Taxonomy in a Phylogenetic Framework
Julia Clarke
OUTLINE
1. Taxonomy in historical context
2. Incorporating an evolutionary perspective
3. Species in a phylogenetic framework
4. Concerns about and misunderstanding of phylogenetic nomenclature
5. The future of phylogenetic nomenclature
In biology, naming of groups of organisms is a separate but linked enterprise to determining relationships among them. Although, historically, an array of properties of interest were considered relevant for clustering organisms and applying names, today most biologists are interested specifically in discovering and naming phylogenetic groups: organisms related by virtue of descent from common ancestry. Because named groups of organisms, or taxa, figure prominently in our evolutionary theories, many biologists are deeply invested in how names are applied. A major focus has been on naming clades, all the descendants of a common ancestor and that ancestor. Some workers have focused on adapting the ranked Linnean system of taxonomy, while others have proposed a new phylogenetic system. Names for taxa defined phylogenetically utilize specimen or species specifiers and refer explicitly to clades of the tree of life. A taxon name may be tied explicitly to a clade (see chapter II.1) through definitions of three basic forms: node-based, stem-based, or apomorphy-based. These phylogenetic definitions for clade names can be described algorithmically, which may help address informatics needs in the face of increasingly dense taxonomic sampling and assembly of larger and larger sections of the tree of life. Debate concerning the format for the definitions of species names is linked to ongoing controversy over the reality and nature of species.
GLOSSARY
Binomen. A two-part species name comprising a generic name and a specific name (ICZN) or epithet (ICNB, ICBN) under the rank-based codes, or of a praenomen and a species name under the PhyloCode.
Clade. A monophyletic group; a group of organisms including an ancestor and all its descendants.
Phylogenetic System of Nomenclature. An integrated set of rules and principles governing the naming of taxa and application of taxon names that is based on the principle of common descent and formalized in the PhyloCode.
Specifier. A species, specimen, or apomorphy in a phylogenetic definition of a name that serves to specify the clade to which the name is applied. An internal specifier is a part of the clade to be named, and an external specifier is outside that clade and used in stem-based definitions of names.
Taxon (Pl., Taxa). A named group(s) of organisms.
Taxonomic Definition. A statement specifying the meaning of a name (i.e., the taxon to which the name refers).
1. TAXONOMY IN HISTORICAL CONTEXT
The discipline of taxonomy is concerned with identifying significant groups of organisms, or taxa, and giving them scientific names that can be used to facilitate communication about these organisms and their features. Whether in evolutionary theory, public policy, or conservation, there is no doubt that what groups of organisms we recognize as taxa matters. Taxa are routinely discussed not only with reference to conservation status but also in patents for biotic compounds or in the assessment and governance of public health risks. Assessment of phylogenetic diversity is important to developing conservation priorities. This is one of the main reasons taxonomy now strives to ensure that the groups recognized as taxa are those united by evolutionary relatedness—but this was not always the case.
In the eighteenth and nineteenth centuries, especially in Europe, there was a penchant for ordering (and reordering) the natural world as a way to organize knowledge of living organisms, many of which were newly known to Western science. The resulting classification systems emphasized particular attributes or ecological factors used to determine taxon membership. At this time, taxa were viewed as static and divinely determined groups of organisms. It was in this environment that Carl Linnaeus (1707–1778) proposed and refined an all-encompassing system of classification or taxonomy. Linnaeus’s taxonomy was composed of named groups of organisms, taxa (singular = taxon), arranged in relation to one another. In addition to proposing a taxonomic system (the organization of organisms into ranked categories by different character systems), Linnaeus also developed a system of nomenclature: a system of rules governing taxon names.
Linnaeus recognized five ranks of taxa from species up to class and brought into broad use a binomial (two-part) name for species, consisting of a genus name combined with a species epithet. The composition of taxa was determined by the presence of characteristics considered to define the taxon. Different character systems were thought to naturally distinguish different categories of taxa at distinct ranks. The hope was that, by using a limited number of key features or clusters of features, it would be possible to classify all known life, at that time a few thousand described species.
