NOTES
2 Ralf Kittler, Manfred Kayser, and Mark Stoneking, “Molecular Evolution of
Pediculus Humanus and the Origin of Clothing,”
Current Biology 13:1414-1417 (2003). Other researchers have challenged a technical aspect of the paper. The challenge, if sustained, would suggest a considerably older date, perhaps up until 500,000 years ago, for the evolution of the body louse and the invention of clothing. David L. Reed et al., “Genetic Analysis of Lice Supports Direct Contact between Modern and Archaic Humans,”
Public Library of Science Biology 2:1972-1983 (2004); Nicholas Wade, “What a Story Lice Can Tell,”
New York Times, October 5, 2004, p. F1.
3 Feng-Chi Chen and Wen-Hsiung Li, “Genomic Divergences between Humans and Other Hominoids and the Effective Population Size of the Common Ancestor of Humans and Chimpanzees,”
American Journal of Human Genetics 68:444-456 (2001).
4 Pascal Gagneux et al., “Mitochondrial Sequences Show Diverse Evolutionary Histories of African Hominoids,”
Proceedings of the National Academy of Sciences 96:5077-5082 (1999).
5 Richard G. Klein,
The Human Career, 2nd edition, University of Chicago Press, 1999, p. 251. Unless otherwise specified, the paleoanthropological and archaeological facts in this chapter are mostly drawn either from this broad and lucid textbook, or from a more popular book that is based on it,
The Dawn of Human Culture by Richard G. Klein and Blake Edgar, John Wiley & Sons, 2002.
6 Chen and Li, “Genomic Divergences.” The number of DNA differences between two species depends on the size of the parent population and the number of generations for which the two species have been separate. If the generation time and the number of years since the split are known, geneticists can estimate what they call the “effective” population size. This is a theoretical population, which must be multiplied by a factor of two to five to get the census-size population.
7 P. S. Rodman, in
Adaptations for Foraging in Non-human Primates, Columbia University Press 1984, pp. 134-160, cited in Robert Foley,
Humans before Humanity, Blackwell, 1995, p. 140.
8 Richard G. Klein,
The Human Career, 2nd edition, University of Chicago Press, 1999, figure 8.3, p. 580.
9 Roger Lewin and Robert A. Foley,
Principles of Human Evolution, 2nd ed., Blackwell, 2004, p. 450.
10 Robert Foley,
Humans before Humanity, Blackwell, 1995, p. 170.
11 Richard G. Klein,
The Human Career, p. 292.
12 Richard Wrangham, “Out of the Pan, Into the Fire,” in Frans B. M. DeWaal, ed.,
Tree of Origin, Harvard University Press, 2001, p. 137.
13 Richard G. Klein and Blake Edgar,
The Dawn of Human Culture, Wiley, 2002, p. 100; Robert A. Foley, “Evolutionary Perspectives,” in W. G. Runciman, ed.,
The Origin of Human Social Institutions, Oxford University Press, 2001, pp. 171-196.
14 Richard G. Klein,
The Human Career, p. 292.
15 Charles Darwin,
The Descent of Man and Selection in Relation to Sex, 2nd edition, 1874, p. 58.
16 Mark Pagel and Walter Bodmer, “A Naked Ape Would Have Fewer Parasites,”
Proceedings of the Royal Society B (Suppl.) 270:S117-S119 (2003).
17 Rosalind M. Harding et al., “Evidence for Variable Selective Pressures at MC1R,”
American Journal of Human Genetics 66:1351-1361 (2000).
18 Nina G. Jablonski and George Chaplin, “The Evolution of Human Skin Coloration,”
Journal of Human Evolution 39:57-106 (2000).
19 Alan R. Rogers, David Iltis, and Stephen Wooding, “Genetic Variation at the MC1R Locus and the Time Since Loss of Human Body Hair,”
Current Anthropology 45:105-108 (2004).
20 Nina G. Jablonski and George Chaplin, “Skin,”
Scientific American 74:72-79 (2002).
21 Arthur H. Neufeld and Glenn C. Conroy, “Human Head Hair Is Not Fur,”
Evolutionary Anthropology 13:89 (2004); B. Thierry, “Hair Grows to Be Cut,”
Evolutionary Anthropology 14:5 (2005); Alison Jolly, “Hair Signals,”
Evolutionary Anthropology 14:5 (2005).
22 Hermelita Winter et al., “Human Type I Hair Keratin Pseudogene phihHaA Has Functional Orthologs in the Chimpanzee and Gorilla: Evidence for Recent Inactivation of the Human Gene After the Pan-Homo Divergence.”
Human Genetics 108:37-42 (2001).
23 R. X. Zhu et al., “New Evidence on the Earliest Human Presence at High Northern Latitudes in Northeast Asia,”
Nature 431:559-562 (2004).
24 Robert Foley,
Humans before Humanity, Blackwell, 1995, p. 75.
25 Richard G. Klein, “Archeology and the Evolution of Human Behavior,”
Evolutionary Anthropology 9(1):17-36 (2000).
26 Richard Klein and Blake Edgar,
The Dawn of Human Culture, p. 192.
27 Richard Klein,
The Human Career, p. 512.
28 Ian McDougall et al., “Stratigraphic Placement and Age of Modern Humans from Kibish, Ethiopia,”
Nature 433:733-736 (2005).
29 From a list of fifteen modern behaviors described by Paul Mellars in “The Impossible Coincidence—A Single-Species Model for the Origins of Modern Human Behavior in Europe,”
Evolutionary Anthropology 14:12-27 (2005).
30 Richard Klein,
The Human Career, p. 492.
31 Sally McBrearty and Alison S. Brooks, “The Revolution That Wasn’t: A New Interpretation of the Origin of Modern Human Behavior,”
Journal of Human Evolution 39:453-563 (2000).
32 Christopher Henshilwood et al., “Middle Stone Age Shell Beads from South Africa,”
Science 304:404 (2004).
33 James M. Bowler et al., “New Ages for Human Occupation and Climatic Change at Lake Mungo, Australia,”
Nature 421:837-840 (2003).
34 Michael C. Corballis, “From Hand to Mouth: The Gestural Origins of Language,” in Morten H. Christiansen and Simon Kirby,
Language Evolution, Oxford University Press, 2003, pp. 201-219.
35 Marc D. Hauser,
The Evolution of Communication, MIT Press, 1996, p. 309.
37 Steven Pinker,
The Language Instinct, William Morrow, 1994, p. 339.
38 Marc D. Hauser, Noam Chomsky, and W. Tecumseh Fitch, “The Faculty of Language: What Is It, Who Has It, and How Did It Evolve?”
Science 298:1569-1579 (2002).
39 Derek Bickerton,
Language and Species, University of Chicago Press, 1990, p. 105.
40 Ray Jackendoff,
Foundations of Language, Oxford University Press, 2002, p. 233.
41 Frederick J. Newmeyer, “What Can the Field of Linguistics Tell Us about the Origins of Language?” in
Language Evolution, by Morten H. Christiansen and Simon Kirby, Oxford University Press, 2003, p. 60.
42 Nicholas Wade, “Early Voices: The Leap of Language,”
New York Times, July 15, 2003, p. F1.
43 Steven Pinker, e-mail, June 16, 2003, quoted in part in Wade, “Early Voices.”
44 Steven Pinker and Paul Bloom, “Natural Language and Natural Selection,”
Behavioral and Brain Sciences 13(4):707-784 (1990).
45 Derek Bickerton,
Language and Species, p. 116.
46 Ray Jackendoff,
Foundations of Language, p. 240.
47 Ann Senghas, Sogaro Kita, and Asli Özyürek, “Children Creating Core Properties of Language: Evidence from an Emerging Sign Language in Nicaragua,”
Science 305:1779-1782 (2004).
48 Wendy Sandler et al., “The Emergence of Grammar: Systematic Structure in a New Language,”
Proceedings of the National Academy of Sciences 102:2661-2665 (2005).
49 Nicholas Wade, “Deaf Children’s Ad Hoc Language Evolves and Instructs,”
New York Times, September 21, 2004, p. F4.
50 Michael C. Corballis, “From Hand to Mouth,” p. 205.
51 Louise Barrett, Robin Dunbar, and John Lycett,
Human Evolutionary Psychology, Princeton University Press, 2002.
52 Geoffrey Miller,
The Mating Mind, Random House, 2000.
53 Steven Pinker, “Language as an Adaptation to the Cognitive Niche,” in Morten H. Christiansen and Simon Kirby,
Language Evolution, Oxford University Press, 2003, p. 29.
