Concepts in Character Macroevolution: Adaptation, Homology, and Evolvability
Allan Larson
OUTLINE
1. Darwinism and character variation
2. Evolutionary analysis of character homology
3. Testing hypotheses of character adaptation
4. Character evolvability
Evolutionary analysis requires deconstructing an organism into separately measurable parts that we call characters. This operation succeeds if the characters have biological validity, representing semiautonomous units of evolutionary change within the context of the organism as a whole. All empirical tests of Darwinian evolutionary theory rely on the biological validity of the characters constructed to test it. This article presents a critical analysis of the evolutionary character concepts used to test Darwinian evolutionary theory in a macroevolutionary framework (comparisons among species encompassing millions of years of evolutionary time). The term macroevolution often carries connotations of a rejection of Darwinian evolutionary theory or at least a perception that Darwinian theory is inadequate to explain major features of evolution. I briefly review the major conjectures of Darwinian evolutionary theory and the ways in which construction of characters and analysis of their variation can test these conjectures.
GLOSSARY
Adaptation (as a Process). Evolution of a population by natural selection in which hereditary variants most favorable to organismal survival and reproduction are accumulated and less advantageous forms discarded; includes character adaptation and exaptation.
Character Adaptation. A character that evolved gradually by natural selection for a particular biological role; character adaptation contrasts with disaptation, exaptation, and nonaptation.
Convergence. Evolution of superficially similar characters by different developmental means in different population lineages. In diagnostic tests of homology, it passes the conjunction test but fails tests of similarity and congruence.
Deep Homology. Relationship between similar morphological characters that evolved in parallel by separate evolutionary activations of homologous developmental pathways.
Developmental Constraint. A bias in the morphological forms that a population can express, caused by the mechanisms and limitations of organismal growth and morphogenesis.
Disaptation. A character that reduces fitness relative to contrasting conditions evident in a population’s evolutionary history. A primary disaptation is disadvantageous within the populational context in which it first appears; a secondary disaptation acquires a selective liability not present at its origin as a consequence of environmental or evolutionary change.
Evolvability. Ability of a population to produce new morphological characters by mutation or genetic recombination, often by activating latent developmental modules.
Exaptation. Co-option of a character by natural selection for a biological role other than one through which the character evolved by natural selection.
Function. Biological role through which an adaptive character evolved by natural selection.
Genetic Assimilation. By experimentally selecting individuals most susceptible to an environmentally induced change of development, a formerly latent developmental module comes to be expressed even without the environmental treatment formerly needed to activate it.
Gradualism. Accumulation of individually small quantitative changes in organismal form in a population leads over many generations to qualitative change in organismal structure; contrasts with saltation, in which a single genetic change induces a large qualitative change in organismal structure.
Homology. Two characters are homologous if they derive, with or without some modification, from an equivalent character of a common ancestor. Diagnostic tests of character homology include similarity (physical resemblance), conjunction (alternative states do not occur together in the same organism at the same developmental stage), and congruence (sharing of homologies among species forms a nested hierarchy of groups within groups that can be summarized as a cladogram). Homology contrasts with serial homology (fails conjunction test), parallelism (fails congruence test), and convergence (fails similarity and congruence).
Modularity. As applied to development, a process of pattern formation or morphogenesis that is semiautonomous with respect to other aspects of organismal development, and which produces a characteristic arrangement of morphological substructures in the adult body. Developmental modules often feature characteristic patterns of gene expression. Ectopic expression of a module during organismal development can lead to evolution of new structures.
Nonaptation. A character selectively indistinguishable from contrasting conditions present in a population’s evolutionary history.
Orthology. Homology relationship between DNA sequences whose genealogies coalesce to a common ancestral molecule with no intragenomic gene duplication (intragenomic here referring to a haploid genome) and no horizontal transfer between genomes of different organisms.
Parallelism. Origins of similar characters independently in two different population lineages, usually because these lineages share homologous developmental constraints that channel production of morphological variation in similar directions. It is diagnosed by failure of the congruence test of homology but passing tests of similarity and conjunction; reversal of a derived character to an ancestral condition is a special case of parallelism using this diagnosis.
Paralogy. Homology relationship between DNA sequences whose genealogical coalescence to a common ancestral molecule includes at least one intragenomic gene duplication (intragenomic here referring to a haploid genome) but no horizontal transfer between genomes of different organisms.
Saltation. Evolution of a large, qualitative change in phenotype in a single mutational step; contrasts with gradualism. Also, genetic assimilation of a qualitative change in organismal structure initially caused by an environmental treatment.
Xenology. Homology relationship between DNA sequences whose genealogical coalescence features at least one horizontal transfer between genomes of different organisms.
