The Puzzle of Immunoglobulin Gene Structure
The immune system relies on a vast array of B-cell receptors (BCRs) that possess the ability to bind specifically to a correspondingly large number of potential pathogens. The first indication of the immense size of the antibody repertoire was provided by immunologists using synthetic organic molecules to stimulate antibody production. Changing the position of an amino or a nitro group on a phenyl ring was sufficient to alter the capacity of an antibody to bind. Investigators reasoned that if the immune system can discriminate in this exquisitely specific manner between small molecules that it had presumably never encountered during evolutionary selection, then the number of potential antibodies must be very large indeed. A series of experiments conducted in the late 1970s and early 1980s estimated the number of different BCRs generated in a normal mouse immune system to be at least 107; we now know that estimate was many orders of magnitude too small.
Investigators trying to understand immunoglobulin (Ig) genetics were also faced with an additional puzzle: protein sequencing of mouse and human antibody heavy and light chains revealed that approximately the first 110 amino-terminal amino acids of antibody heavy and light chains are extremely variable among different antibody molecules. This region was therefore defined as the variable (V) region of the heavy and light chains. In contrast, the remainder of the light and heavy chains could be classified into one of only a few sequences and was therefore named the constant (C) region of heavy and light chains (Figure 6-1a). So immunologists of this era were now faced with two seemingly intractable problems: First: how can a finite amount of genetic information encode a vast number of different antigen-binding sites? And second: how can an immunoglobulin heavy or light chain be identical to many other heavy or light chains in one part of its sequence, but be extraordinarily variable in another? Pertinent to the latter point, geneticists at the time pointed out that, if each antibody chain were separately encoded in the genome, the process of evolutionary drift would result in the accumulation of silent mutations in the constant regions, even in the absence of any particular evolutionary pressures toward diversity.
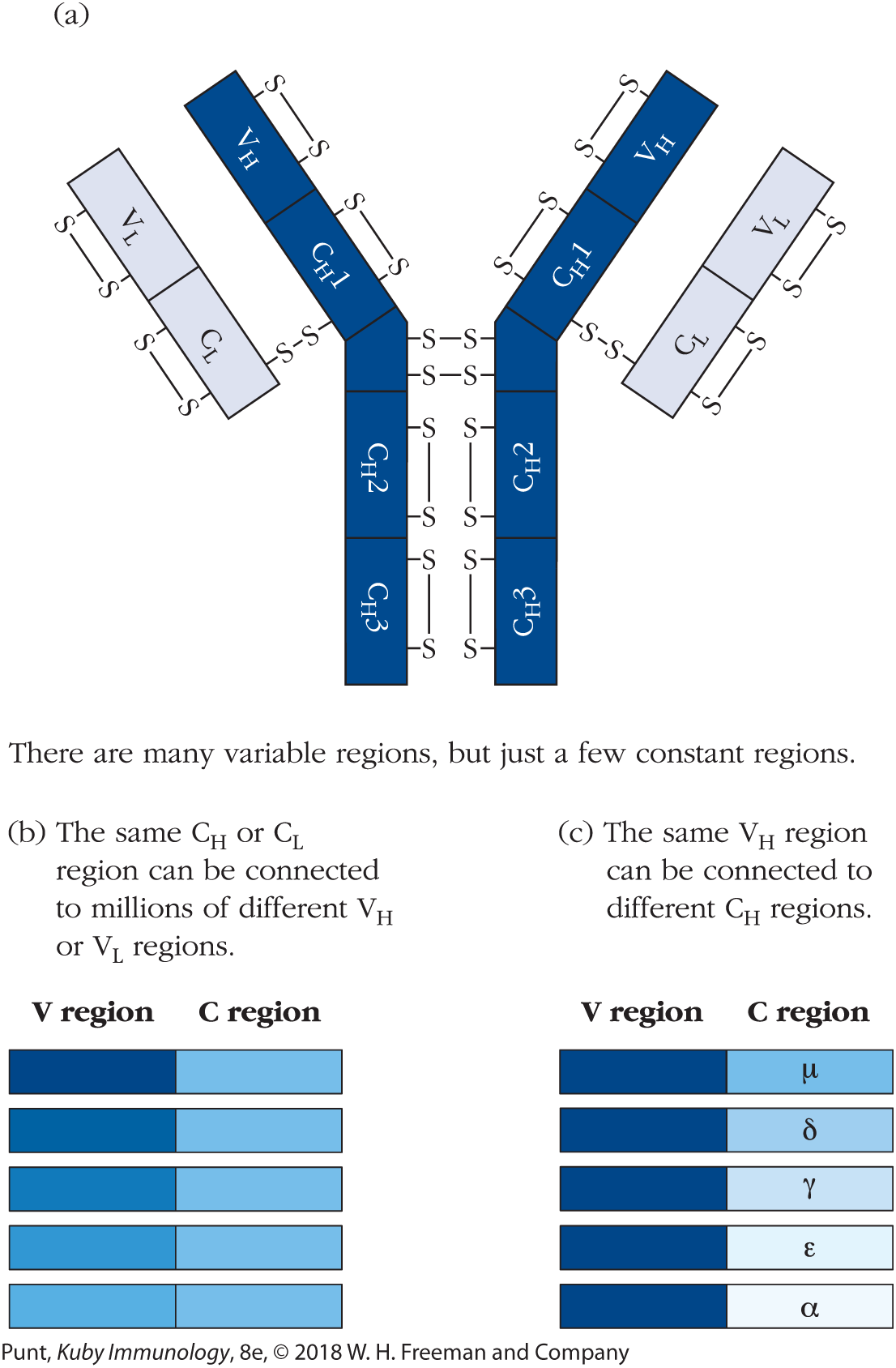
FIGURE 6-1 Early sequencing studies indicated that the variable and constant regions of both the immunoglobulin heavy and light chains are encoded separately in the germ-line genome. (a) An IgG antibody molecule consists of two heavy chains and two light chains. Each chain consists of an amino-terminal variable (V) region and a carboxy terminal constant (C) region. (b) For any given constant region there are millions of variable regions that can be associated with it. (c) For any given variable region there are only a few constant regions that can be associated with it. CH = heavy-chain constant region; CL = light-chain constant region; VH = heavy-chain variable region; VL = light-chain variable region.