54 Paul Mellars, “Neanderthals, Modern Humans and the Archaeological Evidence for Language,” in Nina G. Jablonski and Leslie C. Aiello, eds.,
The Origin and Diversification of Language, California Academy of Sciences, 1988, p. 99.
55 Faraneh Vargha-Khadem, interview, October 1, 2001.
56 Faraneh Vargha-Khadem, Kate Watkins, Katie Alcock, Paul Fletcher, and Richard Pass ingham, “Praxic and Nonverbal Cognitive Deficits in a Large Family with a Genetically Transmitted Speech and Language Disorder
,” Proceedings of the National Academy of Sciences 92:930-933 (1995).
57 Faveneh Vargha-Khadem et al., “Neural Basis of an Inherited Speech and Language Disorder,”
Proceedings of the National Academy of Science 95:12659-12700 (1998).
58 Many genes were first discovered by biologists working with fruit flies, among whom it is a point of pride to give genes colorful names. These odd names are often adopted for the equivalent gene when it is discovered in humans, apart from names like
dunce or
rutabaga that are deemed too colorful. The forkhead gene was so named because when it is mutated, the fly larvae develop spiky structures on their head. Other genes of similar structure were found, and all turned out to have a signature section of DNA, called the forkhead box, which specified a region in the forkhead protein that binds to specific sequence of DNA. This is because the forkhead proteins control the activity of other genes by binding to regions of DNA just upstream of genes. Humans have turned out to possess a large family of forkhead box genes, the subgroups of which are named by letters of the alphabet. FOXP2 is the second member of the P group of the family of forkhead box genes. Gary F. Marcus and Simon F. Fisher, “FOXP2 in Focus: What Can Genes Tell Us About Speech and Language?”
Trends in Cognitive Sciences 7:257-262 (2003).
59 Cecilia S. L. Lai, Simon E. Fisher, Jane A. Hurst, Faraneh Vargha-Khadem, and Anthony P. Monaco, “A Forkhead-Domain Gene Is Mutated in a Severe Speech and Language Disorder,”
Nature 413:519-523 (2001).
60 Faraneh Vargha-Khadem et al., “FOXP2 and the Neuroanatomy of Speech and Language,”
Nature Reviews Neuroscience 6:131-138 (2005).
61 Wolfgang Enard et al., “Molecular Evolution of FOXP2, a Gene Involved in Speech and Language,”
Nature 418:869-872 (2002).
62 Svante Pääbo, interview, August 10, 2002.
63 Richard Klein,
The Human Career, University of Chicago Press, 1999, p. 492.
64 The reason is probably that a rivalry between mitochondria inside the cell would be too disruptive. The sperm’s mitochondria are made to carry a special chemical tag that says “kill me.” As soon as the sperm enters the egg, its mitochondria are destroyed. The egg possesses about 100,000 mitochondria of its own, and has no need for the mere 100 or so contributed by the sperm. Douglas C. Wallace, Michael D. Brown, and Marie T. Lott, “Mitochondrial DNA Variation in Human Evolution and Disease,”
Gene, 238:211-230 (1999).
65 Peter A. Underhill et al., “Y Chromosome Sequence Variation and the History of Human Populations,”
Nature Genetics, 26:358-361 (2000).
66 S. T. Sherry, M. A. Batzer, and H. C. Harpending, “Modeling the Genetic Architecture of Modern Populations,”
Annual Review of Anthropology 27:153-169 (1968).
67 Jonathan K. Pritchard, Mark T. Seielstad, Anna Perez-Lezaun, and Marcus W. Feldman, “Population Growth of Human Y Chromosomes: A Study of Y Chromosome Micro-satellites,”
Molecular Biology and Evolution 16:1791-1798 (1999).
68 P. A. Underhill, G. Passarino, A. A. Lin, P. Shen, M. Mirazon-Lahr, R. A. Foley, P. J. Oefner, and L. L. Cavalli-Sforza, “The Phylogeography of Y Chromosome Binary Haplotypes and the Origins of Modern Human Populations,”
Annals of Human Genetics 65:43-62 (2001).
69 S. T. Sherry et al., “Modeling the Genetic Architecture of Modern Populations,” p. 166.
70 Richard Borshay Lee,
The !Kung San, Cambridge University Press, 1979, p. 31.
71 Tom Güldemann and Rainer Vossen, “Khoisan,” in Bernd Heine and Derek Nurse, eds.,
African Languages, Cambridge University Press, 2000.
72 Yu-Sheng Chen et al., “mtDNA Variation in the South African Kung and Khwe—and Their Genetic Relationships to Other African Populations,”
American Journal of Human Genetics 66:1362-1383 (2000). The team sampled a group of !Kung from the northwestern Kalahari Desert known as the Vasikela !Kung.
73 Alec Knight et al., “African Y Chromosome and mtDNA Divergence Provides Insight into the History of Click Languages,”
Current Biology 13:464-473 (2003).
74 Nicholas Wade, “In Click Languages, an Echo of the Tongues of the Ancients,”
New York Times, March 18, 2003, p. F2.
75 Ornella Semino et al., “Ethiopians and Khoisan Share the Deepest Clades of the Human Y-Chromosome Phylogeny,”
American Journal of Human Genetics 70:265-268 (2002).
76 Donald E. Brown,
Human Universals, McGraw-Hill, 1991, p. 139.
77 Henry Harpending and Alan R. Rogers, “Genetic Perspectives on Human Origins and Differentiation,”
Annual Review of Genomics and Human Genetics 1:361-385 (2000).
78 Richard Borshay Lee,
The !Kung San, Cambridge University Press, 1979, p. 135.
79 Jon de la Harpe et al., “Diamphotoxin, the Arrow Poison of the !Kung Bushmen,”
Journal of Biological Chemistry 258:11924-11931 (1983).
80 Richard Borshay Lee,
The !Kung San, p. 440.
81 Nancy Howell,
Demography of the Dobe !Kung, Academic Press, 1979, p. 119.
82 Steven A. LeBlanc,
Constant Battles, St. Martin’s Press, 2003, p. 116.
83 Lawrence H. Keeley,
War before Civilization, Oxford University Press, 1996, p. 134.
84 Richard Borshay Lee,
The !Kung San, p. 399. The odd symbols represent different kinds of click.
85 Frank W. Marlowe, “Hunter-Gatherers and Human Evolution,”
Evolutionary Anthropology 14:54-67 (2005).
86 M. Siddall et al., “Sea-Level Fluctuations during the Last Glacial Cycle,”
Nature 423:853-858.
88 Sarah A. Tishkoff and Brian C. Verrelli, “Patterns of Human Genetic Diversity: Implications for Human Evolutionary History and Disease,”
Annual Review of Genomics and Human Genetics 4:293-340 (2003).
89 Jeffrey I. Rose, “The Question of Upper Pleistocene Connections between East Africa and South Arabia,”
Current Anthropology 45:551-555 (2004).
90 Luis Quintana-Murci et al., “Genetic Evidence of an Early Exit of Homo sapiens from Africa through Eastern Africa,”
Nature Genetics 23:437-441 (1999).
91 Martin Richards et al., “Extensive Female-Mediated Gene Flow from Sub-Saharan Africa into Near Eastern Arab Populations,”
American Journal of Human Genetics 72:1058-1064 (2003).
92 John D. H. Stead and Alec J. Jeffreys, “Structural Analysis of Insulin Minisatellite Alleles Reveals Unusually Large Differences in Diversity between Africans and Non-Africans,”
American Journal of Human Genetics 71:1273-1284 (2002).
93 Nicholas Wade, “To People the World, Start with 500,”
New York Times, November 11, 1997, p. F1.
94 Vincent Macaulay et al., “Single, Rapid Coastal Settlement of Asia Revealed by Analysis of Complete Mitochondrial Genomes,”
Science 308:1034-1036 (2005). The geneticists decoded the full mitochondrial DNA of people belonging to the lineages M and N, which are the earliest ones found outside Africa, and compared them with L3, the lineage inside Africa from which M and N are derived. Knowing the rate at which mutations occur in mitochondrial DNA, they calculated it would have taken 826 generations, or 20,650 years, for the M and N lineages to have evolved from L3. The time for this process to occur depends on the population size, and knowledge of the time allows the initial population to be estimated. The answer is 550 women of breeding age if the population remained the same size throughout the 20,000 years, and less than that if the population expanded, as was doubtless the case.