1. DARWINISM AND CHARACTER VARIATION
My favorite concise account of Darwinian evolutionary theory is by Mayr (1985). He consolidates the many connotations acquired by the term Darwinism into five principal theories testable by measuring character variation.
The first and most fundamental theory is that life has a long history of irreversible change with hereditary continuity from past to present life. Mayr (1985) calls this theory Evolution as Such. Cambrian organisms of the Burgess Shale, for example, would not be mistaken for any organisms alive today because their characters contrast with those of living forms, yet those characters reveal homologies critical for establishing historical continuity between extinct and living forms.
Darwin’s “second” theory states that all past and present forms descend from a shared common ancestor of life on earth (called Common Descent by Mayr). Life’s history thus takes the form of a branching tree of population lineages. This theory makes the prediction that sharing of characters among species forms a nested hierarchy of groups within groups. Common descent of species was firmly established during Darwin’s lifetime by studies of morphological characters, and we now measure its details with great precision using character variation revealed by DNA sequence data.
Multiplication of Species denotes the spatial dimension of evolution in Darwinian theory, geographical processes by which population lineages branch to form two or more descendant lineages. A lineage is an unbranched series of ancestor-descendant populations through time. Geographic isolation of two populations typically precedes evolution of genetic differences that prevent them from merging should they make secondary geographic contact. Character variation across geographic space is the primary means by which species lineages are diagnosed, based on Darwin’s principle of divergence of character among geographically isolated populations.
The remaining two Darwinian theories pertain specifically to populational processes of evolution, typically measured as change in organismal morphology. Gradualism states that quantitative change in organismal characters leads to qualitative change; by accumulating, over many generations, hereditary variants that individually have very small effects on organismal appearance, diverging populations eventually acquire sharply contrasting morphological characters. This is Darwin’s theory of gradualism, whose alternative, traditionally called saltation, is that sharply contrasting characters arise as such within a generation. Saltation is compatible with but not identical to punctuated equilibrium, which postulates that morphological change occurs in geologically brief (fewer than 1 million years) events of branching speciation, followed by morphological evolutionary stasis within species maintained over a much longer interval of evolutionary time. Saltation associated with branching speciation is a special case of punctuated equilibrium, as would be phenotypically continuous divergence that leads to qualitative change at the formation of a new species. A saltation occurring within a species and not associated with branching speciation does not constitute punctuated equilibrium. Punctuated phyletic evolution denotes a punctuated pattern of character evolution not associated with branching speciation.
Darwin’s fifth theory, Natural Selection, is itself a composite of many subtheories. New variation occurs at random with respect to its potential utility to an organism that possesses it. Variation is “heritable,” in the sense that organisms resemble their parents more closely than they do individuals drawn at random from their population. Variant forms that enhance their possessors’ fitness are thereby transmitted to the next generation at a higher rate than are contrasting characters less conducive to survival and reproduction. By accumulating many such changes across many generations, a population can gradually construct a new character qualitatively different from ancestral conditions.
If a new character arises by saltation, natural selection might increase its frequency in the population if the new character enhances fitness, but natural selection does not formally explain the character’s origin. The mechanistic explanation lies alternatively in specifying how a genetic change and its interaction with environmental conditions during development produced a discontinuous phenotype. Developmental constraint denotes the hypothesis that the structure of organismal development, particularly specific interactions between proliferating cells at critical stages, makes some morphological forms more accessible to a population than are various conceivable alternatives.
Developmental constraint is compatible with gradualism if its main consequence is to make certain directions of continuous character change more accessible than others. Hypotheses of developmental constraint nonetheless often include an argument that disparate morphologies often lack developmentally accessible intermediate conditions. If the physical processes of cell proliferation and differentiation during development preclude a gradual transition between disparate states of a character, then the developmental-constraint hypothesis is saltational.
Darwin and many of his followers have argued that single variants of large phenotypic effect are inevitably detrimental and that natural selection would eliminate them. This assumption underlies Darwin’s commitment to gradualism, but numerous discoveries in evolutionary developmental biology now challenge this assumption.
2. EVOLUTIONARY ANALYSIS OF CHARACTER HOMOLOGY
Darwin’s theory of common descent is the foundation for testing the biological validity of character constructs through the concept of evolutionary homology: two characters are homologous if they derive, with or without some modification, from an equivalent character of a common ancestor.
Evolutionary homology was applied first to organismal anatomy and form. The forelimbs of a human and an orangutan are homologous as vertebrate forelimbs because they descend, with much modification, from the forelimbs of a common ancestral form. Wagner (1989) elaborated in his concept of biological homology the properties that we expect of homologous organismal structures. First, homologies are historically unique; they arise in a particular population lineage at a particular place and time and occur only in the descendants of that lineage. Second, they have evolutionary continuity; two characters are homologous to each other only if there is an unbroken chain of lineal descent connecting them to each other and to their common ancestral origin. Third, homologies are individuated; they exist as semiautonomous components within the context of the organism as a whole. A vertebrate forelimb, for example, has an individual evolutionary history and semiautonomous developmental dynamic within an organism.