The taxonomic endeavor started by Linnaeus quickly took hold, becoming the focal point of natural history for at least 200 years, although his particular taxonomic scheme was largely revised. The nomenclatural system he initiated was accepted but greatly expanded to accommodate the great diversity of species found by explorers in the nineteenth century. Separate nomenclatural codes eventually formalized the rules and governance of taxonomic systems for the naming of bacteria (BC), plants (ICBN), and animals (ICZN). All took the Linnaean system as their base and were thus focused on ways to name ranked taxa whose membership was determined by defining characters.
2. INCORPORATING AN EVOLUTIONARY PERSPECTIVE
The publication of The Origin by Charles Darwin in 1859 signaled a profound revolution in natural history with the realization that living organisms are linked by descent from common ancestry. Darwin articulated the view that while taxa may be identified based on their distinctive characteristics, the taxa we wish to discuss share characteristics by virtue of evolutionary history. Two members of a taxon should be more closely related to each other than to any organism not a part of that taxon. The centrality of defining characters present in all members of a class is incompatible with the mutability fundamental to evolution; however, a shift in nomenclatural approach did not immediately occur.
The hierarchical aspects of the Linnaean system as a whole seemed to fit well with the nested relationships implied by a single tree of life; however, the Linnaean system was built on taxonomic rank and the idea that distinct kinds of characters (e.g., reproductive, locomotory) characterize different ranks. Subsequent naturalists determined that taxa at each rank in the Linnaean hierarchy do not share any essential properties that could allow ranks to be recognized as natural entities. A family of plants, for example, does not share special family-category properties with a family of birds, or even with other families of plants.
In the twentieth century, evolutionary taxonomy, which arose with the modern synthesis of the 1940s, advocated the application of names with consideration of shared history but also an emphasis on certain characteristics. By contrast, Willi Hennig (1913–1976) and others working around the same time emphasized that discovering and naming of monophyletic groups (= clades) of organisms should supersede the emphasis on characteristics. Such a perspective resulted from an interest in discovering and communicating about groups of organisms related by virtue of common descent (monophyletic groups or clades).
As discussed in chapter II.1, monophyletic groups have the property that its members share a more recent common ancestor with each other than with any organism outside the group. The phylogenetic approach contrasts with evolutionary taxonomy and older approaches that would allow for the recognition of groups unified by collections of characters not due to shared ancestry (polyphyletic groups) or taxa that exclude descendants that have lost or transformed particular features (paraphyletic groups). For example, evolutionary taxonomists accepted the utility of a concept of a taxon Reptilia that included crocodiles, lizards, snakes, and extinct apparently “reptile-like” dinosaurs but excluded birds. While this concept may seem intuitive, it actually communicates less about the natural world than a taxon Reptilia that includes one complete branch of the tree of life, a monophyletic group, rather than artificially excluding birds. Recognizing that birds are nested within Reptilia, specifically as most closely related to Crocodylia, has explanatory power and is useful for identifying biological questions of interest. For example, this relationship was recognized in large part based on bony characteristics (e.g., aspects of the skull such as an antorbital fenestra). The later recognition that both crocodilians and birds share parental care among Reptilia, could have been anticipated by a taxonomy that reflected monophyletic groups. Specifically knowing that dinosaurs include birds, and that crocodilians are most closely related to that clade, makes the discovery of parental care in dinosaurs not a surprise but predicted.
Over the decades following Willi Hennig’s seminal 1966 book, the importance of naming monophyletic groups of organisms (clades) became largely the consensus view. At first, the nested hierarchy of the Linnaean taxonomic system was thought to be easily translatable into a nested hierarchy, a taxonomy, communicating a particular phylogenetic hypothesis represented by more or less inclusive clades with ranks; however, such rank-based systems (or variants that used numerical annotations, or indented lists) did not allow a set of taxon names to be unambiguously adjusted in response to a new hypothesis of relationships. Also, there remained misleading nonequivalency of ranks and a lack of sufficient ranks to represent the tree of life. There were taxonomies based on phylogenies that used ranked names for clades of organisms but no system of nomenclature that was explicitly built on a phylogenetic framework.