95 T. Kivisild et al., “The Genetic Heritage of the Earliest Settlers Persists Both in Indian Tribal and Caste Populations,”
American Journal of Human Genetics 72:313-332 (2003).
96 Richard G. Roberts et al., “New Ages for the Last Australian Megafauna: Continent-Wide Extinction About 46,000 Years Ago,”
Science 292:1888-1892 (2001).
97 Vincent Macaulay et al., “Single, Rapid Coastal Settlement of Asia.”
98 Kirsi Huoponen, Theodore G. Schurr, Yu-Sheng Chen, and Douglas C. Wallace, “Mitochondrial DNA Variation in an Aboriginal Australian Population: Evidence for Genetic Isolation and Regional Differentiation,”
Human Immunology 62:954-969 (2001).
99 Max Ingman and Ulf Gyllensten, “Mitochondrial Genome Variation and Evolutionary History of Australian and New Guinean Aborigines,”
Genome Research 13:1600-1606 (2003).
100 Steven A. LeBlanc,
Constant Battles, St. Martin’s Press, 2003, p. 121.
101 Manfred Kayser et al., “Reduced Y-Chromosome, but Not Mitochondrial DNA, Diversity in Human Populations from West New Guinea,”
American Journal of Human Genetics 72:281-302 (2003).
102 Steven A. LeBlanc,
Constant Battles, p. 151.
103 Nicholas Wade, “An Ancient Link to Africa Lives On in the Bay of Bengal,”
New York Times, December 10, 2002, p. A14.
104 Kumarasamy Thangaraj et al., “Genetic Affinities of the Andaman Islanders, a Vanishing Human Population,”
Current Biology 13:86-93 (2003).
105 Philip Endicott et al., “The Genetic Origins of the Andaman Islanders,”
American Journal of Human Genetics 72:1590-1593 (2003).
106 Madhusree Mukerjee,
The Land of Naked People, Houghton Mifflin, 2003, p. 240.
107 Stephen Oppenheimer,
The Real Eve, Carroll & Graf, 2003, p. 218.
108 David L. Reed et al., “Genetic Analysis of Lice Supports Direct Contact Between Modern and Archaic Humans,”
Public Library of Science Biology 2:1972-1983 (2004); Nicholas Wade, “What a Story Lice Can Tell,”
New York Times, October 5, 2004, p. F1. The second cluster of lice is found only in the Americas, suggesting that modern people contracted them from
Homo erectus in East Asia or Siberia before crossing the Pleistocene-epoch land bridge that joined Siberia to North America. This lineage of lice may have been the dominant kind in the Americas until European colonists brought over the lineage standard in the rest of the world.
109 P. Brown et al., “A New Small-Bodied Hominin from the Late Pleistocene of Flores, Indonesia,”
Nature 431:1055-1091 (2004); Nicholas Wade, “New Species Revealed: Tiny Cousins of Humans,”
New York Times, October 28, 2004, p. A1; Nicholas Wade, “Miniature People Add Extra Pieces to Evolutionary Puzzle,”
New York Times, November 9, 2004, p. F2. M. J. Morwood et al., “Further evidence for small-bodied hominims from the Late Pleistocene of Flores, Indonesia,”
Nature 437, 1012-1017, 2005.
110 Roger Lewin and Robert A. Foley,
Principles of Human Evolution, Blackwell Science, 2004, p. 387.
111 Richard G. Klein,
The Human Career, 2nd ed., University of Chicago Press, 1999, p. 470.
112 Christopher Stringer and Robin McKie,
African Exodus, Henry Holt, 1996, 1999, p. 106.
113 Richard G. Klein,
The Human Career, p. 477.
114 Matthias Krings et al., “Neanderthal DNA Sequences and the Origin of Modern Humans,”
Cell 90:19-30, 1997.
115 David Serre et al., “No Evidence of Neanderthal mtDNA Contribution to Early Modern Humans,”
Public Library of Science Biology 2:1-5 (2004).
116 A new theory of human origins proposes that the modern humans leaving Africa initially interbred with archaic humans as the emigrants’ wave of advance engulfed the archaic populations. The theory predicts that a majority of sites on the nuclear genome may have some archaic alleles. Vinyarak Eswaran, Henry Harpending, and Alan R. Rogers, “Genomics Refutes an Exclusively African Origin of Humans,” J
ournal of Human Evolution 49:1-18 (2005).
117 The Aurignacian tools at this site now seem to have been made by modern humans who occupied the site in between periods of Neanderthal occupation. Brad Gravina, Paul Mellars and Christopher Bronk Ramsey, “Radiocarbon Dating of Interstratified Neanderthal and Early Modern Human Occupations at the Chatelperronian Type-Site,”
Nature 438:51-56 (2005).
118 Martin Richards, “The Neolithic Invasion of Europe,”
Annual Review of Anthropology 32:135-162 (2003).
119 Ofer Bar-Yosef, “The Upper Paleolithic Revolution,”
Annual Review of Anthropology 31:363-393 (2002).
120 Agnar Helgason et al., “An Icelandic Example of the Impact of Population Structure on Association Studies,”
Nature Genetics 37:90-95, 2005; Nicholas Wade, “Where Are You From? For Icelanders, the Answer Is in the Genes,”
New York Times, December 28, 2004, p. F3.
121 Patrick D. Evans et al., “Microcephalin, a Gene Regulating Brain Size, Continues to Evolve Adaptively in Humans,”
Science 309:1717-1220 (2005); Nitzan Mekel-Bobrov, “Ongoing Adaptive Evolution of ASPM, a Brain Size Determinant in Homo sapiens,”
Science 309:1720-1722 (2005).
122 Nicholas Wade, “Brain May Still Be Evolving, Studies Hint,”
New York Times, September 9, 2005, p. A14.
123 Steve Dorus et al., “Accelerated Evolution of Nervous System Genes in the Origin of Homo sapiens,”
Cell 119:1027-1040 (2004).
124 Richard G. Klein,
The Human Career, 2nd edition, University of Chicago Press, 1999, p. 540.
125 Martin Richards et al., “Tracing European Founder Lineages in the Near Eastern mtDNA Pool,”
American Journal of Human Genetics 67:1251-1276 (2000); Martin Richards, “The Neolithic Invasion of Europe,”
Annual Review of Anthropology 32:135-162 (2003).
126 Antonio Torroni et al., “A Signal, from Human mtDNA, of Postglacial Recolonization in Europe,”
American Journal of Human Genetics 69:844-852 (2001).
127 Ornella Semino et al., “The Genetic Legacy of Paleolithic
Homo sapiens sapiens in Extant Europeans: A Y Chromosome Perspective,”
Science 290:1155-1159, 2000.
128 Colin Renfrew, “Commodification and Institution in Group-Oriented and Individualizing Societies,” in
The Origin of Human Social Institutions, Oxford University Press, 2001, p. 114.
129 Carles Vilà et al., “Multiple and Ancient Origins of the Domestic Dog,”
Science 276:1687-1689 (1997).
130 Peter Savolainen et al., “Genetic Evidence for an East Asian Origin of Domestic Dogs,”
Science 298:1610-1616 (2002).
131 Lyudmila N. Trut, Early Canid Domestication: The Farm-Fox Experiment,”
American Scientist 17:160-169 (1999).
132 Brian Hare, Michelle Brown, Christine Williamson, and Michael Tomasello, “The Domestication of Social Cognition in Dogs,”
Science 298:1634-1636 (2002).
133 Nicholas Wade, “From Wolf to Dog, Yes, but When?”
New York Times, November 22, 2002, p. A20.
134 Jennifer A. Leonard et al., “Ancient DNA Evidence for Old World Origin of New World Dogs,”
Science 298:1613-1616 (2002).
135 Joseph H. Greenberg,
Language in the Americas, Stanford University Press, 1987, p. 43.
136 Vivian Scheinsohn, “Hunter-Gatherer Archaeology in South America,”
Annual Review of Anthropology 32:339-361 (2003).
137 Yelena B. Starikovskaya et al., “mtDNA Diversity in Chukchi and Siberian Eskimos: Implications for the Genetic History of Ancient Beringia and the Peopling of the New World,”
American Journal of Human Genetics 63:1473-1491 (1998).
138 Mark Seielstad et al., “A Novel Y-Chromosome Variant Puts an Upper Limit on the Timing of First Entry into the Americas,”
American Journal of Human Genetics 73:700-705 (2003).
