Chapter 4
The chemistry of life
It was once believed that organic compounds were unique to life and could not be synthesized in the laboratory. We now know that this is not the case, but it is clear that organic molecules are fundamental to the existence of life on this planet. Chapter 3 described how organic chemists synthesize organic molecules from smaller molecular building blocks. This might seem very clever, but nature has been doing the same thing for much longer, and does it far more efficiently. From very simple molecular building blocks, life has created an astonishing diversity of molecules, some of which are extremely complex structures that prove very difficult to synthesize in a laboratory. Not only does life produce complex molecules, it does so under mild conditions in an aqueous environment.
Amino acids and proteins
Proteins are large molecules (macromolecules) which serve a myriad of purposes, and are essentially polymers constructed from molecular building blocks called amino acids (Figure 40). In humans, there are twenty different amino acids having the same ‘head group’, consisting of a carboxylic acid and an amine attached to the same carbon atom (Figure 41). The simplest amino acid is glycine which has a hydrogen atom in place of a side chain, but all the other amino acids have some form of side chain present.
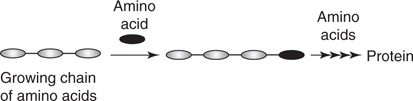
40. Biosynthesis of proteins formed by linking amino acids one at a time.
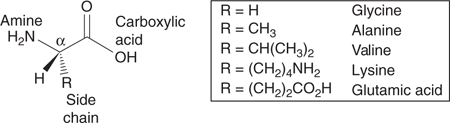
41. Structures of selected α-amino acids.
The amino acids are linked up by the carboxylic acid of one amino acid reacting with the amine group of another to form an amide link. Since a protein is being produced, the amide bond is called a peptide bond, and the final protein consists of a polypeptide chain (or backbone) with different side chains ‘hanging off’ the chain (Figure 42). The sequence of amino acids present in the polypeptide sequence is known as the primary structure. Once formed, a protein folds into a specific 3D shape, which is determined by the intramolecular interactions occurring between different side chains and peptide bonds, as well as intermolecular hydrogen bonding to surrounding water molecules. Life’s method of synthesizing proteins can be mimicked in the laboratory. For example, HIV protease is a viral enzyme that has been synthesized in the laboratory.
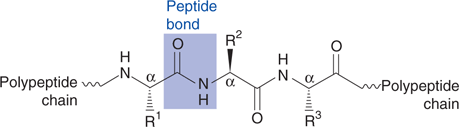
42. The polypeptide backbone of a protein with substituents  etc.) attached to each α-carbon.
etc.) attached to each α-carbon.
Nucleotides and nucleic acids
Nucleic acids (Figure 43) are another form of biopolymer, and are formed from molecular building blocks called nucleotides. These link up to form a polymer chain where the backbone consists of alternating sugar and phosphate groups. There are two forms of nucleic acid—deoxyribonucleic acid (DNA) and ribonucleic acid (RNA). In DNA, the sugar is deoxyribose 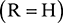 , whereas the sugar in RNA is ribose
, whereas the sugar in RNA is ribose 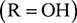 . Each sugar ring has a nucleic acid base attached to it. For DNA, there are four different nucleic acid bases called adenine (A), thymine (T), cytosine (C), and guanine (G) (Figure 44). These bases play a crucial role in the overall structure and function of nucleic acids.
. Each sugar ring has a nucleic acid base attached to it. For DNA, there are four different nucleic acid bases called adenine (A), thymine (T), cytosine (C), and guanine (G) (Figure 44). These bases play a crucial role in the overall structure and function of nucleic acids.