In recent years a number of systematists have argued that the rank-based, Linnaean system of nomenclature has undesirable features if we equate taxa with monophyletic groups. The primary goal of biological nomenclature is to allow names to be assigned to taxa in such a way as to minimize ambiguity about content of the taxon and maximize stability over time. However, finding that one taxon is embedded within another of the same rank would, for example, require a change in name (at least the suffix used to indicate rank in the Linnaean system) of one or both taxa. Also, Linnaean systems require that new species be placed in sets of higher taxa regardless of the actual degree of known phylogenetic resolution. For example, perhaps new species L is known only to be a member of a large clade M previously identified as a class, but its specific relationships to subclades of M is unresolved based on the data available. Regardless, in order to avoid creating paraphyletic groups, L would need to be placed in an existing order, family, etc., or new taxa at these ranks would need to be named, all of which would imply more phylogenetic structure than the data supported.
3. SPECIES IN A PHYLOGENETIC FRAMEWORK
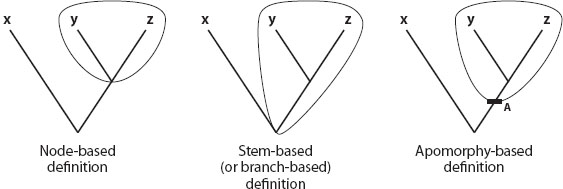
Although there were important precursors in preceding decades, proposal of a formal phylogenetic system of taxonomy dates to the early 1990s. De Queiroz and Gauthier proposed that what we want to name in the tree of life are clades, and that definitions of taxon names should explicitly reference ancestor-descendant relationships. To aid this enterprise they argued that the definitions of the names of taxa should be phylogenetic, proposing three main kinds of phylogenetic definitions of taxon names: node-based, stem-based, and apomorphy-based. All required that certain tips of the tree of life be specifiers, species or specimens that serve as referents in the definition of a name. For example, tips X-Y-Z (figure 1) are specifiers. Node-based definitions of clade names take the form: the most recent common ancestor of specifiers Y and Z and all of its descendants. Stem-based or branch-based definitions take the form: all taxa more closely related to Y (or Y and Z) than to X. Apomorphy-based definitions take the form: the most recent common ancestor that shares apomorphy A with Y and Z and all its descendants. For example, one proposed node-based phylogenetic definition of the taxon name Aves linking it to the most recent common ancestor of all extant birds and all of its descendants takes the general form: Aves is the name for the most recent common ancestor of carefully chosen species specifiers (the Andean Condor, Vultur gryphus; Great Tinamou, Tinamus major; and Ostrich, Struthio camelus) and all its descendants.
Figure 1. The three basic forms of a clade name definition in a phylogenetic frame (after De Queiroz and Gauthier 1992). Species or specimens X–Z and apomorphy A are specifiers in the (left to right) node-based, stem-based (or branch-based), and apomorphy-based definitions of the taxon names.
The PhyloCode (ICPN: International Code of Phylogenetic Nomenclature), under development since the late 1990s, is a code that formalizes the rules and recommendations of phylogenetic nomenclature and establishes an organizational structure overseeing the implementation of this practice. As of this writing, it is not yet formally in effect. The fundamental form of definitions for the names of taxa first outlined by De Queiroz and Gauthier is retained in the PhyloCode. As with other nomenclatural codes, the PhyloCode is not expected to dictate taxonomic practice, which groups should be recognized as taxa, but to provide rules to govern the names of taxa that are recognized. Under the Phylo-Code, a practitioner can name any group, even those that are nonmonophyletic. Furthermore, ranks can be used in conjunction with names defined phylogenetically, although unlike the traditional codes, the ranks are not part of the definitions of those names.
The PhyloCode includes rules governing publication of new names, conversion of existing names that were previously established (i.e., validly published and named) under the rank-based codes, as well as rules for priority (which of two names for the same taxon is correct) and synonymy (when two names apply to the same taxon). Other fundamentals of the system include requiring the formal registration of names and their associated phylogenetic definitions in a database, RegNum. A current draft of the code is downloadable from www.ohiou.edu/phylocode/ (or from phylocode.org).