We construct characters and test hypotheses of their homology also at the cellular level. Among the many cellular-level characteristics used in evolutionary studies are the detailed structures of chromosomes as they appear during cell division. Chromosomes are homologous to each other if they descend with some modification from a common ancestral chromosome. Chromosomes of an orangutan each have homologous chromosomes in human cells despite some minor rearrangements of chromosomal contents. Perhaps most effective for testing precise hypotheses of common descent of species is homology at the level of DNA sequences. For example, the gene encoding hemoglobin β in humans is homologous to the gene encoding hemoglobin β in orangutans; these DNA segments descend with modification from an ancestral gene encoding hemoglobin β.
Having defined the concept of homology, we must establish principles for testing hypotheses of character homology, evaluating whether the general principle of homology explains our comparisons for a particular set of characters. There are three diagnostic tests of homology that can be applied at the organismal, cellular, and molecular levels (Patterson 1988). Whether a set of structures passes or fails these tests separates homology from contrasting explanations for character variation and resemblance.
The first diagnostic test is that of character similarity. This test is the definitive one at the molecular level. If one compares the DNA base sequences of two pieces of DNA and finds greater than 70 percent sequence similarity, then those sequences undoubtedly trace to a common ancestral DNA sequence. The hereditary pathway that connects them to their common ancestral sequence is nonetheless sometimes a contorted one, as revealed by the second and third diagnostic tests.
For characters of organismal morphology, similarity testing implies a nontrivial structural correspondence that transcends differences in exact form. For example, the forelimbs of humans, horses, bats, birds, lizards, and frogs are all homologous to each other as vertebrate forelimbs despite enormous dissimilarities in overall form; the homologies reveal themselves in the major bones present and the patterns by which the bones connect to each other and to the rest of the body; specific homologies of the limb bones and their developmental genetic origins are evident across all four-limbed vertebrates and even among bony fishes. Note that although the forelimbs of bats and birds are homologous as vertebrate forelimbs, they are not homologous as wings. The most recent common ancestor of birds and bats had forelimbs that did not form wings, and the structural modifications that make a bird’s forelimb a wing are very different from those that make the bat’s forelimb a wing. Bird wings and bat wings thus pass similarity testing as vertebrate forelimbs, but they fail similarity testing as wings.
The second diagnostic test for homology is the conjunction test; if two organismal structures are hypothesized to be homologous to each other, a single organism should not express both structures at the same stage of development. The hypothesis that human arms and bird wings are homologous as vertebrate forelimbs would be rejected were we to find angels as often depicted in Italian Renaissance artwork; no living or fossil forms, however, have the characteristic arms of humans and wings of birds present in the same organism.
Duplication of body parts is nonetheless common in evolutionary history. Vertebrate forelimbs and hindlimbs have sufficient similarity in skeletal structures that they would perhaps pass similarity testing, but they clearly fail conjunction testing. We apply the term serial homology (or homonomy) to structurally similar features that pass similarity testing but fail conjunction testing. Intraorganismal duplication of structures separates serially homologous structures from homologous ones. In tracing the evolutionary history of serially homologous structures, one must traverse at least one event of intraorganismal duplication of the structure. For example, multiple intraorganismal duplications separate a neck vertebra from a trunk vertebra.
For molecular characters, the unit of comparison is not the organism but the haploid genome as transmitted through a sperm or egg. DNA segments passing the similarity test must be present one time only per haploid genome to pass the conjunction test. The conjunction test separates molecular sequence homology into the contrasting subcategories of orthology and paralogy. A pair of sequences is orthologous if their coalescence to a common ancestral molecule features no intragenomic gene duplication (intragenomic here referring to a haploid genome). A pair of sequences is paralogous if its path of coalescence to a common ancestral molecule includes at least one event of intragenomic duplication.
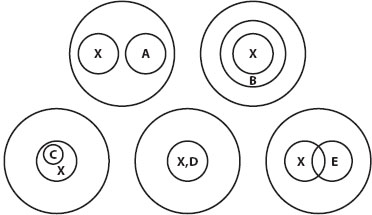
The third diagnostic criterion is the congruence test (figure 1). A homology has by definition only a single origin on the tree of life, and we hypothesize that a homology is transmitted from its lineage of origin to all descendant lineages (unless secondarily lost). If we assume that a new homology spreads throughout its population shortly after arising and is not subsequently lost, the homology should characterize all and only the descendants of that ancestral population. Hence, a homology should characterize a particular clade of the phylogenetic tree. Different homologies often characterize different branches of the phylogenetic tree of species, but the tree structure restricts the relationships between the set of species having homology A and the set of species having homology X to one of three possible relationships: (1) sets A and X are identical, (2) one set is nested within the other one, or (3) sets A and X are mutually exclusive. Each of these conditions passes the congruence test. If the two sets intersect but do not meet conditions (1) or (2), that is, if they partially overlap in a non-nested way, then at least one character, A or X, fails the congruence test.