139 Maria-Catira Bortolini et al., “Y-Chromosome Evidence for Differing Ancient Demographic Histories in the Americas,”
American Journal of Human Genetics 73:524-539 (2003).
140 Michael D. Brown et al., “mtDNA Haplogroup X: An Ancient Link between Europe/ Western Asia and North America?”
American Journal of Human Genetics 63:1852-1861 (1998).
141 Ripan S. Malhi and David Glenn Smith, “Haplogroup X Confirmed in Prehistoric North America,”
American Journal of Physical Anthropology 119:84-86 (2002).
142 Eduardo Ruiz-Pesini et al., “Effects of Purifying and Adaptive Selection on Regional Variation in Human mtDNA,”
Science 303:223-226; Dan Mishmar et al., “Natural Selection Shaped Regional mtDNA Variation in Humans.”
Proceedings of the National Academy of Sciences 100:171-176 (2003).
143 Mark T. Seielstad, Erich Minch, and L. Luca Cavalli-Sforza, “Genetic Evidence for a Higher Female Migration Rate in Humans,”
Nature Genetics 20:278-280 (1998).
144 Kennewick Man, the 9,000-year-old, non-Mongoloid-looking skeleton discovered in Washington state and claimed by sinodont American Indians as their intimate ancestor, is a sundadont.
145 Marta Mirazón Lahr,
The Evolution of Modern Human Diversity, Cambridge University Press, 1996, p. 318.
146 Richard G. Klein,
The Human Career, p. 502.
147 Ofer Bar-Yosef, “On the Nature of Transitions: The Middle to Upper Paleolithic and the Neolithic Revolution,”
Cambridge Journal of Archaeology 8:(2):141-163 (1998).
148 Ofer Bar-Yosef, “From Sedentary Foragers to Village Hierarchies,”
The Origin of Human Social Institutions, Oxford University Press, 2001, p. 7.
149 Peter M. M. G. Akkermans and Glenn M. Schwartz,
The Archaeology of Syria, Cambridge University Press, 2003, p. 45.
150 Brian M. Fagan,
People of the Earth, 10th ed., Prentice Hall, 2001, p. 226.
151 The evidence included a Natufian skeleton with a Natufian type arrow point embedded in the thoracic vertebrae. Fanny Bocquentin and Ofer Bar-Yosef, “Early Natufian Remains: Evidence for Physical Conflict from Mt. Carmel, Israel,”
Journal of Human Evolution 47:19-23 (2004).
152 Akkermans and Schwartz,
The Archaeology of Syria, p. 96.
153 Colin Renfrew, “Commodification and Institution in Group-Oriented and Individualizing Societies,” in W. G. Runciman, ed.,
The Origin of Social Institutions, Oxford University Press, 2001, p. 95.
154 Natalie D. Munro, “Zooarchaeological Measures of Hunting Pressure and Occupation Intensity in the Natufian,”
Current Anthropology 45:S5-33 (2004).
155 Akkermans and Schwartz,
The Archaeology of Syria, p. 70.
156 Daniel Zohary and Maria Hopf,
Domestication of Plants in the Old World, 3rd ed., Oxford University Press, 2000, p. 18.
157 Robin Allaby, “Wheat Domestication,” in
Archaeogenetics: DNA and the Population Prehistory of Europe, McDonald Institute for Archaeological Research, 2000, pp. 321-324.
158 Francesco Salamini et al., “Genetics and Geography of Wild Cereal Domestication in the Near East,”
Nature Reviews Genetics 3:429-441 (2002).
159 Manfred Heun et al., “Site of Einkorn Wheat Domestication Identified by DNA Fingerprinting,”
Science 278:1312-1314 (1997); Daniel Zohary, and Maria Hopf,
Domestication of Plants in the Old World, p. 36.
160 Francesco Salamini et al., “Genetics and Geography of Wild Cereal Domestication in the Near East.”
161 Christopher S. Troy et al., “Genetic Evidence for Near-Eastern Origins of European Cattle,”
Nature 410:1088-1091 (2001).
162 Carlos Vilà et al., “Widespread Origins of Domestic Horse Lineages,”
Science 291:474-477 (2001).
163 Ornella Semino et al., “The Genetic Legacy of Paleolithic
Homo sapiens sapiens in Extant Europeans: A Y Chromosome Perspective,”
Science 290:1155-1159 (2000).
164 Roy King and Peter A. Underhill, “Congruent Distribution of Neolithic Painted Pottery and Ceramic Figurines with Y-Chromosome Lineages,”
Antiquity 76:707-714 (2002).
165 Albano Beja-Pereira et al., “Gene-Culture Coevolution between Cattle Milk Protein Genes and Human Lactase Genes,”
Nature Genetics 35:311-315 (2003).
166 Nabil Sabri Enattah et al., “Identification of a Variant Associated with Adult-type Hypolactasia,”
Nature Genetics 30:233-237 (2002).
167 Todd Bersaglieri et al., “Genetic Signatures of Strong Recent Positive Selection at the Lactase Gene,”
American Journal of Human Genetics 74:1111-1120 (2004).
168 Charlotte A. Mulcare et al., “The T Allele of a Single-Nucleotide Polymorphism 13.9 kb Upstream of the Lactase Gene (LCT) (C-13.9kbT) Does Not Predict or Cause the Lactase-Persistence Phenotype in Africans,”
American Journal of Human Genetics 74:1102-1110 (2004).
169 Napoleon Chagnon, “Life Histories, Blood Revenge, and Warfare in a Tribal Population,”
Science 239:985-992 (1988).
170 Anne E. Pusey, in Frans B. M. de Waal, ed.,
Tree of Origin, Harvard University Press 2001, p. 21.
171 John C. Mitani, David P. Watts, and Martin N. Muller, “Recent Developments in the Study of Wild Chimpanzee Behavior,”
Evolutionary Anthropology 11: 9-25 (2002).
172 Anne E. Pusey,
Tree of Origin, p. 26
. 173 Julie L. Constable, Mary V. Ashley, Jane Goodall, and Anne E. Pusey, “Noninvasive Paternity Assignment in Gombe Chimpanzees,”
Molecular Ecology 10:1279-1300 (2001).
174 Anne Pusey, Jennifer Williams, and Jane Goodall, “The Influence of Dominance Rank on the Reproductive Success of Female Chimpanzees,”
Science 277:828-831 (1997).
175 A. Whiten et al., “Cultures in Chimpanzees,”
Nature 399:682 (1999).
176 W. C. McGrew, “Tools Compared,” in Richard W. Wrangham et al., eds.,
Chimpanzee Cultures, Harvard University Press, 1994, p. 25.
177 Richard W. Wrangham et al.,
Demonic Males, Houghton Mifflin, 1996, p. 226.
178 Some experts argue that chimps are derived from bonobos, but the direction of change makes no difference here.
179 Frans de Waal,
Our Inner Ape, Riverhead Books, 2005, p. 221.
180 John C. Mitani, David P. Watts, and Martin N. Muller, “Recent Developments in the Study of Wild Chimpanzee Behavior,”
Evolutionary Anthropology 11:9-25 (2002).
181 Napoleon A. Chagnon,
Yanomamo, 5th ed., Wadsworth, 1997, p. 189.
183 Lawrence H. Keeley,
War before Civilization, Oxford University Press, 1996, p. 174.
185 Steven A. LeBlanc,
Constant Battles, St. Martin’s Press, 2003, p. 8.
186 Simon Mead et al., “Balancing Selection at the Prion Protein Gene Consistent with Prehistoric Kurulike Epidemics,”
Science 300:640-643 (2003).
187 Edward O. Wilson,
On Human Nature, Harvard University Press, 1978, p. 114.
188 Louise Barrett, Robin Dunbar, and John Lycett,
Human Evolutionary Psychology, Princeton University Press, 2002, p. 64.
189 Napoleon A. Chagnon,
Yanomamo, 5th ed., Wadsworth, 1997, p. 76.
190 Napoleon Chagnon, “Life Histories, Blood Revenge, and Warfare in a Tribal Population,”
Science 239:985-992 (1988).
191 Napoleon A. Chagnon,
Yanomamo, p. 77.
192 David M. Buss,
Evolutionary Psychology, 2nd ed., Pearson Education, 2004, p. 257.
193 Robert L. Trivers, “The Evolution of Reciprocal Altruism,”
Quarterly Review of Biology 46:35-57, 1971.
194 Following Steven Pinker,
How the Mind Works, W. W. Norton, 1997, pp. 404-405.
195 Matt Ridley,
The Origins of Virtue, Viking, 1996, p. 197.