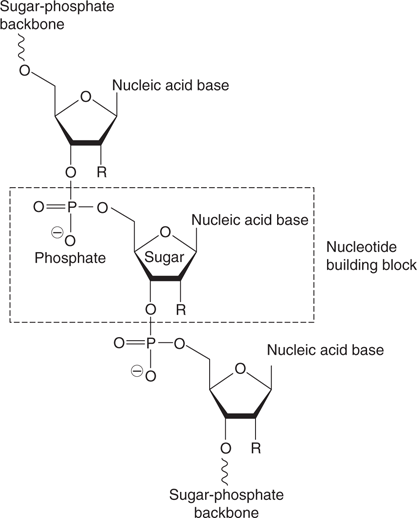
43. General structure of nucleic acids (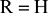 or OH).
or OH).
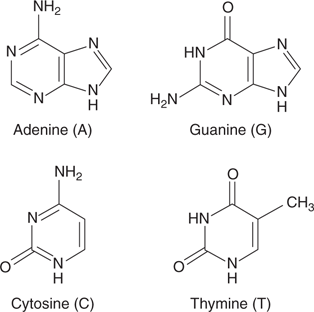
44. The nucleic acid bases present in DNA.
DNA is actually made up of two DNA strands (Figure 45) where the sugar-phosphate backbones are intertwined to form a double helix. The nucleic acid bases point into the centre of the helix, and each nucleic acid base ‘pairs up’ with a nucleic acid base on the opposite strand through hydrogen bonding. The base pairing is specifically between adenine and thymine, or between cytosine and guanine. This means that one polymer strand is complementary to the other, a feature that is crucial to DNA’s function as the storage molecule for genetic information.
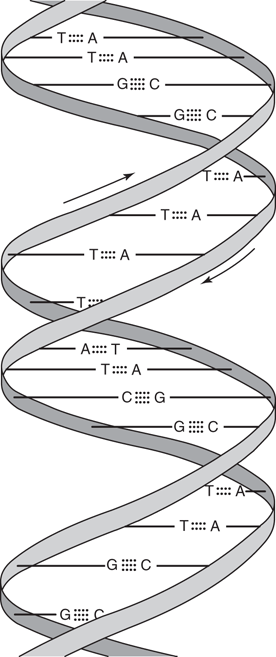
45. The double helical structure of DNA.
Other biosynthetic processes
Polymerization processes are involved in the biosynthesis of many other natural products. For example, fatty acids are formed from 2-C building blocks. In the case of cyclic compounds, a polymerization process creates a linear polymer chain, which then undergoes cyclization reactions. For example, the biosynthesis of steroids creates a polymer chain made up of 5-C building blocks, which is cyclized to form a tetracyclic structure (Figure 46). The same general strategy is used in different organisms. For example, penicillin G is a fungal metabolite that is created from linking up two amino acids and one fatty acid, then carrying out cyclization reactions (Figure 47).
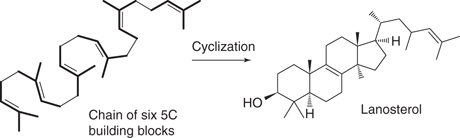
46. The process involved in steroid biosynthesis.
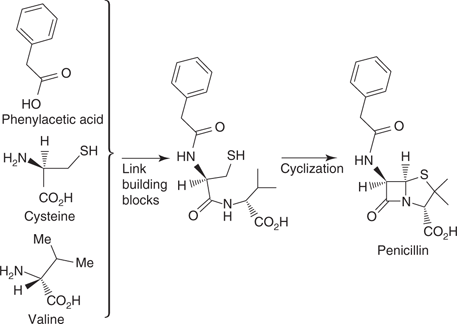
47. The general biosynthetic process for penicillin G.
Function of proteins
Proteins have a variety of functions. Some proteins, such as collagen, keratin, and elastin, have a structural role. Others catalyse life’s chemical reactions and are called enzymes. They have a complex 3D shape, which includes a cavity called the active site (Figure 48). This is where the enzyme binds the molecules (substrates) that undergo the enzyme-catalysed reaction. The resulting product is then released.