Further sequencing studies uncovered yet more complexity. Not only were different variable regions found to be associated with the same constant regions, as described above (Figure 6-1b), but additional studies identified antibodies in which the same heavy-chain variable (VH) region was associated with more than one heavy-chain constant (CH) region (Figure 6-1c). Together, these findings strongly implied that for each antibody chain, expression of the variable region and expression of the constant regions are independently controlled.
Investigators Proposed Two Early Theoretical Models of Antibody Genetics
The earliest theories of how antibody light- and heavy-chain genes were arranged in the genome were called germ-line theories. Germ-line theories suggest, as their name implies, that the genetic information for each antibody is separately encoded within the germ-line (inherited) genome. However, a quick calculation demonstrates that if there are 107 or more antibodies, each of which needs approximately 2000 nucleotides to specify both its heavy and light chains, encoding each antibody individually would require 2 × 1010 nucleotides. Since the size of the mouse genome is 2.8 × 109 nucleotides, this is clearly an impossibility. In addition, as more accurate estimates of the size of the antibody repertoire became available, it quickly became clear that 107 was a minimal estimate of the number of antibodies that could be made by an individual mouse. Immunologists had to face the fact that there simply was not enough DNA to encode all the relevant B-cell receptors, using the classical one gene–one polypeptide chain rule, and that new ideas had to be found.
In 1965, William Dreyer and J. Claude Bennett proposed that antibody heavy and light chains are each encoded in two separate segments in the germ-line genome (Figure 6-2a). To specify a complete heavy or light chain polypeptide, one of each of the V region– and C region–encoding segments must then be brought together in B-cell DNA to form one complete polypeptide-encoding gene (Figure 6-2b). The idea that DNA in somatic cells might engage in recombination was revolutionary—previously, only cells in meiosis had been shown to undergo recombination. However, Dreyer and Bennett’s theory fitted the data so elegantly that germ-line theorists began to modify their ideas to embrace the idea that two genes might be necessary to encode one antibody polypeptide chain.