196 Paul Seabright, “The Company of Strangers: A Natural History of Economic Life,” Princeton University Press, 2004, p. 28.
197 Michael Kosfeld et al., “Oxytocin Increases Trust in Humans,”
Nature 435:673-676 (2005).
198 Roy A. Rappaport, “The Sacred in Human Evolution,”
Annual Review of Ecology and Systematics 2:23-44 (1971).
199 Joyce Marcus and Kent V. Flannery, “The Coevolution of Ritual and Society: New
14C Dates from Ancient Mexico,”
Proceedings of the National Academy of Sciences 18252-18261 (2004).
200 Richard Sosis, “Why Aren’t We all Hutterites?”
Human Nature 14:91-127 (2003).
201 Edward O. Wilson,
On Human Nature, Harvard University Press, 1978, p. 175.
202 Frank W. Marlowe, “A Critical Period for Provisioning by Hadza Men: Implications for Pair Bonding,”
Evolution and Human Behavior 24:217-229 (2003).
203 Tim Birkhead,
Promiscuity, Harvard University Press, 2000, p. 41.
204 Nicholas Wade, “Battle of the Sexes is Discerned in Sperm,”
New York Times, February 22, 2000, p. F1.
205 Alan F. Dixson,
Primate Sexuality, Oxford University Press, 1998, p. 218.
206 Gerald J. Wyckoff, Wen Wang, and Chung-I Wu, “Rapid Evolution of Male Reproductive Genes in the Descent of Man,”
Nature 403:304-309 (2000).
207 Sperm generally survive for less than forty-eight hours in the human female reproductive tract, although survival times up to five days occasionally occur. Tim Birkhead,
Promiscuity, p. 67.
208 Robin Baker,
Sperm Wars, Basic Books, 1996, p. 38. Baker has contributed several novel findings to this field but some have not survived challenge by other researchers; see Birkhead, as cited, pp. 23-29.
209 W. H. James, “The Incidence of Superfecundation and of Double Paternity in the General Population,”
Acta Geneticae Medicae et Gemellologiae 42:257-262 (1993).
210 R. E. Wenk et al., “How Frequent is Heteropaternal Superfecundation?”
Acta Geneticae Medicae et Gemellologiae 41:43-47 (1992).
211 Louise Barrett, Robin Dunbar, and John Lycett,
Human Evolutionary Psychology, Princeton University Press, 2002, p. 181.
212 Bobbi S. Low,
Why Sex Matters, Princeton University Press, 2000, p. 80.
213 David M. Buss,
Evolutionary Psychology, 2nd ed., Pearson Education, 2004, p. 112.
215 Geoffrey Miller,
The Mating Mind: How Sexual Choice Shaped the Evolution of Human Nature, Doubleday, 2000.
216 Marta Mirazón Lahr,
The Evolution of Modern Human Diversity: A Study of Cranial Variation, Cambridge University Press, 1996, p. 263.
218 Marta Mirazón Lahr and Richard V. S. Wright, “The Question of Robusticity and the Relationship Between Cranial Size and Shape in
Homo sapiens,” Journal of Human Evolution 31:157-191 (1996).
219 Helen M. Leach, “Human Domestication Reconsidered,”
Current Anthropology 44:349-368 (2003).
222 Allen W. Johnson and Timothy Earle,
The Evolution of Human Societies, 2nd ed., Stanford University Press, 2000.
224 Derek V. Exner et al., “Lesser Response to Angiotensin-converting-enzyme Inhibitor Therapy in Black as Compared with White Patients with Left Ventricular Dysfunction,
New England Journal of Medicine 344:1351-1357 (2001).
225 Anne L. Taylor et al., “Combination of Isosorbide Dinitrate and Hydralazine in Blacks with Heart Failure,”
New England Journal of Medicine 351:2049-2057, 2004; Nicholas Wade, “Race-Based Medicine Continued,”
New York Times November 14, 2004, Section 4, p. 12.
226 Robert S. Schwartz, “Racial Profiling in Medical Research,”
New England Journal of Medicine 344:1392-1393 (2001).
227 “Genes, Drugs and Race,”
Nature Genetics 29:239 (2001).
229 Agnar Helgason et al., “An Icelandic Example of the Impact of Population Structure on Association Studies,”
Nature Genetics 37:90-95 (2005); Nicholas Wade, “Where Are You From? For Icelanders, the Answer Is in the Genes,”
New York Times, December 28, 2004, p. F3.
230 Rebecca L. Lamason et al., “SLC24A5, a Putative Cation Exchanger, Affects Pigmentation in Zebrafish and Humans,”
Science 310:1782-1786 (2005).
231 Noah A. Rosenberg et al., “Genetic Structure of Human Populations,”
Science 298:2381-2385 (2002).
232 Nicholas Wade, “Gene Study Identifies 5 Main Human Populations, Linking Them to Geography,”
New York Times, December 20, 2002, p. A37.
233 In the forensic system used in the United States, a suspect’s genome is analyzed at 13 specific sites and the number of repeats is counted. At any one site, there may be many people who possess the same number of repeats. Far fewer people have the same number at two sites. And the likelihood of any two people in the U.S. population having the same repeat number at all 13 sites is so small that everyone save identical twins is assumed to have their own unique set of repeats. (The number of repeats at each site is in fact a pair of numbers, one from the chromosome inherited from the mother, the other from the father’s, but they are often the same.)
234 Nicholas Wade, “For Sale: A DNA Test to Measure Racial Mix,”
New York Times, October 1, 2002, p. F4.
235 Nicholas Wade, “Unusual Use of DNA Aided in Serial Killer Search,”
New York Times, June 2, 2003, p. A28.
236 Richard Lewontin,
Human Diversity, W. H. Freeman, 1995, p. 123.
237 Statement adopted by the council of the American Sociological Association, August 9, 2002; available on
www.asanet.org.
238 American Anthropological Association statement on “Race,” May 17, 1998;
www.aaanet.org.
239 Quoted by Henry Harpending and Alan R. Rogers in “Genetic Perspectives on Human Origins and Differentiation,”
Annual Review of Genomics and Human Genetics 1:361-385 (2000).
240 David L. Hartl and Andrew G. Clark,
Principles of Population Genetics, 3rd ed., Sinauer Associates, 1997, p. 119.
241 Patrick D. Evans et al., “Microcephalin, a Gene Regulating Brain Size, Continues to Evolve Adaptively in Humans,”
Science 309:1717-1720 (2005).
242 Nitzan Mekel-Bobrov, “Ongoing Adaptive Evolution of ASPM, a Brain Size Determinant in
Homo sapiens,”
Science 309:1720-1722 (2005).
243 John H. Relethford, “Apportionment of Global Human Genetic Diversity Based on Craniometrics and Skin Color,”
American Journal of Physical Anthropology 118:393-398 (2002).
244 Henry Harpending and Alan R. Rogers, “Genetic Perspectives on Human Origins and Differentiation,” 1:380.
245 Nicholas Wade, “Race-Based Medicine Continued,”
New York Times, November 14, 2004, Section 4, p. 12.
246 Jon Entine,
Taboo, Public Affairs, 2000, p. 34.
248 Jon Entine, “The Straw Man of ‘Race,’”
World & I, September 2001, p. 309.
251 Jared Diamond,
Guns, Germs, and Steel, W. W. Norton, 1997, p. 25.
252 Richard Klein,
The Human Career, 2nd ed., University of Chicago Press, 1999, p. 502.
253 Robin I. M. Dunbar, “The Origin and Subsequent Evolution of Language,” in Morten H. Christiansen and Simon Kirby,
Language Evolution, Oxford University Press, 2003, p. 231.
255 Denis Mack Smith,
Medieval Sicily, Chatto & Windus, 1968, p. 71.
256 William A. Foley, “The Languages of New Guinea,”
Annual Review of Anthropology 29:357-404 (2000).
257 Jonathan Adams and Marcel Otte, “Did Indo-European Languages Spread before Farming?”
Current Anthropology 40:73-77 (1999).
258 Jared Diamond and Peter Bellwood, “Farmers and Their Languages: The First Expansions,”
Science 300:597-603 (2003).
259 Christopher Ehret, O. Y. Keita, and Paul Newman, “Origins of Afroasiatic,”
Science 306:1680-1681 (2004).
260 Christopher Ehret, “Language Family Expansions: Broadening Our Understandings of Cause from an African Perspective,” in Peter Bellwood and Colin Renfrew, eds.,
Examining the Farming/Language Dispersal Hypothesis, McDonald Institute for Archaeological Research, 2002, p. 173.