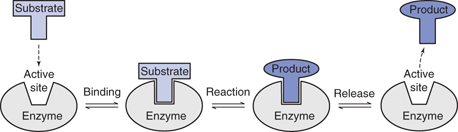
48. The overall process involved in an enzyme-catalysed reaction.
A substrate has to have the correct shape to fit an enzyme’s active site, but it also needs binding groups to interact with that site (Figure 49). These interactions hold the substrate in the active site long enough for a reaction to occur, and typically involve hydrogen bonds, as well as van der Waals and ionic interactions. When a substrate binds, the enzyme normally undergoes an induced fit. In other words, the shape of the active site changes slightly to accommodate the substrate, and to hold it as tightly as possible.
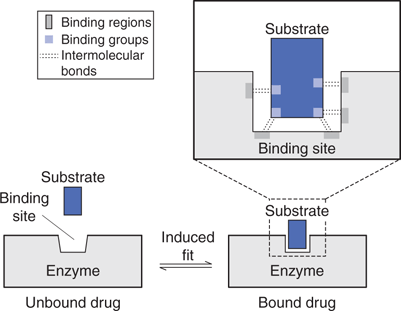
49. The process by which a substrate is recognized by an enzyme’s active site.
Once a substrate is bound to the active site, amino acids in the active site catalyse the subsequent reaction. One example of an enzyme-catalysed reaction is the hydrolysis of a neurotransmitter called acetylcholine (Figure 50). In water, this reaction is very slow, but it is several million times faster if the enzyme is present.
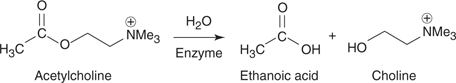
50. The enzyme-catalysed hydrolysis of acetylcholine.
Proteins called receptors are involved in chemical communication between cells and respond to chemical messengers called neurotransmitters if they are released from nerves, or hormones if they are released by glands. Most receptors are embedded in the cell membrane, with part of their structure exposed on the outer surface of the cell membrane, and another part exposed on the inner surface. On the outer surface they contain a binding site that binds the molecular messenger. An induced fit then takes place that activates the receptor. This is very similar to what happens when a substrate binds to an enzyme (Figure 49). However, the receptor has no catalytic activity. The molecular messenger stays bound for a certain period of time, then departs unchanged. Once that happens, the protein receptor returns to its inactive state.
The induced fit is crucial to the mechanism by which a receptor conveys a message into the cell—a process known as signal transduction. By changing shape, the protein initiates a series of molecular events that influences the internal chemistry within the cell. For example, some receptors are part of multiprotein complexes called ion channels. When the receptor changes shape, it causes the overall ion channel to change shape. This opens up a central pore allowing ions to flow across the cell membrane. The ion concentration within the cell is altered, and that affects chemical reactions within the cell, which ultimately lead to observable results such as muscle contraction. Not all receptors are membrane-bound. For example, steroid receptors are located within the cell. This means that steroid hormones need to cross the cell membrane in order to reach their target receptors.
Transport proteins are also embedded in cell membranes and are responsible for transporting polar molecules such as amino acids into the cell. They are also important in controlling nerve action since they allow nerves to capture released neurotransmitters, such that they have a limited period of action. Transport proteins contain an extracellular binding site that binds the target molecule. Once bound, the molecule is passed through the protein and released into the cell.
Function of nucleic acids
DNA is the molecule responsible for storing genetic information, and transmitting it from one generation to another. However, for many years, DNA was considered an unlikely candidate for these roles. The backbone was a regular sequence of sugar and phosphate groups, and there were only four different types of nucleic acid base present, which appeared to be randomly arranged along the chain. However, attitudes changed dramatically when it was discovered that DNA was a double helix held together by interactions between specific base pairs. This meant that the two chains were complementary and demonstrated how DNA could pass on genetic information from one generation to another. By unwinding the double helix, each strand could act as the template for the creation of a new strand to produce two identical ‘daughter’ DNA double helices (Figure 51). But this still left the question of how a genetic alphabet of four letters (A, T, G, C) could code for twenty amino acids. The answer lies in the triplet code, where an amino acid is coded, not by one nucleotide, but by a set of three. The number of possible triplet combinations using four ‘letters’ is more than enough to encode all the amino acids.