Figure 1. Congruence test of homology as applied to morphological characters. Circles in the Venn diagram denote sets of organisms or species possessing a particular homology or hypothetical homology. Assume that the outer circle denotes all organisms having vertebrae and that X denotes organisms possessing hair or fur. Character X is tested for congruence with characters A–E. Character X passes the test if it forms with others a nested hierarchy of groups within groups. Character A, presence of feathers, illustrates one way to pass congruence testing: the two hypothetical homologies characterize mutually exclusive subgroups of the larger group. Character B, presence of an amniotic membrane in the egg, illustrates another way to pass congruence testing: X characterizes a nested subset of amniotic vertebrates. Character C, presence of a placenta, identifies a nested subset of the organisms that have character X, again passing the congruence test. Character D, presence of mammary glands, identifies an equivalent set of organisms to character X, also passing the congruence test. Character E, presence of wings, fails congruence testing because it partially overlaps with organisms possessing X (bats), but it is not a nested subset of X because organisms possessing feathers (A) also have wings and are outside the group possessing X. Character E must be separated into two different homologies, E1 = bat wings and E2 = bird wings. As noted in the text, character E “wings” also fails similarity testing; although bat and bird wings are homologous as vertebrate forelimbs, the forelimb modifications that make them wings are very different. (Modified from Patterson.)
For example, the hypothetical homology of bird wings and bat wings failed similarity testing but passed conjunction testing. The hypothesis that vertebrate wings are homologous fails congruence testing with at least three very strong morphological homologies: feathers, hair, and mammary glands. Feathers characterize all living birds and some fossil forms more closely related to birds than to other living vertebrates. Mammary glands and hair characterize mammals but no other vertebrates. Wings characterize all birds and a subset of mammals (bats), thus failing the congruence test. If we treat bird wings and bat wings as separate homologous characters not homologous to each other, then each one passes congruence testing with each other and with feathers, mammary glands, and hair.
Note that the nested hierarchy of groups within groups corresponds to the pattern of a branching tree of common descent of the species being studied. A tree of life, termed a cladogram or phylogenetic tree, is a way to depict results of congruence testing of many characters (see chapter II.1). Phylogenetic analysis in all its complexities is fundamentally a congruence test of homologies.
In DNA sequence comparisons that pass the similarity test, failure of the congruence test implies horizontal transfer of DNA between organisms that typically are not familial relatives and often not of the same species. Such transfer may involve viral transmission of DNA from one organism to another. Domestic cats and their closest wild relatives have in their genomes a viral gene absent from other felids and carnivores but that has strong sequence similarity to viral genes in rat genomes. A hypothesis that cats and their closest wild relatives are more closely related to rats than to other felids and carnivores is contradicted by many strong homologies. A better hypothesis is that an ancestor of cats and their closest wild relatives acquired the rat viral gene while feeding on rats, and that the gene inserted into the germ line, became fixed in the ancestral population, and was passed to the descendants of that lineage.
Morphological characters that fail the congruence test but pass the other two (conjunction and similarity) are parallelisms. A major discovery of evolutionary developmental biology is that species often share a latent developmental potential to produce characteristics that are not actively expressed in the species at a given time (Carroll 2008). A developmental switch, controlled by interactions among proliferating cells at a critical developmental stage, may determine which pathway is taken. Interactions between genetic and environmental factors influence whether a genetic switch follows one pathway or a contrasting one.
Conrad H. Waddington demonstrated one such developmental switch in the fruit fly Drosophila melanogaster. Flies are unusual among insects in having a pair of halteres, also called balancers, in the position where most insects have a pair of hindwings. Although physically very different from hindwings, halteres are homologous to hindwings and represent an evolutionary change that occurred in a common ancestor of all flies more than 200 million years ago. Expression of a pair of hindwings has not been a normal condition in flies since that time, yet a latent developmental potential to produce hindwings remains and can be activated either by genetic mutation (bithorax and related mutations) or by treating the developing eggs with ether at a critical stage. Waddington experimentally selected for increased susceptibility of genetically normal flies to respond to ether treatment of the egg by developing hindwings rather than halteres. Many generations of selection produced lineages that developed hindwings at a high rate even without ether treatment or the bithorax mutation. He called this phenomenon genetic assimilation. A trait originally induced by environmental means (ether treatment) had been stabilized by selective accumulation of the genetic variants most conducive to expressing a developmental pathway that had been latent for more than 200 million years.
