___________________________________
It did not begin with Neanderthals, but with ancient Egyptian mummies. Ever since my mother took me to Egypt when I was thirteen, I had been fascinated with its ancient history. But when I started to pursue this study in earnest, at the University of Uppsala in my native Sweden, it became increasingly clear that my fascination with Pharaohs, pyramids, and mummies was the romantic dream of an adolescent. I did my homework; I memorized the hieroglyphs and the historical facts; I even worked two consecutive summers cataloging pottery shards and other artifacts at the Mediterranean Museum in Stockholm, which might well have become my future workplace, were I to become an Egyptologist in Sweden. I found that the same people did very much the same things the second summer as they had the first summer. Moreover, they went to lunch at the same time, to the same restaurant, ordered the same meals, discussed the same Egyptological puzzles and academic gossip. In essence, I came to realize that the discipline of Egyptology was moving too slowly for my tastes. It was not the kind of professional life I imagined for myself. I wanted more excitement, and more relevance to the world I saw around me.
This disenchantment threw me into a crisis of sorts. In response, and inspired by my father, who had been an MD and later became a biochemist, I decided to study medicine, with a view to doing basic research. I entered medical school at the University of Uppsala and after a few years surprised myself by how much I enjoyed seeing patients. It seemed to be one of the few professions in which you not only met all sorts of people but could also play a positive role in their lives. This ability to engage with people was an unexpected talent, and after four years of medical studies I had another mini-crisis: Should I become a clinician or move, as I had originally meant to do, into basic research? I opted for the latter, thinking that I could—and most likely would—come back to the hospital after my PhD. I joined the lab of one of the then-hottest scientists in Uppsala, Per Pettersson. Not long before, his group had been the first to clone the genetic sequence of an important class of transplantation antigens, protein molecules that sit on the surface of immune cells and mediate their recognition of viral and bacterial proteins. Not only had Pettersson produced exciting biology insights with relevance to clinical practice, but his lab was one of the few in Uppsala that had mastered the then-novel methods of cloning and manipulating DNA by introducing it into bacteria.
Pettersson asked me to join his group’s efforts to study a protein encoded by an adenovirus, a virus that causes diarrhea, cold-like symptoms, and other unpleasant features of our lives. It was thought that this viral protein became bound by the transplantation antigens inside the cell, so that, once transported to the cell surface, it could be recognized by immune-system cells, which would then become active and kill other infected cells in the body. Over the next three years, I and the others working on this protein came to realize that this idea of what the protein did was utterly wrong. We found that rather than becoming a hapless target of the immune system, the viral protein seeks out the transplantation antigens inside the cell, binds to them, and blocks their transport out to the cell surface. Since the infected cell thus ends up having no transplantation antigens on its surface, the immune system cannot recognize that it is infected. This protein camouflages the adenovirus, so to speak. In fact, it leads to the creation of a cell within which the adenovirus can probably survive for a long time, perhaps even as long as the infected person lives. That viruses could foil the immune system of their hosts in this way was a revelation, and our work resulted in a number of high-profile papers in the best journals. Indeed, it turns out that other viruses, too, use similar mechanisms to evade the immune system.
This was my first taste of cutting-edge science, and it was fascinating. It was also the first (but not the last) time I saw that progress in science often entails a painful process of realizing that your ideas and those of your peers are wrong, and an even longer struggle to persuade your closest associates and then the world at large to consider a new idea.
But somehow, in the midst of all the biological excitement, I could not quite shake off my romantic fascination with ancient Egypt. Whenever I had time, I went to lectures at the Institute of Egyptology, and I continued to take classes in Coptic, the language of pharaonic Egypt as spoken during the Christian era. I befriended Rostislav Holthoer, a jovial Finnish Egyptologist with an immense capacity for friendships across social, political, and cultural boundaries. During long dinners and evenings at Rosti’s home in Uppsala in the late 1970s and early ’80s, I often complained that I loved Egyptology but saw little future in it, while I also loved molecular biology, with its apparently boundless promise of advances in the welfare of humankind. I was torn between two equally alluring career paths—a conundrum no less painful because it was doubtless viewed without much sympathy as the fretting of a young man faced with nothing but good choices.