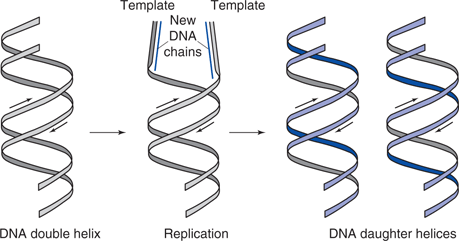
51. Replication of DNA chains.
RNA is the other form of nucleic acid present in cells, and is crucial to protein synthesis (translation). There are three forms of RNA—messenger RNA (mRNA), transfer RNA (tRNA), and ribosomal RNA (rRNA). mRNA carries the genetic code for a particular protein from DNA to the site of protein production. Essentially, mRNA is a single-strand copy of a specific section of DNA. The process of copying that information is known as transcription.
tRNA decodes the triplet code on mRNA by acting as a molecular adaptor. At one end of tRNA, there is a set of three bases (the anticodon) that can base pair to a set of three bases on mRNA (the codon). An amino acid is linked to the other end of the tRNA and the type of amino acid present is related to the anticodon that is present. When tRNA with the correct anticodon base pairs to the codon on mRNA, it brings the amino acid encoded by that codon.
rRNA is a major constituent of a structure called a ribosome, which acts as the factory for protein production. The ribosome binds mRNA then coordinates and catalyses the translation process. Each ribosome attaches to the end of mRNA, and travels along its length. As it does so, only two codons are exposed at any one time allowing the binding of two tRNA molecules to the ribosome. The protein is built up one amino acid at a time, with the growing protein chain being transferred from one tRNA to the amino acid on the other (Figure 52).
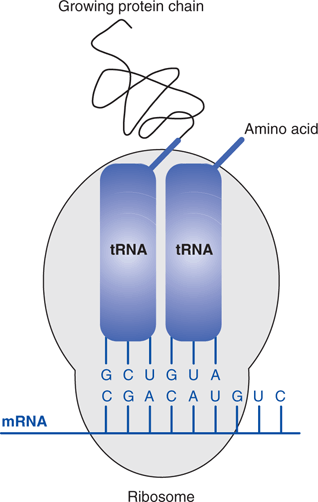
52. Translation.
Chemical evolution
In the 1880s, Charles Darwin proposed that the organic chemicals essential for life might have been formed in a ‘warm little pond’. Since then, organic chemists have postulated how the essential building blocks for life might have been formed on Earth during the one billion-year period before life began. This is a topic known as chemical evolution or prebiotic chemistry.
The first chemists to study chemical evolution seriously were Stanley L. Miller and Harold C. Urey who worked at the University of Chicago. In 1953, they conducted an experiment that was designed to mimic the environmental conditions that might have been present during the prebiotic period. They proposed that Earth’s early atmosphere contained gases such as methane and ammonia rather than oxygen, which only appeared once plant life had become established.
The experiment devised by Miller and Urey involved a round-bottomed flask containing water to mimic the oceans, and a mixture of methane, ammonia, and hydrogen to represent the atmosphere. A couple of electrodes were inserted into the flask, across which electrical charges could be passed to mimic lightning. It was proposed that lightning could provide the energy required to drive reactions between the gaseous molecules, and that the products formed would end up dissolved in the ‘ocean’. The experiment was carried out for a week and, when the water was analysed, a number of naturally occurring amino acids were found to be present. This single experiment convinced many scientists that life could be created, not only on Earth, but elsewhere in the cosmos.