The hindwings of Waddington’s experimental flies are not homologous to hindwings of other insects, but the developmental potential to express hindwings is a separate character likely homologous among all winged insects. Because species share latent developmental pathways, parallelisms for complex characters are more likely than Darwin and his early followers could have anticipated. Because the developmental pathway to produce hindwings, although latent in flies, was presumably constructed by natural selection in an ancient common ancestor of winged insects, activation of this pathway in fly development produces a highly ordered structure with potential utility. This observation negates the traditional Darwinian assumption that mutations of large effect (saltations) are inevitably harmful and eliminated by natural selection.
Evolutionary loss of a homologous body part in different evolutionary lineages is a particularly common parallelism. For example, loss of limbs occurred in parallel in a common ancestor of snakes and in some legless lizard groups, including most amphisbaenians. Thus, absence of limbs is not homologous between snakes and amphisbaenians, as revealed by failure of congruence testing of limblessness with other skeletal homologies. The same is true for the loss of limbs in whales (mammals) and caecilians (amphibians).
Note that parallelism as defined here includes evolutionary reversal as a special case; reversal implies a transformation from an evolutionarily derived character state to an ancestral state, whereas parallelism in a stricter sense features two separate origins of the same derived state. Parallelism is the primary source of error in species taxonomies derived from morphological characters. It is a biologically interesting phenomenon because it usually reveals deep homology of developmental pathways that species share despite variation among them in whether the shared pathway is active or latent during typical organismal development.
Convergence denotes cases where hypothetically homologous morphological structures fail both similarity and congruence testing, but pass conjunction testing. These are typically contrasting states of a more inclusive homology, illustrated above by convergent evolution of wings of bats and birds by very different modifications of their homologous forelimbs. In contrast to parallelisms, convergently evolved characters do not imply deep homology of developmental pathways.
Evolution of new homologies often occurs by serial duplication of structures followed by fusion and modification of the repeated parts to form a new structure. This process is perhaps best illustrated by evolution of insects in the animal phylum Arthropoda. Serial repetition of many nearly identical body segments is most evident in millipedes and centipedes, fellow arthropods that lack structures equivalent or homologous to an insect thorax. The insect thorax is hypothesized to have arisen by a fusion of three originally identical segments, each one bearing a pair of legs, as observed in centipedes and millipedes. Fusion of the three segments produced the insect thorax. Individual segments of the insect thorax are serially homologous to each other and individually homologous to particular segments of other arthropods; however, the thorax produced by developmental fusion and elaboration of these segments has no homologue in millipedes and centipedes.
There is no necessary one-to-one correspondence between homology of morphological structures and homology of the genes whose expression contributes to their development. The product of a particular gene typically contributes to multiple, nonhomologous morphological structures. Although shared patterns of gene expression often characterize homologous structures, and shared patterns of gene expression play a role in similarity testing of hypothetically homologous structures, gene expression is not a sure guide to homology. Homologous morphological structures can undergo evolutionary changes in the developmental genetic processes underlying their formation while maintaining evolutionary continuity and thus homology in the fully formed structure. For example, digit formation in salamanders follows a fundamentally different developmental pathway from the one it follows in other terrestrial vertebrates despite clear homology of the digits among terrestrial vertebrates; following the evolutionary origins of digits, a lineage ancestral to living salamanders evolved a novel developmental mechanism for forming the digits without destroying their homology to digits of other vertebrates. This change is evident in contrasting developmental constraints on loss of digits in salamanders versus those in other terrestrial vertebrates (figure 2): parallelisms for loss of digits typically start with loss of the fifth digit in hindlimbs of salamanders, whereas such loss typically begins with loss of the first digit in other tetrapods.
Figure 2. Empirical evidence for contrasting developmental constraints on loss of digits from the hindlimbs of frogs and salamanders. The inferred ancestral condition in both frogs and salamanders is to have five digits on the hind foot. Digits are numbered I–V based on their characteristic appearance and ordering relative to the fibula and tibia. The evolutionary history of frogs features two evolutionary losses of a single digit; in both cases, digit I is lost. Experimental treatment of frog limb buds with a mitotic inhibitor can cause species that normally have five digits to lose one, always the first digit. In contrast, in salamanders, seven independent evolutionary reductions in digit number from V to IV feature loss of digit V, which is also the digit lost on treatment of Ambystoma mexicanum with a mitotic inhibitor (see figure 4B). The developmental constraint seen in frogs also characterizes other tetrapod vertebrates, whereas the contrasting constraint in salamanders is a uniquely derived condition in that group. (Modified from Alberch 1991.)