But Rosti was patient with me. He listened when I explained how scientists could now take DNA from any organism (be it a fungus, a virus, a plant, an animal, or a human), join it to a plasmid (a carrier molecule made of DNA from a bacterial virus), and introduce the plasmid into bacteria, where it would replicate along with its host, making hundreds or thousands of copies of the foreign DNA. I explained how we could then determine the sequence of the foreign DNA’s four nucleotides and find differences in the sequences between the DNAs of two individuals or two species. The more similar two sequences were—that is, the fewer the number of differences between them—the more closely related they were. In fact, from the number of shared mutations we could infer not only how the particular sequences had evolved from common ancestral DNA sequences over thousands and millions of years but also approximately when those ancestral DNA sequences had existed. For example, in a 1981 study the British molecular biologist Alec Jeffreys analyzed the DNA sequence of a gene that encodes a protein in the red pigment in the blood of both humans and apes and deduced when the genes began evolving independently in humans and apes. This, I explained, could soon be done for many genes, from many individuals of any species. In this way, scientists would be able to determine how different species were related to one another in the past, as well as when they began their separate histories, with much greater accuracy than was possible from the study of morphology or fossils.
As I explained all this to Rosti, a question gradually arose in my mind. Would this kind of investigation necessarily be restricted to DNA from blood samples or tissues from humans and animals that live today? What about those Egyptian mummies? Could DNA molecules have survived in them—and could they, too, be joined to plasmids and made to replicate in bacteria? Could it be possible to study ancient DNA sequences and thereby clarify how ancient Egyptians were related to one another and to people today? If that could be done, then we could answer questions that no one could answer by the conventional means of Egyptology. For example, how are present-day Egyptians related to Egyptians who lived when the Pharaohs ruled, some 2,000 to 5,000 years ago? Did great political and cultural changes, such as the conquest by Alexander the Great in the fourth century BCE, or by the Arabs in the seventh century AD, result in replacement of a large part of the Egyptian population? Alternatively, were these just military and political events that caused the native population to adopt new languages, new religions, and new ways of life? In essence, were the people who lived in Egypt today the same as those who built the pyramids, or had their ancestors mixed so much with invaders that modern Egyptians were now completely different from their country’s ancient population? Such questions were breathtaking. Surely they must have already occurred to someone else.
I went to the university library and searched in journals and books but found no report of any isolation of DNA in ancient materials. No one seemed even to have tried to isolate ancient DNA. Or if they had, they had not succeeded, because if so, surely they would have published their findings. I talked to the more experienced graduate students and postdocs in Pettersson’s lab. Given how sensitive DNA is, they argued, why would you expect it to last for thousands of years? The conversations were discouraging, but I didn’t give up hope. In my forays into the literature, I had found articles whose authors claimed to have detected proteins in hundred-year-old animal hides in museums—proteins that could still be detected by antibodies. I had also found studies claiming to have detected, under the microscope, the outlines of cells in ancient Egyptian mummies. So something did seem to survive, at least sometimes. I decided to do a few experiments.
The first question seemed to be whether DNA could survive for long in tissues after death. I speculated that if the tissue became desiccated, as was the case when a mummy was prepared by the embalmers in ancient Egypt, then DNA might well survive for a long period since the enzymes that degrade DNA need water to be active. This would be the first thing to test. So in the summer of 1981, when not too many people were around in the lab, I went to the supermarket and bought a piece of calf liver. I glued the receipt from the store onto the first page of a new lab book that I would use to record these experiments. I labeled the book with my name but nothing else, since I had decided to keep my experiments as secret as possible. Pettersson might forbid me to pursue them, if they struck him as an unnecessary distraction from the intensely competitive study of the molecular workings of the immune system that I was supposed to be working on. And, in any case, I wanted to keep all this under wraps to spare myself the ridicule of my lab colleagues in the likely event of failure.
To somewhat imitate ancient Egyptian mummification, I decided to artificially mummify the calf liver by sequestering it in an oven in the lab heated to 50°C. The first effect of this was that the secrecy of my project was compromised. By the second day, the repugnant smell elicited considerable comment, and I had to reveal my project before someone found the liver and disposed of it. Fortunately, the smell decreased as the desiccation progressed, and neither the smell nor word of what was putrefying in the lab made it to my professor.
After a few days, the liver had become hard, blackish-brown, and dry—just like an Egyptian mummy. I proceeded to extract DNA from it, with immediate success. The DNA was in small pieces of a few hundred nucleotide pairs instead of the many thousands of nucleotide pairs typical of DNA extracted from fresh tissue, but there was still lots of it. I felt vindicated. It was not totally ridiculous to think that DNA could survive in a dead tissue—at least for some days or weeks. But what about thousands of years? The obvious next step was to try performing the same stunt with an Egyptian mummy. Now my friendship with Rosti came in handy.