261 Colin Renfrew,
Archaeology and Language, Jonathan Cape, 1987.
262 L. Luca Cavalli-Sforza, Paolo Menozzi, and Alberto Piazza,
The History and Geography of Human Genes, Princeton University Press, 1994, pp. 296-299.
264 Marek Zvelebil, “Demography and Dispersal of Early Farming Populations at the Mesolithic-Neolithic Transition: Linguistic and Genetic Implications,” in Peter Bellwood and Colin Renfrew, ed.,
Examining the Farming/Language Dispersal Hypothesis, McDonald Institute for Archaeological Research, 2002, p. 381.
265 Colin Renfrew, April McMahon, and Larry Trask, eds.,
Time Depth in Historical Linguistics , McDonald Institute for Archaeological Research, 2000.
266 Bill J. Darden, “On the Question of the Anatolian Origin of Indo-Hittite,” in
Great Anatolia and the Indo-Hittite Language Family, Journal of Indo-European Studies Monograph No. 38, Institute for the Study of Man, 2001.
267 Mark Pagel, “Maximum-Likelihood Methods for Glottochronology and for Reconstructing Linguistic Phylogenies,” in Colin Renfrew, April McMahon, and Larry Trask, eds.,
Time Depth in Historical Linguistics, McDonald Institute for Archaeological Research, 2000, p. 198.
268 K. Bergsland and H. Vogt, “On the Validity of Glottochronology,”
Current Anthropology 3:115-153 (1962).
269 Russell D. Gray and Quentin D. Atkinson, “Language-Tree Divergence Times Support the Anatolian Theory of Indo-European Origin,”
Nature 426:435-439 (2003).
270 Peter Forster and Alfred Toth, “Toward a Phylogenetic Chronology of Ancient Gaulish, Celtic and Indo European,”
Proceedings of the National Academy of Sciences 100:9079-9084 (2003).
271 Bernd Heine and Derek Nurse, eds.,
African Languages, Cambridge University Press, 2000, p. 1.
272 Christopher Ehret, “Language and History,” in Bernd Heine and Derek Nurse, eds.,
African Languages, Cambridge University Press, 2000, pp. 272-298; “Testing the Expectations of Glottochronology against the Correlations of Language and Archaeology in Africa,” in Colin Renfrew, April McMahon, and Larry Trask, eds.,
Time Depth in Historical Linguistics, McDonald Institute for Archaeological Research, 2000, pp. 373-401.
273 Richard J. Hayward, “Afroasiatic,” in Bernd Heine and Derek Nurse, eds.,
African Languages , Cambridge University Press, 2000, pp. 79-98.
274 Christopher Ehret, “Language and History,” in Bernd Heine and Derek Nurse, eds.,
African Languages, Cambridge University Press, 2000, p. 290.
275 Nicholas Wade, “Joseph Greenberg, 85, Singular Linguist, Dies,”
New York Times, May 15, 2001, p. A23.
276 Joseph H. Greenberg, “Language in the Americas,” Stanford University Press, 1987.
277 L. Luca Cavalli-Sforza, Paolo Menozzi, and Alberto Piazza,
The History and Geography of Human Genes, Princeton University Press, 1994, pp. 317, 96.
279 L. L. Cavalli-Sforza, Eric Minch, and J. L. Mountain, “Coevolution of Genes and Languages Revisited,”
Proceedings of the National Academy of Sciences 89:5620-5624, (1992).
280 M. Lionel Bender, in Bernd Heine and Derek Nurse, eds.,
African Languages, Cambridge University Press, 2000, p. 54.
281 The example is adapted from Lyle Campbell,
Historical Linguistics, MIT Press, 1999, p. 111.
282 Richard Hayward, in Bernd Heine and Derek Nurse, eds.,
African Languages, Cambridge University Press, 2000, p. 86.
283 Nicholas Wade, “Scientist at Work: Joseph H. Greenberg; What We All Spoke When the World Was Young,”
New York Times, February 1, 2000, p. F1.
284 Joseph H. Greenberg,
Indo-European and Its Closest Relatives, Volume 1: Grammar, Stanford University Press, 2000, p. 217.
285 Joseph H. Greenberg,
Indo-European and Its Closest Relatives, Volume 2: Lexicon, Stanford University Press, 2002.
287 Nicholas Wade, “Joseph Greenberg, 85, Singular Linguist, Dies,”
New York Times, May 15, 2001, p. A23; Harold C. Fleming, “Joseph Harold Greenberg: A Tribute and an Appraisal,”
Mother Tongue: The Journal VI:9-27(2000-2001).
288 Johanna Nichols, “Modeling Ancient Population Structures and Movement in Linguistics,”
Annual Review of Anthropology, 26:359-384 (1979).
289 Mark Pagel, “Maximum-Likelihood Methods for Glottochronology and for Reconstructing Linguistic Phylogenies.”
290 Merritt Ruhlen,
The Origin of Language, Wiley, 1994, p. 115.
291 Tatiana Zerjal et al., “The Genetic Legacy of the Mongols,”
American Journal of Human Genetics 72:717-721 (2003).
292 ’Ala-ad-Din ’Ata-Malik Juvaini,
The History of the World Conqueror, trans. J. A. Boyle, Manchester University Press, 1958, vol. 2, p. 594.
293 Nicholas Wade, “A Prolific Genghis Khan, It Seems, Helped People the World,”
New York Times, February 11, 2003, p. F3.
294 Yali Xue et al., “Recent Spread of a Y-Chromosomal Lineage in Northern China and Mongolia,”
American Journal of Human Genetics, 77:1112-1116 (2005).
295 George Redmonds, interview, April 5, 2000.
296 Bryan Sykes,
Adam’s Curse, W. W. Norton, 2004, p. 7.
297 Bryan Sykes and Catherine Irven, “Surnames and the Y Chromosome,”
American Journal of Human Genetics 66:1417-1419 (2000).
298 Nicholas Wade, “If Biology Is Ancestry, Are These People Related?”
New York Times, April 9, 2000, Section 4, p. 4.
299 Bryan Sykes,
Adam’s Curse, p. 18.
300 Cristian Capelli et al., “AY Chromosome Census of the British Isles,”
Current Biology 13:979-984 (2003).
301 Emmeline W. Hill, Mark A. Jobling, and Daniel G. Bradley, “Y-chromosome Variation and Irish Origins,”
Nature 404: 351 (2000); Nicholas Wade, “Researchers Trace Roots of Irish and Wind Up in Spain,”
New York Times, March 23, 2000, p. A13; Nicholas Wade, “Y Chromosomes Sketch New Outline of British History,”
New York Times, May 27, 2003, p. F2.
302 James F. Wilson et al., “Genetic Evidence for Different Male and Female Roles During Cultural Transitions in the British Isles,”
Proceedings of the National Academy of Sciences 98:5078-5083 (2001).
303 Norman Davies
, The Isles, Oxford University Press, 1999, p. 3.
304 Benedikt Hallgrimsson et al., “Composition of the Founding Population of Iceland: Biological Distance and Morphological Variation in Early Historic Atlantic Europe,”
American Journal of Physical Anthropology 124:257-274 (2004).
305 Agnar Helgason et al., “Estimating Scandinavian and Gaelic Ancestry in the Male Settlers of Iceland,”
American Journal of Human Genetics 67:697-717 (2000).
306 Agnar Helgason et al., “mtDNA and the Origin of the Icelanders: Deciphering Signals of Recent Population History,”
American Journal of Human Genetics 66:999-1016 (2000).
307 Benedikt Hallgrimsson et al., “Composition of the Founding Population of Iceland.”
308 Nicholas Wade, “A Genomic Treasure Hunt May Be Striking Gold,”
New York Times, June 18, 2002, p. F1.
309 Agnar Helgason et al., “A Population-wide Coalescent Analysis of Icelandic Matrilineal and Patrilineal Genealogies: Evidence for a Faster Evolutionary Rate of mtDNA Lineages than Y Chromosomes,”
American Journal of Human Genetics 72:1370-1388 (2003).
310 M. F. Hammer et al., “Jewish and Middle Eastern Non-Jewish Populations Share a Common Pool of Y-chromosome Biallelic Haplotypes,”
Proceedings of the National Academy of Sciences 97:6769-6774 (2000).
311 Mark G. Thomas et al., “Founding Mothers of Jewish Communities: Geographically Separated Jewish Groups Were Independently Founded by Very Few Female Ancestors,”
American Journal of Human Genetics 70:1411-1420 (2002).