Beginning in the 1990s, questions were raised about addressing the Linnaean species binomen, or two-part species name (e.g., Homo sapiens) in a phylogenetic system of nomenclature given that the first part of the binomen, genus, is a taxon of rank. Early drafts of the PhyloCode covered only clades and did not address species. It was noted that some species taxa are recognized by utilizing a criterion of monophyly, making them equivalent to clades; however, there was little consensus on how, or even if, species should be incorporated into the code, but it was generally recognized that a complete system of nomenclature based on phylogenetic principles would be expected to formally address species. Species figure centrally in the languages of evolutionary theory and public policy. They are the most numerous named taxa and commonly employed in many metrics of standing biodiversity. A diverse public is accessing knowledge about species daily, whereas it is often only specialists who are invested in the names defined for its major subclades.
The debate over the form that species names and their definitions should take in a phylogenetic system is linked to the extensive debate over the nature and importance of taxa recognized as species. Within any community of biologists there is always heterogeneity with respect to the concept of species (see chapter VI.1). Some taxonomists working in a phylogenetic framework have wanted to consider, discuss, and name only clades, and remove all discussion of species. Several of these authors consider that retention of species in a phylogenetic framework would conflict with the rank-free aspects of the system. Others have proposed that instead of “species,” we should focus on the least-inclusive taxonomic unit or LITU, the smallest clades below which there is no phylogenetic structure. Yet others simply wish to discuss clades that might approximate the content of traditional species, without according those clades any special status or named rank. The latter authors prefer to use Linnaean species epithets for small clades and name them using node- or branch-based definitions.
The community has debated how to convert species names under the PhyloCode, given that epithets or specific names (i.e., the second part of the binomen; e.g., sapiens) were never required to be unique in the rank-based codes. Consequently, species epithets have been used repeatedly in distinct clades of organisms, leading to concerns about homonymy (the same name being used for different taxa). The sheer number of existing species names, more than a million named under the rank-based codes, also presents a challenge if species names are to take a different form in a phylogenetic system, because all these names would have to be converted and registered in RegNum. Such conversion was ultimately deemed nonpracticable and undesirable given the large number of named species.
Based on this reasoning, it was established in a PhyloCode article that all new species names will be required to be validly established under the appropriate existing bacterial, botanical, or zoological rank-based codes; however, this PhyloCode article interprets an established species name within the context of a phylogenetic system. Under the approach adopted, the first part of a species binomen (called in the PhyloCode, the praenomen) is recommended to be a converted clade name. It does not need to be a converted genus name after the species name is validly established compliant with the appropriate rank-based code. After the establishing first publication, clade names can be used in combination with species names (the second part of the binomen) that are not genera in rank-based codes, or a species name can be used alone with further recommended identifying information (e.g., author and publication date).
Named species are heterogeneous entities recognized by an array of criteria, and in many cases they are not clades. The PhyloCode article interprets established species names in a phylogenetic system and provides additional recommendations for increased explicitness in taxonomic practice. Species are recognized as distinct biological entities from clades that can be identified by a broader array of criteria than monophyly alone. While this position does place limits on biologists who equate species only with clades and/or want to apply species epithets to unranked clades without discussing species, this route was adopted in the face of lack of consensus concerning a unified way to accommodate all the diverse interpretations of species within the PhyloCode. Debates are predictably ongoing concerning the equivalency of species taxa, their nature as biological entities (e.g., as taxa or functional units), their boundaries, and how they can be appropriately named.
4. CONCERNS ABOUT AND MISUNDERSTANDING OF PHYLOGENETIC NOMENCLATURE
Concerns about phylogenetic nomenclature have been diverse, including some based on a misunderstanding of the system, perhaps confused by changes in the system from its earliest forms to its ultimate articulation in the PhyloCode. Some authors appear to have been confused by the intentions of the system and its implications for taxon names established under other codes; however, the PhyloCode does not require that all existing taxon names be replaced with new names. Likewise, it does not enforce particular taxon concepts (e.g., require monophyly) or disallow ranks. Other critics have maintained that the PhyloCode intends to replace the rank-based codes. Although at some point in the distant future the phylogenetic community could decide this is the right decision, the PhyloCode is presently designed to function alongside the rank-based codes; indeed, it explicitly requires valid establishment of species names, which are also the most broadly accessed taxon names, under these codes.