Rosti had been primed by my fretting about Egyptology and molecular biology and was happy to abet my attempt to take Egyptology into the molecular age. The small university museum of which he was the curator had some mummies, and he consented to my request to sample them. He was, of course, not about to let me cut them open and remove their livers. But if a mummy was already unwrapped and its limbs had broken off, Rosti allowed me to remove small pieces of skin or muscle tissue from the area where the mummy had already been broken to try my DNA extraction. Three such mummies were available. As soon as I put the scalpel to what had once been the skin and muscles of a person who existed some 3,000 years ago, I realized that the texture of the tissue was different from that of the calf liver I had baked in the oven. The liver had been hard and a bit tough to slice up, whereas the mummies were brittle and their tissues tended to crumble to brown powder when cut. Undeterred, I submitted them to the same extraction procedure I had performed on the liver. The mummy extracts differed from the liver extract in that they were as brown as the mummies themselves, whereas the liver extract had been as clear as water. And when I looked for DNA in the mummy extracts by letting them migrate in a gel in an electric field and staining them with a dye that would fluoresce pink in ultraviolet light if it had bound to DNA, I saw nothing except the brown stuff, which indeed fluoresced in the ultraviolet light, but blue instead of pink, not what was expected if it was DNA. I repeated this process on the two other mummies. Again, there was no DNA; nothing but an undetermined brown substance had ended up in the extracts that I had hoped would contain DNA. My lab colleagues seemed to be right: How could the fragile DNA molecules survive for thousands of years, when even inside a cell it needed constant repair in order not to decompose?
I hid my secret lab book at the bottom of my desk drawer and returned to the virus that tricked the immune system with its clever little protein—but I could not get the mummies out of my mind. How could it be that others had seen what seemed to be remains of cells in some mummies? Perhaps that brown stuff was actually DNA, chemically modified in some way so as to look brown and fluoresce blue in UV light. Perhaps it was naïve to expect DNA to survive in every mummy. Perhaps one needed to analyze many mummies to find the rare good ones. The only way to find out was to convince museum curators to sacrifice pieces of many mummies in the perhaps vain hope that one of them would produce ancient DNA, and I had little idea how to get their permission. It seemed I needed a quick and minimally destructive way to analyze a lot of mummies. My medical education gave me a clue. Very small pieces of tissue, such as those removed with a biopsy needle from a suspected tumor, for example, could be fixed and stained and then studied under a microscope. The level of discernible detail was generally exquisite, allowing a trained pathologist to distinguish normal cells in the lining of the intestine or in a prostate or mammary gland, on the one hand, from cells that had started to change in ways that suggested they were early tumors, on the other. Moreover, there were dyes specific for DNA that could be applied to microscope slides to show whether DNA was present. What I needed to do was to collect small samples from a large number of mummies and analyze them by microscopy and DNA staining. The largest numbers of mummies, obviously, were to be found in the largest museums. But the curators could be expected to be skeptical about letting a slightly overexcited student from Sweden remove even tiny pieces for what seemed a pie-in-the-sky project.
Again, Rosti proved sympathetic; he pointed out that there was one large museum that had huge mummy collections and might be willing to cooperate. It was the Staatliche Museen zu Berlin, a complex of museums in East Berlin, the capital of the German Democratic Republic. Rosti had spent many weeks there working on its ancient Egyptian pottery collection. That Rosti came to East Germany as a professor from Sweden, which at the time was perceived as a country that attempted to find a “third way” between capitalism and communism, probably helped him gain permission to work in the museum. But it was his ability to develop warm friendships across borders that then allowed him to become close friends with several of the curators at the museum. And thus, in the summer of 1983, I found myself on a train that was driven onto a ferry in southern Sweden to arrive the next morning in communist East Germany.
I spent two weeks in Berlin. Every morning I had to pass several security controls to enter the storage facility of the Bode Museum, located on an island in the River Spree near the heart of Berlin. Almost forty years after the war, the museum was still clearly marked by it. On several of the facades, I could see bullet holes in the walls around the windows that had been targeted by machine guns as Berlin fell to the Soviet Army. On the first day, when I was taken to see the prewar Egyptological exhibition, I was handed a hard hat like the ones used by construction workers. It soon became clear why. The exhibition hall had huge holes in the roof from artillery shelling and bombs. Birds were flying in and out, and some were nesting in the pharaonic sarcophagi. Everything that was not of durable material was now sensibly stored elsewhere.