312 Harry Ostrer, “A Genetic Profile of Contemporary Jewish Populations,”
Nature Review Genetics 2:891-898 (2001).
313 Shaye J. D. Cohen,
The Beginnings of Jewishness, University of California Press, 1999, p. 303.
314 Denise Grady, “Father Doesn’t Always Know Best,”
New York Times, July 7, 1988, Section 4, p. 4.
316 Karl Skorecki et al., “Y Chromosomes of Jewish Priests,”
Nature 385:32 (1997).
317 Israel Finkelstein and Neil Asher Silberman,
The Bible Unearthed, Free Press, 2001, p. 98.
318 Mark G. Thomas et al., “Origins of Old Testament Priests,”
Nature 394:138-139 (1998).
319 James S. Boster, Richard R. Hudson, and Steven J. C. Gaulin, “High Paternity Certainties of Jewish Priests,”
American Anthropologist 111(4):967-971 (1999).
320 Doron M. Behar et al., “Multiple Origins of Ashkenazi Levites: Y Chromosome Evidence for Both Near Eastern and European Ancestries,”
American Journal of Human Genetics 73:768-779 (2003).
321 Nicholas Wade, “Geneticists Report Finding Central Asian Link to Levites,”
New York Times, September 27, 2003, p. A2.
322 Harry Ostrer, “A Genetic Profile of Contemporary Jewish Populations.”
323 Jared M. Diamond, “Jewish Lysosomes,”
Nature 368:291-292 (1994).
324 Neil Risch et al., “Geographic Distribution of Disease Mutations in the Ashkenazi Jewish Population Supports Genetic Drift over Selection,”
American Journal of Human Genetics 72:812-822 (2003).
325 Montgomery Slatkin, “A Population-Genetic Test of Founder Effects and Implications for Ashkenazi Jewish Diseases,”
American Journal of Human Genetics 75:282-293 (2004).
326 Gregory Cochran, Jason Hardy, and Henry Harpending, “Natural History of Ashkenazi Intelligence,”
Journal of Biosocial Science, in press (2005).
327 Melvin Konner,
Unsettled: An Anthropology of the Jews, Viking Compass, 2003, p. 198.
328 Merrill D. Peterson,
The Jefferson Image in the American Mind, Oxford University Press, 1960, p. 187.
329 Joseph J. Ellis,
American Sphinx: The Character of Thomas Jefferson, Knopf, 1997.
330 Annette Gordon-Reed,
Thomas Jefferson and Sally Hemings: An American Controversy, University of Virginia Press, 1997, p. 224.
331 Eugene A. Foster et al., “Jefferson Fathered Slave’s Last Child,”
Nature 396:27-28 (1998).
332 Nicholas Wade, “Defenders of Jefferson Renew Attack on DNA Data Linking Him to Slave Child,”
New York Times, January 7, 1999, p. A20.
333 Edward O. Wilson
, Consilience, Alfred A. Knopf 1998, p. 286.
334 The Chimpanzee Sequencing and Analysis Consortium, “Initial Sequence of the Chimpanzee Genome and Comparison with the Human Genome,”
Nature 436:69-87 (2005). The 99 percent agreement rests on a comparison of the human and chimp DNA sequences that directly correspond with one another. The human-chimp genome similarity falls to 96 percent after taking into account the insertions and deletions, i.e., stretches of DNA in one genome that have no counterpart in the other.
335 Louise Barrett, Robin Dunbar, and John Lycett,
Human Evolutionary Psychology, Princeton University Press, 2002, p. 12.
336 Mark Pagel, in
Encyclopedia of Evolution, p. 330.
337 Sarah A. Tishkoff et al., “Haplotype Diversity and Linkage Disequilibrium at Human G6PD: Recent Origin of Alleles That Confer Malarial Resistance,”
Science 293: 455-462 (2001).
338 J. Claiborne Stephens et al., “Dating the Origin of the CCR5-∆32 AIDS-Resistance Allele by the Coalescence of Haplotypes,”
American Journal of Human Genetics 62:1507-1515 (1998).
339 Alison P. Galvani and Montgomery Slatkin, “Evaluating Plague and Smallpox as Historical Selective Pressures for the CCR5-∆32 HIV-Resistance Allele,”
Proceedings of the National Academy of Sciences, 100:15276-15279 (2003).
340 Hreinn Stefansson et al., “A Common Inversion under Selection in Europeans,”
Nature Genetics 37:129-137 (2005); Nicholas Wade, “Scientists Find DNA Region That Affects Europeans’ Fertility,”
New York Times, January 17, 2005, p. A12.
341 Yoav Gilad et al., “Human Specific Loss of Olfactory Receptor Genes,”
Proceedings of the National Academy of Sciences 100:3324-3327 (2003).
342 Patrick D. Evans et al., “Microcephalin, a Gene Regulating Brain Size, Continues to Evolve Adaptively in Humans,”
Science 309:1717-1720 (2005).
343 Nitzan Mekel-Bobrov et al., “Ongoing Adaptive Evolution of ASPM, a Brain Size Determinant in
Homo sapiens,”
Science 309:1720-1722 (2005).
344 Li Zhisui,
The Private Life of Chairman Mao, Random House, 1994.
345 Quoted in Bobbi S. Low,
Why Sex Matters, Princeton University Press, 2000, p. 57.
346 Elizabeth A. D. Hammock and Larry J. Young, “Microsatellite Instability Generates Diversity in Brain and Sociobehavioral Traits,”
Science 308:1630-1634 (2005).
347 Richard E. Nisbett,
The Geography of Thought, Free Press (2003).
348 Victor Davis Hanson,
Carnage and Culture, Doubleday, 2001, p. 54.
349 Nicholas Wade, “Can It Be? The End of Evolution?”
New York Times, August 24, 2003, Section 4, p. 1.
350 Bobbi S. Low,
Sex, Wealth, and Fertility, in
Adaptation and Human Behavior, edited by Lee Cronk, Napeoleon Chagnon and William Irons, Walter de Gruyter, 2000, p. 340.
351 The reason is that before the generation of eggs or sperm, the chromosome inherited from the mother must align itself with the counterpart chromosome inherited from the father. For the alignment to take place successfully, the DNA of the two chromosomes must match fairly well throughout their length. If the chromosomes are too diverse, with too many different DNA units, they will not pair up correctly; viable sperm or eggs will not be created and the individual will be infertile. M. A. Jobling et al.,
Human Evolutionary Genetics, Garland, 2004, p. 434.
352 “Harmless” is one interpretation of the Ju|’hosansi’s name for themselves, as Elizabeth Marshall Thomas notes in a new book,
The Old Way. Thomas argues that their way of life was “the most successful culture that our kind has ever known,” as judged by its ecological stability and its endurance for at least 35,000 years. Her book’s vivid personal account of the !Kung’s hunter-gatherer lifestyle complements the anthropological study by Richard Borsay Lee. Elizabeth Marshall Thomas,
The Old Way—
A Story of the First People, Farrar Strauss Giroux, 2006.
353 Argument about which species the little Floresians should be assigned to has continued to seethe, with several researchers suggesting the skull is a pathologically small modern human. A new position, developed by researchers at the Australian National University and the University of Sydney, is that that Floresians are neither a pathological version of
Homo sapiens nor a downsized version of
Homeo erectus, but stem from an independent and much earlier migration out of Africa, perhaps before the island of Flores separated from the mainland. The argument is based on the skull’s similarity in brain size and other features to
Homo ergaster, the predecessor of
erectus. Debbie Argue, Denise Donlon, Colin Groves, and Richard Wright, “
Homo floresiensis: Microcephalic, pygmoid,
Australopithecus, or
Homo?,”
Journal of Human Evolution 51, 360-374 (2006).
354 The roots of the Aurignacian culture are still obscure. If modern humans reached Europe from India, as seems likely from the genetic evidence, signs of predecessor culture might be expected in the Indian subcontinent. But so far the archaeological evidence from India shows little evidence of the sophisticated behaviors possessed by the Aurignacians. (Hannah V. A. James and Michael D. Petraglia, “Modern Human Origins and the Evolution of Behavior in the Later Pleistocence Record of South Asia,”
Current Anthropology; 46 [Supplement]: 3-27 [2005].) Possibly the elements of the Aurignacian culture were formed at some point during the migration from India to Europe, as the first modern humans adapted from a subtropical climate to that of the European ice age.