Other objections to phylogenetic taxonomy and the PhyloCode have been philosophical. One argument in favor of rank-based codes is that the imprecision of the definitions of taxon names under these codes yields flexibility. By precisely tying a name to a particular clade with particular specifiers, the argument goes, we may discover that we have not applied a widely used name to the particular biological entity we most wish to discuss. Other phylogeneticists value the stability of knowing that a name will always refer to one specific set of ancestor-descendant relationships, even if the list of species it contains changes with new data.
5. THE FUTURE OF PHYLOGENETIC NOMENCLATURE
In some systematic communities, phylogenetic taxonomy is in broad use. These communities, however, are heterogeneous in the way they tend to deploy phylogenetic nomenclature. Some systematists prefer not to use apomorphy-based definitions; some reject the recommendation in the PhyloCode of applying widely used names to the living members (the “crown” group) of major clades. To some, larger questions remain contentious; for example, are complete definitions possible for biological entities that may have, by their nature, imprecise edges and boundaries? There has been much debate over how recognition of a proposed temporal framework for biological kinds may affect their properties.
While the formal publication of the PhyloCode and its start date are not yet firmly set, the community of phylogenetic taxonomists continues to increase. Some authors have noted the fit between this system of phylogenetic nomenclature and computer-based methods for tracking biodiversity (phyloinformatics). A given set of specifiers is sufficient for a computer to apply the definition of a name unambiguously with the input of a current estimate of phylogenetic relationships. To some in the systematic community, these properties are desirable; to others, the flexibility/imprecision of the rank-based codes is preferable, even though it means no way to automate taxonomic practice. It will be interesting to see how the differences of opinion are resolved in the future. The one thing we can be sure of is that systems of taxonomy and nomenclature will also continue to evolve as phylogenetic methods and the scope of the questions asked with them continue to expand.
FURTHER READING
Baum, D. A., and S. D. Smith. 2013. Tree Thinking: An Introduction to Phylogenetic Biology. Denver, CO: Roberts & Company. A recent systematic textbook that includes a chapter on taxonomy and nomenclature.
Barkley, T. M., P. DePriest, V. Funk, R. W. Kiger, W. J. Kress, and G. Moore. 2004. Linnaean nomenclature in the 21st century: A report from a workshop on integrating traditional nomenclature and phylogenetic classification. Taxon 53: 153–158. A recent attempt at reconciling phylogenetic systematics with rank-based codes (see further discussion in Laurin 2008)
Bryant, H. N., and P. D. Cantino. 2002. A review of criticisms of phylogenetic nomenclature: Is taxonomic freedom the fundamental issue? Biological Reviews of the Cambridge Philosophical Society 77: 39–55. A response to criticisms of phylogenetic nomenclature.
Cantino, P. D., and K. De Queiroz. 2010. International Code of Phylogenetic Nomenclature, version 4c. Downloadable from www.ohiou.edu/phylocode/. The current draft of the PhyloCode, with a preface describing its development and glossary.
De Queiroz, K., and J. Gauthier. 1992. Phylogenetic taxonomy. Annual Reviews of Ecology and Systematics 23: 449-480. The first in-depth address of the motivations for the proposal of a phylogenetic system of nomenclature and description of what such a system would look like.
Ereshefsky, M. 2002. The Poverty of the Linnaean Hierarchy. A Philosophical Study of Biological Taxonomy. Cambridge: Cambridge University Press. A nuanced yet accessible treatment of the philosophical issues with Linnaean taxonomy.
Hennig, W. 1966. Phylogenetic Systematics. Urbana: University of Illinois Press. A landmark contribution to systematics discussing the necessary centrality of phylogenetics in taxonomy.
Laurin, M. 2008. The splendid isolation of biological nomenclature. Zoologica Scripta 37: 223–233. A response to critiques of phylogenetic nomenclature.
Pleijel, F., and G. W. Rouse. 2003. Ceci n’est pas une pipe: Names, clades and phylogenetic nomenclature. Journal of Systematics and Evolutionary Research 41: 162–174. A discussion of phylogenetic nomenclature and one early proposal that species not be recognized but only named clades (includes LITU concept).
Rieppel, O. 2006. The PhyloCode: A critical discussion of its theoretical foundation. Cladistics 22: 186–197. A critical view of phylogenetic nomenclature from a philosophical perspective.