Over the following days, the curator in charge of Egyptian antiquities showed me all his mummies. For a few hours before lunch in his dusty run-down office I removed small snippets of tissues from mummies that were unwrapped and broken. Lunch was a long affair that required exiting through all the security checks to reach a restaurant across the river, where we ate greasy food that needed lubrication with copious amounts of beer and schnapps. Back in the collections, we spent the afternoon over more schnapps, lamenting the fact that the only foreign travel the curator had ever been allowed was visits to Leningrad. It soon became clear that my host dreamed of visiting the capitalist West and that if he got the chance he would probably defect. To provide some perspective on working life in the West I suggested, as diplomatically as I could, that in the west if you drank on the job, you were likely to be fired—an unknown concept in socialism. Such sobering thoughts seemed not to detract from the allure of the opportunities my host imagined to abound in capitalism. In spite of the hours spent on these theoretical discussions, I managed to collect more than thirty mummy samples to take back to Sweden.
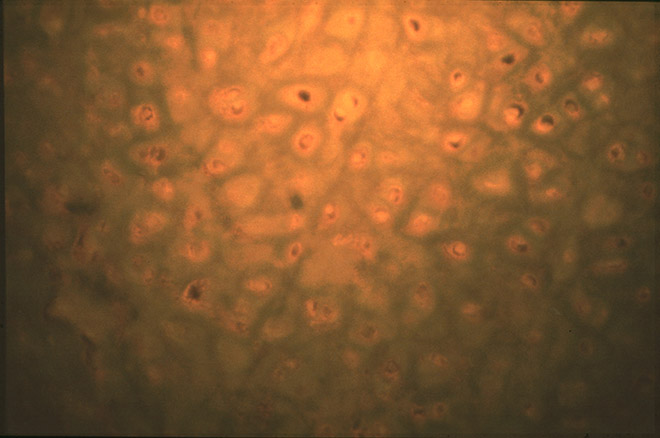
At Uppsala, I prepared the samples for microscopy by soaking them in a salt solution to rehydrate them, then mounting them on glass slides and staining them with dyes that permitted visualization of cells. Then I looked for preserved cells in the tissues. I did this work on weekends and late at night, so as not to let it be widely known what I was doing. As I peered through the microscope, the appearance of the ancient tissues depressed me. In muscle sections, I could barely discern the fibers, let alone any traces of cell nuclei where DNA might be preserved. I was almost despairing, until one night I looked at a section of cartilage from a mummified outer ear. In cartilage, as in bone, cells live in small holes, or lacunae, inside the compact, hard tissue. When I looked at the cartilage, I saw what appeared to be the remains of cells inside the lacunae. Excited, I stained the section for DNA. My hands were trembling as I put the slide under the microscope. Indeed, there was staining within the cellular remains in the cartilage (see Figure 2.1). There seemed to be DNA preserved inside!
With renewed energy, I went on to process all of the remaining samples from Berlin. A few looked promising. In particular, the skin from the left leg of the mummy of a child showed what were clearly cell nuclei. When I stained a section of the skin for DNA, the cell nuclei lit up. Since this DNA was in the cell nuclei, where the cellular DNA is stored, it could not possibly be from bacteria or fungi because such DNA would appear at random in the tissue where the bacteria or fungi were growing. This was unambiguous evidence that DNA from the child herself was preserved. I took many photos through the microscope.

Figure 2.1. Microscopic picture of cartilage tissue from an Egyptian mummy from Berlin. In some lacunae, cell remains light up suggesting that DNA may be preserved. Photo: S. Pääbo, Uppsala University.
I found three mummies with staining of the cell nuclei showing the presence of DNA. The child seemed to have the largest number of well-preserved cells. But now doubt started to gnaw at me. How could I be sure that this mummy was really old? Modern corpses were sometimes falsified to look like ancient Egyptian mummies so that the perpetrators might earn a few dollars from tourists and collectors. Some of these mummies might later be donated to museums. The staff of the museum in Berlin had been unable to give me any records of the provenance of this particular mummy, perhaps because the relevant parts of the catalog had been destroyed in the war. The question of its age could be resolved only through carbon dating. Fortunately, Göran Possnert, an expert on carbon dating, worked at Uppsala University. He used an accelerator to determine the ages of tiny samples of ancient remains by measuring the ratios of carbon isotopes present. I asked him how much it would cost to date my mummy, worrying that I would not be able to afford it on my meager student stipend. He took pity on me and offered to date it for free, considerately not even mentioning the price, doubtless because it would have been well out of my range. I delivered a small piece of the mummy to Göran and waited to hear the results. For me, this exemplified one of the most frustrating situations in science, when your work depends crucially on the work of someone else and you can do nothing to expedite it—just wait for a phone call that seems to never come. But finally, a few weeks later, I got the call I had been waiting for. The news was good. The mummy was 2,400 years old; it dated from about the time of the Alexandrian conquest of Egypt. I drew a sigh of relief. First I went out and bought a big box of chocolates, which I delivered to Göran. Then I started to think about publishing my findings.