Subsequent experiments demonstrated that similar reactions took place with heat or ultraviolet radiation as the energy source. Experiments were also carried out with different gases in the ‘atmosphere’, which demonstrated that oxygen prevented the formation of amino acids. If hydrogen cyanide was present, adenine was formed. Adenine is one of the nucleic acid bases, and appears in several biochemicals, suggesting that it may have been formed early on during chemical evolution. Indeed, the structure of adenine is made up entirely of C–N segments that could be derived from five cyanide molecules (Figure 53). When formaldehyde was included in the ‘atmosphere’, ribose was formed. Many scientists now believe that Earth’s prebiotic atmosphere contained carbon dioxide, carbon monoxide, and nitrogen. The Miller–Urey type experiments carried out under this mixture of gases also produces life’s molecular building blocks.
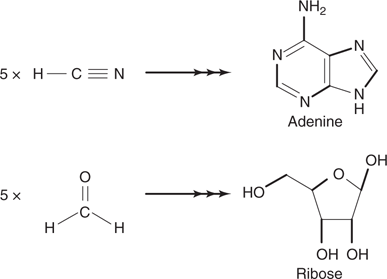
53. Possible building blocks for adenine and ribose.
An alternative theory of chemical evolution is that the building blocks of life were formed in deep-sea hydrothermal vents. These vents spew forth the mixtures of chemicals required for prebiotic reactions, and also provide heat as an energy source. Moreover, the products formed would be sheltered from the hostile environment on the planet’s surface and have a greater chance of forming biopolymers such as proteins and nucleic acids.
Yet another theory proposes that life’s building blocks were synthesized in outer space, then brought to Earth by meteorite showers. Astrochemistry is a branch of chemistry that has been investigating such a possibility by searching for extraterrestrial organic molecules. For example, methane has been detected as a major component of the atmospheres of Jupiter, Saturn, Uranus, and Neptune. NASA’s Curiosity Rover has been searching for organic molecules on the surface of Mars, and, in November 2014, the European space agency’s Rosetta probe landed a robotic detector called Philae on to a meteorite. Initial data revealed organic molecules in the comet’s atmosphere. There is also interest in finding out whether organic molecules are present in the atmosphere of Titan (one of Saturn’s moons). The Cassini probe has been orbiting the moon since 2004 and it is now known that Titan has an atmosphere containing polyaromatic hydrocarbons. Titan’s methane lakes might also have nitrogen-containing organic molecules. However, a lack of oxygen, and Titan’s extremely cold temperatures, make the evolution of life unlikely.
On Earth, a variety of experiments have been carried out to mimic conditions in outer space, and to see what synthetic reactions could take place. One possibility is that reactions are occurring on meteorites as a result of the solar wind acting on formamide—a simple organic molecule that has been detected in significant quantities in outer space. Simulation experiments, where a proton beam has been shone on to a mixture of formamide and meteorite dust, have produced a variety of life’s molecular building blocks. There is a particular interest in meteorites called chondrites. These are the oldest meteorites in space and it is believed that they may help to catalyse reactions. If such meteorites then ‘seed’ planets with the chemical building blocks for life, it is possible that life found elsewhere in the universe would have molecular similarities to life on Earth. One objection to the theory of meteorite ‘seeding’ is whether organic molecules could survive the extreme heat experienced when a meteorite enters a planet’s atmosphere.
One of evolution’s mysteries is how single enantiomers of amino acids and carbohydrates came to be selected as the preferred building blocks for proteins and nucleic acids. One theory, called the Vester–Ulbricht hypothesis, is based on the fact that electrons have left- and right-handed spins. Polarized radiation composed of either left-handed or right-handed electrons might have degraded one enantiomer of a chiral molecule more quickly than the other. There is some experimental evidence for this. However, it is yet to be demonstrated for amino acids or carbohydrates.
Evolution of life’s biopolymers
Amino acids, carbohydrates, and nucleic acid bases can be formed under a variety of prebiotic environmental conditions, but they fail to polymerize into proteins and nucleic acids. For polymerization to take place, some form of catalysis is required. In living cells, DNA synthesis is catalysed by proteins, but the synthesis of those proteins relies on the genetic information provided by DNA. The two synthetic processes are interdependent. Moreover, life depends on genetic information being passed on from one generation to another. How could this have evolved?
