Note that our system obtained a 20% to 60% performance improvement over the best systems.
System | Factoid (MRR) | List (F1) | Yes/no (Macro F1) |
|---|---|---|---|
Best System | 27.84% | 27.21% | 62.05% |
Ours | 48.41% | 43.16% | 75.87% |
Performance comparison between pre-trained models.
Pre-trained models | Factoid | List | ||||
|---|---|---|---|---|---|---|
SAcc | LAcc | MRR | Prec | Recall | F1 | |
BERTBASE+BioASQ Finetune | 24.84% | 36.03% | 28.76% | 42.41% | 35.88% | 35.37% |
BioBERT+BioASQ Finetune | 34.16% | 47.83% | 39.64% | 44.62% | 39.49% | 38.45% |
BioBERT+SQuAD+BioASQ Finetune | 42.86% | 57.14% | 48.41% | 51.58% | 43.24% | 43.16% |
Performance comparison between pre-processing methods. Scores on the BioASQ 6b dataset.
Strategy | Factoid | List | Yes/no | ||||
|---|---|---|---|---|---|---|---|
SAcc | LAcc | MRR | Prec | Recall | F1 | MacroF1 | |
Snippet | 40.99 | 55.90 | 47.38 | 51.58 | 43.24 | 43.16 | 75.10 |
Full Abstract | 42.86 | 57.14 | 48.41 | 42.66 | 32.58 | 33.52 | 66.76 |
Appended Snippet | 39.75 | 58.39 | 48.00 | 44.04 | 41.26 | 39.36 | – |
Ablation study on the post-processing methods. Scores for list type questions in the BioASQ 6b dataset.
Strategy | Precision | Recall | F1 |
|---|---|---|---|
Baseline (Snippet) | 51.58 | 43.24 | 43.16 |
Baseline without filter | 50.79 | 43.24 | 42.64 |
Baseline without answer # extraction | 50.01 | 44.32 | 42.58 |
Predictions by our BioBERT based QA system on the BioASQ 6b factoid dataset
No | Type | Description |
|---|---|---|
1 | Question | What causes “puffy hand syndrome?” |
Passage | Puffy hand syndrome is a complication of intravenous drug abuse, which has no current available treatment | |
Ground Truth | “intravenous drug abuse" | |
Predicted Answer | “intravenous drug abuse", | |
“drug addiction", | ||
“Intravenous drug addiction", | ||
“staphylococcal skin infection", | ||
“major depression" | ||
2 | Question | In which syndrome is the RPS19 gene most frequently mutated? |
Passage | A transgenic mouse model demonstrates a dominant negative effect of a point mutation in the RPS19 gene associated with Diamond-Blackfan anemia | |
Ground Truth | “Diamond-Blackfan Anemia", | |
“DBA" | ||
Predicted Answer | “Diamond-Blackfan anemia", | |
“Diamond-Blackfan anemia (DBA)", | ||
“DBA", | ||
“Diamond Blackfan anemia", | ||
“Diamond-Blackfan anemia. Diamond-Blackfan anemia" | ||
3 | Question | What protein is the most common cause of hereditary renal amyloidosis? |
Passage | We suspected amyloidosis with fibrinogen A alpha chain deposits, which is the most frequent cause of hereditary amyloidosis in Europe, with a glomerular preferential affectation | |
Ground Truth | “Fibrinogen A Alpha protein" | |
Predicted Answer | “fibrinogen", | |
“fibrinogen alpha-chain. Variants of circulating fibrinogen", | ||
“fibrinogen A alpha chain (FGA)", | ||
“Fibrinogen A Alpha Chain Protein. Introduction: Fibrinogen", | ||
“apolipoprotein AI" |
Qualitative Analysis. In Table 7, we show three predictions generated by our system on the BioASQ 6b factoid dataset. Due to the space limitation, we show only small parts of a passage, which contain the answers (predicted answers might be contained in other parts of the passage). We show the top five predictions generated by our system which can also be used for list type questions. In the first example, our system successfully finds the answer and other plausible answers. The second example shows that most of the predicted answers are correct and have only minor differences. In the last example, we observe that the ground truth answer does not exist in the passage. Also, the predicted answers are indeed correct despite the incorrect annotation.
Prediction by our BioBERT based QA system on the BioASQ 6b list dataset
No | Type | Description |
|---|---|---|
1 | Question | Which enzymes are responsible for base J creation in Trypanosoma brucei? |
Passage | JBP1 and JBP2 are two distinct thymidine hydroxylases involved in J biosynthesis in genomic DNA of African trypanosomes | |
Here we discuss the regulation of hmU and base J formation in the trypanosome genome by JGT and base J-binding protein | ||
Ground Truth | “JBP1”, | |
“JBP2”, | ||
“JGT” | ||
Predicted Answer | “JBP1", | |
“JBP", | ||
“thymidine hydroxylase", | ||
“JGT", | ||
“hmU", | ||
“JBP2" |
5 Conclusion
In this paper, we proposed BioBERT based QA system for the BioASQ biomedical question answering challenge. As the size of the biomedical question answering dataset is very small, we leveraged pre-trained language models for biomedical domain which effectively exploit the knowledge from large biomedical corpora. Also, while existing systems for the BioASQ challenge require different structures for different question types, our system uses almost the same structure for various question types. By exploring various pre-/post-processing strategies, our BioBERT based system obtained the best performance in the 7th BioASQ Challenge, achieving state-of-the-art results on factoid, list, and yes/no type questions. In future work, we plan to further systematically analyze the incorrect predictions of our systems, and develop biomedical QA systems that can eventually outperform humans.
We appreciate Susan Kim for editing the manuscript. This work was funded by the National Research Foundation of Korea (NRF-2017R1A2A1A17069645, NRF-2016M3A9A7916996) and the National IT Industry Promotion Agency grant funded by the Ministry of Science and ICT and Ministry of Health and Welfare (NO. C1202-18-1001, Development Project of The Precision Medicine Hospital Information System (P-HIS)).