During my time in East Germany, I had developed some understanding of the sensitivities of people living under socialism. In particular, I knew that the museum curator and other museum officials who hosted my visit would be very disappointed with just a perfunctory expression of gratitude at the end of my paper. I wanted to do the right thing, so after talking to Rosti and conferring with Stephan Grunert, a young and ambitious East German Egyptologist whom I had befriended in Berlin, I decided to publish my first paper on mummy DNA in an East German scientific journal. Struggling with my high school German, I wrote up my findings, including photographs of the mummy itself and of the tissue stained for DNA. In the meantime, I had also extracted DNA from the mummy. This time, the extracts contained DNA that I could demonstrate in a gel, and I included a picture of such an experiment in the paper. Most of the DNA was degraded, but a small fraction of it was several thousand nucleotides long, similar in length to the DNA one could extract from fresh blood samples.
This, I wrote, seemed to indicate that some DNA molecules from ancient tissues might well be large enough to allow the study of individual genes. I speculated wildly about what might be possible if DNA from ancient Egyptian mummies could be systematically studied. The paper ended on a hopeful note: “Work over the next few years will show if these expectations will be fulfilled.” I sent the manuscript to Stephan in Berlin. He fixed up my German, and in 1984 the article appeared in Das Altertum, a journal published by the East German Academy of Sciences.{3} And nothing happened. Not a single person wrote to me about it, much less asked for a reprint. I was excited, but no one else seemed to be.
Having realized that the world at large did not make a habit of reading East German publications, I had written up similar results from the fragment of the mummified head of a man and, in October of the same year, had sent them to a Western journal that seemed appropriate—the Journal of Archaeological Science. But here the frustration turned out to be the unbelievable slowness of the journal, even compared with the delay my manuscript had experienced in East Germany, where it needed to be fixed up linguistically by Stephan and then presumably scrutinized by the political censors. This was, I felt, a reflection of the glacial speed with which the disciplines concerned with ancient things were moving. The Journal of Archaeological Science finally published my paper at the end of 1985{4}—by which time the results it described had been largely overtaken by events.
The next step—now that I had some mummy DNA—was obvious. I needed to clone it in bacteria. So I treated it with enzymes that make the ends of the DNA amenable to being joined to other pieces of DNA, mixed it with a bacterial plasmid, and added an enzyme that joins DNA fragments together. If successful, this would create hybrid molecules in which pieces of DNA from the mummy were joined to the plasmid DNA. When these plasmids were introduced into bacteria, they would not only allow the hybrid molecules to replicate to high copy numbers in bacterial cells but would also make the bacteria resistant to an antibiotic I would add to my culture medium, so that the bacteria would survive only if they contained a functioning plasmid. When seeded on growth plates containing the antibiotic, colonies of bacteria would appear if the experiment was successful. Each such colony would derive from a single bacterium that now carried one particular piece of mummy DNA. To check on my experiment, I did controls—an essential thing in any laboratory experiment. For example, I repeated the exact process in parallel but added no mummy DNA to the plasmid, and also repeated the process but added modern human DNA. After making the bacteria take up the DNA solutions from these experiments, I plated them on agar plates containing the antibiotic and put them in an incubator at 37°C overnight. The next morning I opened the incubator and, with anticipation, inhaled the puff of moist air smelling of rich culture media. The plate with the modern DNA yielded thousands of colonies, so many that it was almost totally covered with bacteria. This showed that my plasmid had worked: the bacteria survived because they had taken up the plasmid. The plate where no DNA had been added to the plasmid yielded hardly any colonies, indicating that I did not have DNA from some unknown source in my experiment. The experiment itself, where I had added the DNA from the Berlin mummy, yielded several hundreds of colonies. I was ecstatic. I had apparently replicated 2,400-year-old DNA! But could it have come from bacteria in the child’s tissues, rather than from the child herself? How could I show that at least some of the DNA I had cloned in the bacteria was human?