One theory is that a molecule was formed under prebiotic conditions that could catalyse its own replication. There have been debates over whether this was an early protein or DNA structure. However, there are problems with both possibilities. Amino acids can certainly link up to form peptides in the presence of a simple catalyst called cyanamide, which can be formed from methane and ammonia under prebiotic conditions. However, there is no control over the order in which the amino acids are linked together, or any mechanism by which a useful peptide could be copied.
As far as DNA is concerned, small DNA chains called oligonucleotides can be formed under prebiotic conditions. Such molecules could potentially act as templates for identical molecules, but they are too short to contain useful genetic information, and there is no prebiotic catalytic mechanism to promote their replication.
Most scientists now believe that the key molecule that sparked the evolution of life was RNA. This is a reasonable proposal considering the central role that RNA now plays in protein synthesis. An early form of RNA may have evolved that acted as a template for the synthesis of identical copies, in which case RNA would have been the original storage molecule for genetic information. It has also been proposed that this early RNA could have catalysed a self-replication process. Recent research has demonstrated that some current-day RNA molecules do indeed have a catalytic property. Such molecules are called ribozymes. Early life might well have relied on ribozymes to serve the functions currently carried out by DNA and proteins. It has even been suggested that the ribosomes in current-day cells might have evolved from an early ribozyme. The central regions of ribosomes from different organisms are relatively similar, which might suggest a common molecular ancestor from 3.8 billion years ago. That ancestor could well have marked the crucial transition from prebiotic chemistry to life itself.
Making use of proteins and nucleic acids
Enzymes and nucleic acids are increasingly being used in commercial applications. For example, enzymes are used routinely to catalyse reactions in the research laboratory, and for a variety of industrial processes involving pharmaceuticals, agrochemicals, and biofuels. In the past, enzymes had to be extracted from natural sources—a process that was both expensive and slow. But nowadays, genetic engineering can incorporate the gene for a key enzyme into the DNA of fast growing microbial cells, allowing the enzyme to be obtained more quickly and in far greater yield. Genetic engineering has also made it possible to modify the amino acids making up an enzyme. Such modified enzymes can prove more effective as catalysts, accept a wider range of substrates, and survive harsher reaction conditions. For example, a modified enzyme was used to catalyse a key synthetic step in the synthesis of sitagliptin—an agent used to treat diabetes. The natural enzyme was unable to catalyse this reaction because the substrate involved was too big to fit the active site. Genetic engineering produced a modified enzyme with a larger active site.
Companies such as Novozymes and Du Pont specialize in designing modified enzymes. For example, biological washing powders contain a variety of enzymes that catalyse the removal of persistent stains caused by food, blood, or sweat. Proteases cleave peptide bonds in proteins, lipases cleave ester groups in lipids and fats, and amylases degrade starches. Cellulases partially degrade the cellulose fibres in clothes to help free trapped dirt particles.
New enzymes are constantly being discovered in the natural world as well as in the laboratory. Fungi and bacteria are particularly rich in enzymes that allow them to degrade organic compounds. It is estimated that a typical bacterial cell contains about 3,000 enzymes, whereas a fungal cell contains 6,000. Considering the variety of bacterial and fungal species in existence, this represents a huge reservoir of new enzymes, and it is estimated that only 3 per cent of them have been investigated so far. Microorganisms that survive extreme climates can be particularly useful sources for enzymes that will operate under harsh conditions. For example, microorganisms that survive in the Arctic have provided enzymes that are effective at room temperature and avoid the need for heating reactions. A protease that acts at high pH was discovered from a microorganism growing in a cemetery. This enzyme has proved a useful addition in detergents.
A number of enzymes including amylase have proved important in the production of biofuels such as ethanol. The enzymes catalyse the breakdown of starch and glycogen found in sugar cane and maize. Unfortunately, there are significant drawbacks to producing biofuels from food crops, since it means that less food reaches the shops and prices increase. A better approach would be to make use of plant material that is currently wasted. Therefore, research is being carried out to find cellulase enzymes that break down the cellulose present in plant leaves and stalks. For example, Novozymes has produced a mixture of cellulase enzymes called Cellic which could be used for bioethanol production.