Experimental study of deep homology reveals modularity in patterns of development and associated gene expression. Modularity features coordinated changes in separately measurable components of morphology arising from shared developmental processes and patterns of gene expression (see Wagner et al. 2007 for a thorough discussion of modularity). Genetic and developmental modules are often called toolkits. A given developmental module presumably evolved by natural selection in the context of one morphological structure, but this does not prevent its later being activated at a new location in the body and at an atypical time during development to generate a new structure. Most biologists would not consider these new structures homologous to the original structure despite their using the same module. For example, patterns of expression of homeobox genes in vertebrate limbs reveal modules homologous to ones used in the development of the caudal axial skeleton of the same organism. The best evolutionary hypothesis is that the module evolved originally as a means for forming the caudal skeleton, followed by a separate expression of the module in the pectoral and pelvic body regions to induce formation there of bony, paired appendages. Nonhomologous characters thus can share homologous modules of gene expression.
3. TESTING HYPOTHESES OF CHARACTER ADAPTATION
Adaptation, the observation that the form of a character serves its biological utility, was at one time thought to contradict biological evolution. The apparent correspondence between form and function implied teleology in the origin of characters, that a preexisting purpose guided their design. Darwin’s theory showed, however, that by accumulating the most favorable of randomly produced variants while discarding less favorable ones, a population would evolve useful structures without a predetermined goal. Since the acceptance of Darwin’s theory, the term adaptation has come to denote a character that evolved gradually by natural selection because successively accumulated variations contributed incrementally to a biological role evident in the resulting composite structure. Adaptation is not necessarily the most parsimonious explanation for all characters, and it requires a sequential testing of relevant hypotheses.
Investigating the role of natural selection in evolution requires that we expand our testing of homology to include not only presence of the character in question but also its utility to organisms possessing it. Assessing the biological role of a character requires identifying the environmental contexts in which the character occurs and inferring environmental contexts of its past evolutionary history.
To argue empirically that a character evolved by natural selection, one must first reject the null hypothesis that the character in question and its historically antecedent condition are equivalent in their utility to the organisms bearing them (Baum and Larson 1991). Adaptation is always relative. Darwin argued that the characters constructed by natural selection are not expected to be perfect in any role, only slightly better than the alternatives against which they have had to compete in their evolutionary history. Depending on the character whose utility is being studied, appropriate tests might exploit the existence of alternative forms segregating in the same population, for instance the sickle-cell allele of hemoglobin β and the contrasting allele called hemoglobin A in various human populations. In other cases, the test might involve comparing different living species that share similar morphological and ecological characteristics except for the character in question; for example, populations of Xiphophorus fishes that have a sword on the caudal fins of males might be compared with those retaining the ancestral condition of lacking the sword. In yet other cases, physical manipulation and experimental analysis or biomechanical modeling might be used to assess the relative utility of ancestral and derived characters. In all such tests, phylogenetic analysis of character origins is a critical tool for identifying the contrasting conditions to be compared (Baum and Larson 1991).
Gould and Vrba (1982) introduced the term nonaptation to denote a character that has no detectable utility, one for which the null hypothesis of the preceding paragraph cannot be rejected. The contrasting term aptation denotes any character that enhances survival or reproduction relative to its antecedent condition. They divided aptation into two subcategories, adaptation and exaptation. An adaptation is defined as an aptation that originated by natural selection for a biological role that the character continues to serve. An exaptation, in contrast, is an aptation that serves a role other than one for which the character might be inferred to have evolved by natural selection. Gould and Vrba (1982) use as an example the utility of feathers for flight in birds. Phylogenetic analysis of birds and their fossil relatives shows that the origin of feathers preceded the origin of flight, thus rejecting the hypothesis that feathers could have originated by natural selection for enhanced flight. Feathers arose coincidentally with evolution of homeothermy in birds, favoring an explanation that feathers are an adaptation for thermoregulation but an exaptation for flight.
Baum and Larson (1991) expanded this terminology to include the possibility that a character is deleterious relative to its antecedent condition. A primary disaptation is one whose lowered utility pertains to the environmental conditions under which the character arose, whereas a secondary disaptation is one that became relatively unfavorable following a subsequent change in environmental conditions. A primary disaptation usually signals processes analogous to natural selection that operate at the level of gene replication and transmission. Selfish DNA is the hypothesis that some DNA sequences, and also possibly their consequences on organismal morphology, become prominent in a population because selection at the genic level favors their proliferation. For example, occurrence of a tailless condition in some mouse populations is associated with a genetic allele that produces tailless mice in the heterozygous condition and lethality when homozygous. The morphological condition is undoubtedly detrimental relative to the antecedent condition (normal development of the tail) in relevant populations. Nonetheless, during spermatogenesis in heterozygous individuals, gametes containing the tailless allele physically destroy those containing the contrasting allele (reviewed by Burt and Trivers 2006). Most offspring of heterozygous males receive the tailless allele. Occurrence of the tailless allele thus represents an interaction between natural selection, which acts to decrease the frequency of the allele, and selfish DNA, which acts to increase its frequency in the population.