I needed to determine the DNA sequence from some of the DNA in order to show that it was human rather than bacterial. But if I merely sequenced random clones, they would be likely to contain DNA sequences that could have come either from the human genome—which in 1984 was not yet decoded, except for some tiny parts that had been sequenced with great effort—or from some microorganism whose DNA sequences were even less likely to be known. So instead of sequencing random clones, I needed to identify some clone of interest. The answer lay in a technique whereby one could identify clones that carried DNA similar in sequence to something one wanted to find. This technique involved transferring some of the bacteria from each of hundreds of colonies to cellulose filters, where the bacteria were broken open and their DNA were bound to the filter. I then used a radioactively labeled piece of DNA, a “probe,” that was single-stranded and then hybridized to complementary sequences from the single-stranded DNA on the filters. I chose to use a piece of DNA that contains a repeated DNA element—the so-called Alu element—of about 300 nucleotides that occurs almost a million times in the human genome and in no organisms besides humans, apes, and monkeys. In fact, these Alu elements are so numerous that more than 10 percent of the human genome is made up of them. If I could find an Alu element among my clones, it would show that at least some of the DNA I had extracted from the mummy came from a human being.
I got a piece of a gene I’d studied in the lab that contained an Alu element, incorporated radioactivity in it, and hybridized it to my filters. Several of the clones took up the radioactivity, as one would expect if some of the DNA was human. I picked the most strongly hybridizing clone. It contained a piece of DNA consisting of about 3,400 nucleotides. With the help of Dan Larhammar, a graduate student who was the master of DNA sequencing in our group, I sequenced a part of the clone. It did indeed contain an Alu element. I was very happy. There was human DNA among my clones, and it could be cloned in bacteria.
As I was grappling with my sequencing gels in November 1984, a paper appeared in Nature that was of great relevance for me. Russell Higuchi, who worked at UC Berkeley with Allan Wilson, the primary architect of the out-of-Africa theory of modern human origins and one of the most famous evolutionary biologists of the time, had extracted and cloned DNA from the 100-year-old skin of a quagga, an extinct subspecies of zebra that had existed in southern Africa until about a hundred years ago. Russell Higuchi had isolated two fragments of mitochondrial DNA and shown that the quagga was, as expected, more closely related to zebras than to horses. This work inspired me greatly. If Allan Wilson was studying ancient DNA, and if Nature considered an article about 120-year-old DNA interesting enough to publish, then surely what I was doing was neither crazy nor uninteresting.
For the first time, I sat down to write a paper of my own that I believed many people in the world would be interested in. Inspired by Allan Wilson’s example, I wrote it for Nature. I described what I had done with the mummy from Berlin. One of my first references was to the paper that had appeared in the East German journal. However, before I sent the manuscript off to London, where Nature had its office, there was something I needed to do. I needed to talk to my thesis adviser, Per Pettersson, and show him the manuscript, now ready to submit. With some trepidation, I entered his office and told him what I had done. I asked if he might perhaps want to be a co-author with me on the paper, in his capacity as my adviser. Obviously, I had underestimated the man. Rather than scolding me for what could have been seen as misappropriation of research funds and valuable time, he seemed amused. He promised to read the manuscript and said that, no, obviously he should not be the co-author of work that he hadn’t even been aware of.
A few weeks later, I received a letter from Nature, with a promise from the editor to publish my manuscript if I could respond to some minor comments from reviewers. Shortly thereafter, the proofs arrived. At that point, I thought about how to approach Allan Wilson—a demigod, in my view—to ask if I might work with him at Berkeley after my PhD defense. Not knowing exactly how to broach this topic, I mailed him a copy of the proofs without any comment whatsoever, thinking he might appreciate seeing the paper before it appeared. I thought that I would then later write to him about job opportunities in his laboratory. Nature progressed rapidly toward publication and even solicited a cover illustration of a mummy with DNA sequences artfully wrapped around it. Even more rapidly, I received a response from Allan Wilson, who addressed me as “Professor Pääbo”—this was before both the Internet and Google, so there was no obvious way for him to find out who I was. The rest of his letter was even more amazing. He asked if he could spend his upcoming sabbatical year in “my” laboratory! This was a hilarious misunderstanding, resulting from my insecurity about knowing what to write to him. I joked with my lab mates that I would have Allan Wilson, perhaps the most famous molecular evolutionist of the time, wash gel plates for me for a year. Then I settled down to write him back—explaining that I was not a professor, not even a PhD, and certainly did not have a lab where he could spend his sabbatical. Rather, I wondered if there might be a chance for me to spend my postdoc in his Berkeley lab.