Enzymes will also prove important in providing many of the reagents and chemicals currently obtained from oil. For example, 5.5 million tons of adipic acid—a key constituent needed in the synthesis of nylon—needs to be produced each year. Oil has been the traditional source, but conventional oil production is predicted to drop by over 50 per cent in the next two decades, and fracking is not a long-term answer. Enzymes will play a crucial role in producing these important chemicals from alternative sources.
There are some unusual applications for enzymes. For example, enzymes are being considered as components in batteries. The idea is to attach enzymes to the electrodes, and to use glucose as a fuel. The enzymes catalyse the oxidation of glucose, producing electrons that are transferred to the electrode. It is predicted that such batteries could eventually power mobile phones, pacemakers, and other small devices.
There are potential applications for other types of protein. For example, a protein called reflectin is largely responsible for the method by which squid, octopus, and cuttlefish camouflage themselves. If the protein is phosphorylated, it causes cells called iridophores to reflect light such that the organism mirrors its surroundings. Researchers are considering using a similar process to design new camouflaged materials. Other experiments are being carried out to see if materials based on this technology could reflect heat. If successful, smart clothing could be designed that regulates how much heat is radiated to the environment, such that a jacket could be cool on warm days and warm on cold days.
Several scientific teams have been investigating proteins that allow organisms to survive in freezing conditions. For example, the larvae of Alaskan beetles can survive temperatures as low as −100°C. It has been found that some proteins can act as a natural antifreeze to prevent ice crystals being formed within cells. The protein structure appears to ‘mop up’ microcrystals of ice and prevent them growing any larger. There are potential commercial uses for proteins that behave in this way. Indeed, antifreeze proteins are already being used in ice creams because they limit the size of ice crystals that are formed. This produces a smooth texture and allows the fat content to be cut.
A large amount of research is also taking place into applications for DNA. For example, DNA is being considered as a data storage system. Although there are only four ‘letters’ in DNA, a binary system can be used to store information, and scientists have demonstrated this by storing Martin Luther’s speech ‘I have a dream’ on a DNA system. The number of ‘letters’ incorporated in DNA molecules could also be increased by including synthetic base pairs. For example, synthetic bases called d5SICS and dNAM have been designed that can base pair in DNA through hydrophobic interactions. DNA is actually a remarkable stable molecule, and bacterial DNA has been recovered from Antarctic ice cores that are millions of years old.
Other commercial uses for DNA include a number of diagnostic devices that can identify metals in water, toxic gases, food spoilage, and the virus responsible for Ebola. Another possible use for DNA is as a template for polymer synthesis. Experiments have shown that DNA can bind molecular adaptors that carry the monomers required for a polymer. Once the adaptors are bound to the DNA template, the polymerization is carried out, and the polymer is released. This method has already been used to prepare polyethylene glycol.
DNA from herring sperm has been found to have a possible use as a fire retardant. When DNA is degraded by fire, it produces ammonia which prevents oxygen from fuelling the flames. The flame retardant industry is big business. However, many traditional fire retardants are halogenated molecules that are hazardous to the environment, and can prove toxic to both animals and humans. If DNA proves effective as a fire retardant, it would have the advantages of being abundant, naturally occurring, and biodegradable.
DNA even has potential in the development of lithium-sulphur batteries, which consist of a lithium metal anode and a carbon-sulphur cathode. When the battery is active, lithium ions are released from the anode and react with the sulphur at the cathode to form polysulphides. The reaction is reversed when the battery is recharged. However, some of the sulphur is lost from the cathode when the battery is in operation, and this results in a loss of performance. The nucleic acid bases and phosphate groups of DNA have a strong affinity for sulphur and it is thought that coating the cathode with DNA might prevent the loss of sulphur into the electrolyte.