4. CHARACTER EVOLVABILITY
Evolutionary biologists sometimes try to evaluate the potential of a species for undergoing further evolutionary change and diversification, including production of new species and new characters. Darwinian theory and its population-genetic models make clear that large geographic distributions and large amounts of genetic variation enhance the opportunities for a species lineage to give rise to new species and new characters. Because genetic variation in a population often stabilizes organismal development rather than expressing itself as greater variation in organismal morphology, a species that is relatively uniform in organismal morphology nonetheless can have the potential to produce a great range of organismal morphologies. If genetic variation is reordered, as might occur in the founding of a new population by a small number of individuals drawn from the ancestral one (Carson and Templeton 1984), organismal development can be destabilized to reveal alternative morphologies whose developmental pathways were latent in the ancestral population. Changes in genetic variation in this case act analogously to the environmental challenges (ether treatment of fruit fly eggs) that shifted development in Waddington’s genetic-assimilation experiments discussed in part 2.
Evolvability has emerged over the past 20 years as a concept that encapsulates the potential of a species to produce new organismal characters. Pigliucci (2008) notes that this term carries disparate connotations among authors, a major contrast being whether the term pertains strictly to expressed morphological variation or the potential to produce unexpressed characters by new mutation or recombination: “Variation is a measure of the realized differences within a population, whereas variability is the propensity of characters to vary (whether or not they actually do) and depends on the input of new genetic variation through mutation or recombination.” Evolvability should pertain to “variability” and not just to “variation” as Pigliucci (2008) defines these terms. Mutagenesis experiments, perturbing development by environmental treatments, and founding laboratory strains by small numbers of wild-caught individuals are means of measuring variability. Such experiments potentially reveal the diversity of latent developmental modules whose expression is inducible by genetic change.
Conceptual diagrams used by Alberch (1991) illustrate the concept of evolvability and the ways in which two hypothetical species can differ in evolvability. Figure 3A shows a series of six alternative organismal morphologies (A–F) whose expression depends on interactions between quantitative values of two parameters, X1 and X2. The parameters might be the amount of a gene product, variation in the multilocus genotype of a quantitative developmental character, or environmental conditions, such as temperature, that influence cellular growth or proliferation. An organism can express only one of the six alternative developmental outcomes, depending on the specific inductive interactions that occur among cells during a critical moment in development. Continuous changes in genetic or environmental conditions can switch development from one pathway to another as shown. Species 1 expresses pathway D, which borders in parameter space all the other contrasting pathways. Species 2 expresses pathway A, which borders only pathways B and D in parameter space. Species 1 has a higher evolvability than does species 2 in this diagram because continuous changes in parameters X1 and/or X2 access a greater number of contrasting pathways than would comparable changes in species 2 (figure 3B). Figure 3C summarizes the relative evolvabilities of species expressing each of the contrasting developmental pathways. Relative evolvabilities of these pathways in decreasing order are D (five connections), C (four connections), B = E = F (three connections), and A (two connections). Note that although the parameter space is continuous, differences between the organismal morphologies represented by pathways A–F can be discontinuous (figure 3C).
Figure 3. Alberch’s (1991) conceptual diagram of character evolvability for two species. (A) Parameters X1 and X2 denote continuously varying genetic or environmental variables that influence embryonic induction to specify one of five contrasting developmental pathways (A–F). These alternative pathways could be, for example, those specifying contrasting hindlimb structures as illustrated in figure 4. Species 1 has parameter values specifying developmental pathway D, although it is near the critical thresholds for expressing pathways E and F. Species 2 has parameter values well within the range that specifies pathway A. (B) Evolvability of species 1 and 2 contrasted according to whether alternative developmental pathways are directly accessible by changes in parameters X1 and X2. Thick arrows denote transformational changes more accessible than those depicted by thin arrows. (C) Transformational diagram corresponding to the parameter space in part A. On the basis of direct accessibility of alternative developmental pathways by changes in parameters X1 and X2, species expressing pathway D have the highest evolvability (five connections) and those expressing pathway A have the lowest evolvability (two connections), with pathway C having four connections and the remaining pathways each having three.
Figure 4. Morphological outcomes of three alternative developmental pathways for the structure of the hind foot in salamanders. Morphology A characterizes adult Ambystoma mexicanum. Morphologies B and C represent two alternative pathways activated by treating developing limb buds with the mitotic inhibitor colchicine. Morphology B resembles the normal condition expressed in the species Hemidactylium scutatum (morphology D). Morphology C resembles the normal condition expressed in the species Proteus anguinus (morphology E). The contrasting developmental pathways expressed by Hemidactylium scutatum and Proteus anguinus thus occur in latent form in Ambystoma mexicanum, indicating shared deep homology of developmental pathways among these species. Should one of these species have access by genetic mutation or recombination to a larger number of contrasting developmental pathways for foot skeletal morphology than do the other species, that species would have greater evolvability for this character. If all three species share the same set of accessible developmental pathways, the one capable of activating alternative conditions with the smallest amount of genetic change would be judged to have greater evolvability for this character. F=fibula; T=tibia (Modified from Alberch 1989.)