355 An important revision in radiocarbon dating indicates an earlier and much compressed time scale for the Aurignacians’ spread across Europe. The revision was prompted by a) a new method of filtering out contaminants that have made ancient carbon sources seem younger than they were, and b) a new estimate of the amount of carbon-14 in the atmosphere in the distant part. With these two refinements, Paul Mellars has now revised his timetable for the arrival of modern humans in Europe. He estimates that they had arrived west of the Black Sea by 46,000 years ago, not 40,000-44,000 years ago as shown in Fig. 5.2, and had reached northern Spain by 41,000 years ago, not 36,000 years ago. Mellars argues that the Neanderthal fossils from the Zafarraya cave in Spain and Vindija in Croatia, both at present dated to about 30,000 years ago, will turn out to be much older. If so, the new timetable indicates that the Neanderthals succumbed much more quickly than had been supposed, perhaps in a mere 5,000 years. The moderns’ rapid advance may have been helped by an improvement in climatic conditions that occurred between 43,000 and 41,000 years ago. Paul Mellars, “A New Radiocarbon Revolution and the Dispersal of Modern Humans in Eurasia,”
Nature 439:931-935, 2006.
356 Lahn’s two microcephalin genes did not show up in a genomewide search for recently selected genes performed by Jonathan Pritchard and colleagues (see note 363). That may reflect limitations of the Pritchard test, no test for selection being perfect. Pritchard did detect signals of selection in two other genes involved in microcephaly, and in several other types of brain gene. Some of these brain genes were under selection in Africans, some in Asians, and some in Europeans, confirming Lahn’s view that cognitive evolution may have proceeded independently in the three populations. Many of the genetic changes occurring independently in the major continental races are likely to have been convergent, meaning that evolution was using the different mutations available to it in each population to bring about the same adaptation.
357 A new refinement of the radiocarbon dating method (see note 355 above) indicates the drawings of the Chauvet cave as much older than supposed. The first occupation can now be dated to 36,000 years ago.
358 Increasing evidence suggests that pale skin, a variation on the dark skin of the ancestral human population, arose independently in the populations of west and east Eurasia, even though the cause—adapting to the reduced sunlight of northern latitudes—was presumably the same in both cases. Five genes affecting skin color show signs of recent selection in Europeans but not in East Asians. (See notes 230 and 363.) This implies either that East Asians acquired their pale skin through changes in a different set of genes or that their skins became pale considerably earlier than did those of Europeans. In the latter case, the signs of selection would have faded and the genes in East Asians would not have been flagged by Pritchard’s test.
359 Most people in Africa and Europe have wet earwax. But dry earwax is the rule among East Asians. A team of Japanese researchers has traced the difference to a mutation in a gene called ABCC11. (Koh-ichiro Yoshiura et al., “A SNP in the ABCC11 Gene Is the Determinant of Human Earwax Type,”
Nature Genetics, 38:324-330 [2006].) The mutation seems to have arisen in the northern part of east Asia and to have become common very quickly—it is almost universal in northern Han Chinese and in Koreans. What selection pressure made the new version of the gene spread so rapidly? Earwax serves the very humble role of biological flypaper—it stops insects and dirt from getting into the ear—and a change in earwax consistency seems unlikely to have been much of an advantage. The new gene was probably selected because it seems also to reduce the amount of sweating, and hence of body odor. One or the other quality, or both, may have given the gene its decisive advantage.
360 However, the horse Y chromosome is telling a possibly different story. Unlike the mitochondrial DNA, samples of a small region of the Y chromosome from 15 different breeds of European and Asian horses proved to be identical. Gabriella Lindgren et al., “Limited Number of Patrilines in Horse Domestication,”
Nature Genetics, 36:335-336 (2004). This indicates that ancient horse breeders, like modern ones, often let a single stallion cover many females. It could also allow for the possibility that independent do mestications of the horse were not as common as the mitochondrial DNA evidence suggests.
361 This assumption has been confirmed by a direct study of the first Neolithic farmers to settle in Europe. Mitochondrial DNA was extracted from bones taken from archaeological sites of the earliest farming communities of 7,500 years ago in Germany, Austria, and Hungary. A quarter of the sample belonged to the N1a subbranch of the mitochondrial tree, a type that is now very rare among Europeans. This implies that though Neolithic farming techniques spread rapidly, the farmers themselves did not make much of a genetic impact. The authors favor the possibility “that small pioneer groups carried farming into new areas of Europe, and that once the technique had taken root, the surrounding hunter-gatherers adopted the new culture and then outnumbered the original farmers, diluting their N1a frequency to the low modern value.” Wolfgang Haak et al., “Ancient DNA from the First European Farmers in 7500-Year-Old Neolithic Sites,”
Science, 310: 1016-1018 (2005).
362 I thank a reader, the food writer Anne Mendelson, for pointing out that in many parts of the world milk is consumed only in sour form, such as yoghurt, after its lactose has been converted by bacteria into lactic acid. In such conditions there would be no pressure for lactose tolerance to evolve. Nonetheless, lactose tolerance did arise, so presumably the people of the Funnel Beaker culture must have consumed milk in raw form, perhaps not knowing how to ferment it.
363 The test by Pritchard and his colleagues was based on the fact that a beneficial mutation is inherited in a large block of DNA, which will carry its own signature set of DNA changes. If the gene with the good mutation spreads rapidly, along with its block, the DNA in that region of the chromosome will become less diverse in the population as a whole because so many people now carry the same sequence of DNA units at that location.
Pritchard’s test measures the difference in diversity between those who carry a new version of a gene and those who do not. Lesser diversity in a population is taken as a sign of selection. The difference in diversity disappears as the gene becomes universal, because increasing numbers of people carry the new variant. Thus the test picks up only new gene variants on their way to becoming universal, i.e., recently selected genes.
Pritchard looked for blocks with selected genes in data gathered by the Hap Map project from Africans (specifically, the Yoruba people of Nigeria), East Asians, and Europeans. The time of the selective pressure is about 10,800 years ago for the African genes and 6,600 for the East Asian and European genes.
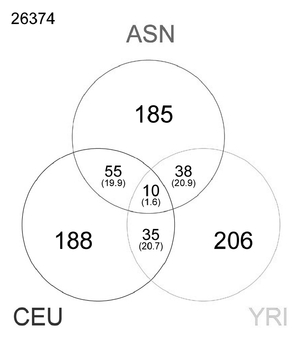
As shown in the figure on page 298, 206 selected genetic regions were identified in Africans, 185 in East Asians, and 188 in Europeans. The fact that the selected genes do not overlap very much indicates that each population evolved independently. The selected genes shared by two races may have arisen by migration or be instances of independent evolution.
The genes under selection clustered in specific categories. Some of the strongest signals of selection were for 4 genes for skin color, found to be under selection in Europeans but not in Asians. Another category of selected genes was in those for skeletal development (possibly reflecting the gracilization of human populations). Other selection categories were genes for fertility, for taste and smell, and genes involved in the metabolism of foodstuffs. The latter two groups may reflect the sharp changes in diet that followed the Neolithic revolution. Benjamin Voight, Sridhar Kudaravalli, Xiao quan Wen, and Jonathan K. Pritchard, “A Map of Recent Positive Selection in the Human Genome,” PloS Biology, 4: 446-458 (2006).
364 A remarkable 20 percent of men in northwestern Ireland carry a particular set of mutations on their Y chromosome, known as the Irish modal haplotype. Many have surnames that are associated with the Ui Neill, a group of dynasties that claimed the high kingship of Ireland and ruled the northwest and other parts of Ireland from about A.D. 600 to 900. Ui Neill means “descendants of Niall.” Historians have tended to regard the Ui Neill as a political construct and its patriarch, Niall of the Nine Hostages, as a probably legendary figure. The genetic evidence provides striking evidence that Niall really existed, a finding as surprising as if the legend of King Arthur turned out to be solid his-tory.
FIGURE 9.1
Genes that have undergone recent evolutionary change in the genomes of East Asians (ASN), Europeans (CEU), and Africans (YRI).
The Irish modal haplotype, the signature of descent from Niall, is most common in northwestern Ireland but is also found in the Irish diaspora, being carried by no less than 2 percent of New Yorkers of European descent. Evidently one should listen less skeptically to Irishmen who declare the blood of Irish kings runs in their veins. Laoise T. Moore et al., “A Y-Chromosome Signature of Hegemony in Gaelic Ireland,”
American Journal of Human Genetics, 78: 334-338 (2006).