An example from Alberch’s work on foot morphology in salamanders illustrates morphological outcomes of different developmental pathways for the hind foot (figure 4). Figure 4A shows the skeletal structure of the hind foot characteristic of Ambystoma mexicanum. Figures 4B and 4C show alternative morphologies produced experimentally by treating the developing limb buds with the mitotic inhibitor colchicine, which alters inductive interactions in the developing limb. Both experimentally produced abnormal limbs match the normal limbs of distantly related salamander species. Hemidactylium scutatum (figure 4D) normally expresses the developmental pathway produced experimentally in A. mexicanum shown in figure 4B. Proteus anguinus normally expresses the developmental pathway induced in A. mexicanum shown in figure 4E. These results show that Ambystoma mexicanum shares with each of the other species as a “deep homology” a developmental pathway normally expressed in the other species but latent in A. mexicanum.
It is possible, although not yet empirically demonstrated, that the various salamander species in figure 4 differ from each other with respect to evolvability of the skeletal structure of the hind foot. If genetic modification of A. mexicanum can more easily access a larger number of alternative developmental pathways than are available to either H. scutatum or P. anguinus, then A. mexicanum would have higher evolvability for this character. Quantitative comparisons of species for evolvability of characters is a new research endeavor that promises to reveal the developmental and genetic factors underlying evolution of new homologies and parallelisms in character evolution.
FURTHER READING
Alberch, P. 1989. The logic of monsters: Evidence for internal constraint in development and evolution. Geobios: Mémoire spécial 12: 21–57. A good summary of the theory and phenomenology underlying the hypothesis of developmental constraint in evolution.
Alberch, P. 1991. From genes to phenotype: Dynamical systems and evolvability. Genetica 84: 5–11. Explains the conceptual transition from developmental constraint to evolvability.
Baum, D. A., and A. Larson. 1991. Adaptation reviewed: A phylogenetic methodology for studying character macroevolution. Systematic Zoology 40: 1–18. A thorough discussion of empirical tests of character adaptation, disaptation, exaptation, and nonaptation.
Burt, A., and R. Trivers. 2006. Genes in Conflict: The Biology of Selfish Genetic Elements. Cambridge, MA: Harvard University Press. Selfish genetic elements provide a mechanism by which a primary disaptation can persist in a population despite natural selection against it.
Carroll, S. B. 2008. Evo-devo and an expanding evolutionary synthesis: A genetic theory of morphological evolution. Cell 134: 25–36. Explains how evolution of genetic regulatory systems can stabilize developmental pathways.
Carson, H. L., and A. R. Templeton. 1984. Genetic revolutions in relation to speciation phenomena: The founding of new populations. Annual Review of Ecology and Systematics 15: 97–131. Explains how founding of new populations by a small number of individuals can reveal previously unexpressed phenotypes, thereby providing a means of measuring evolvability.
Gould, S. J., and E. S. Vrba. 1982. Exaptation: A missing term in the science of form. Paleobiology 8: 4–15. A classic work providing precise terminology for describing the role of natural selection in character evolution.
Larson, A. 2009. Adaptation. In S. Levin, ed., The Princeton Guide to Ecology. Princeton, NJ: Princeton University Press. Discusses testing hypotheses of character adaptation, disaptation, exaptation, and nonaptation in a microevolutionary context.
Mayr, E. 1985. Darwin’s five theories of evolution. In D. Kohn, ed., The Darwinian Heritage. Princeton, NJ: Princeton University Press. An excellent concise summary of Darwinian evolutionary theory.
Patterson, C. 1988. Homology in classical and molecular biology. Molecular Biology and Evolution 5: 603–625. Compares empirical tests for character homology at the levels of organismal morphology and DNA sequences.
Pigliucci, M. 2008. Is evolvability evolvable? Nature Reviews Genetics 9: 75–82. A detailed discussion of the concept of evolvability and how it transcends traditional Darwinian theory.
Wagner, G. P. 1989. The origin of morphological characters and the biological basis of homology. Evolution 43: 1157–1171. A thorough discussion of the biological basis of morphological homology.
Wagner, G. P., M. Pavlicev, and J. M. Cheverud. 2007. The road to modularity. Nature Reviews Genetics 8: 921–931. Explains developmental modules as well as modularity in patterns of character covariation and function.