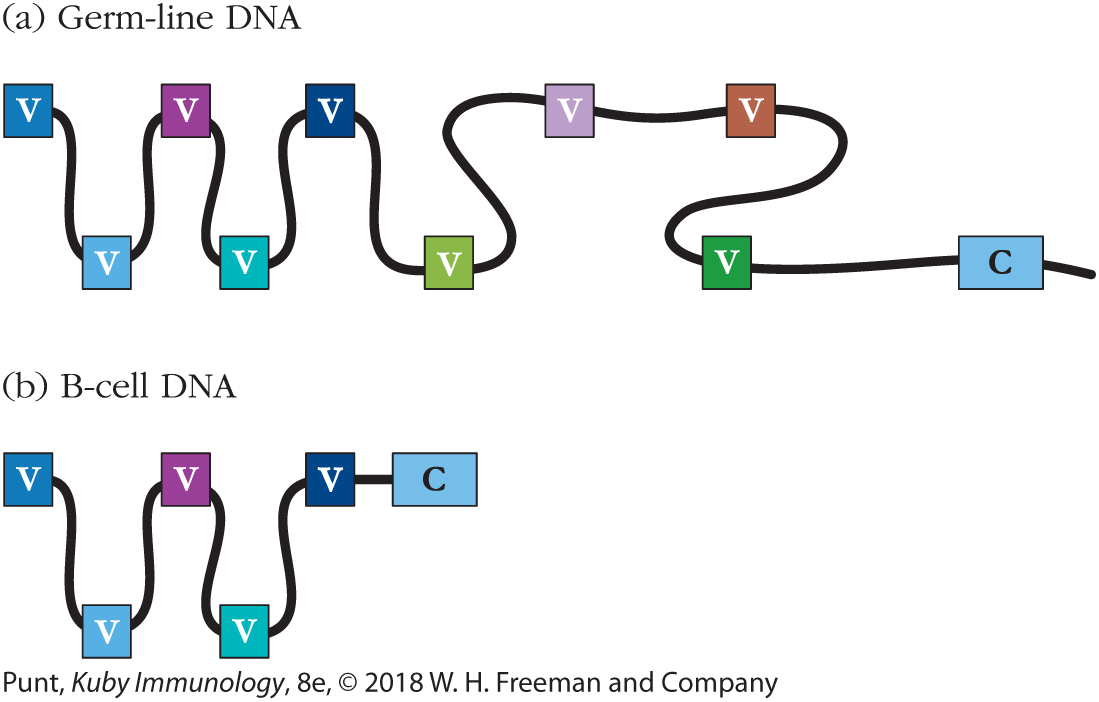
FIGURE 6-2 Dreyer and Bennett hypothesis. (a) Dreyer and Bennett suggested that one constant region (C) gene existed in the germ-line genome along with many different variable (V) region genes. (b) In antibody-producing cells (B cells), they hypothesized that a mechanism existed to bring together one of the variable region genes and the constant region gene. Different B cells would make different variable region genes contiguous with the constant region gene in order to generate a diverse repertoire of B-cell receptors and secreted antibodies.
In the early 1970s, others suggested the innovative somatic hypermutation theory to account for the size of the mature antibody repertoire. The term “somatic” refers to all cells of the body with the exception of the germ-line sperm and egg cells, and therefore the somatic hypermutation theory suggested that a mutational process occurred only in B cells to alter the antibodies in an individual animal and thus increase the total number of different antibodies available to it. Since these mutations are not affecting germ-line genes, such mutations would not be passed on to offspring. According to the somatic mutation hypothesis, therefore, a limited number of germ-line antibody genes is acted on by unknown mutational mechanisms to generate a diverse receptor repertoire, only in mature B lymphocytes. This theory had the advantage of explaining how a large repertoire of antibodies could be generated from a relatively small number of genes, but the disadvantage that such a process had never before been observed.
Heated debates continued between the proponents of modified germ-line versus somatic mutation theories throughout the early 1970s, until a seminal set of experiments revealed that both sides were correct. We now know that multiple germ-line gene segments each encode a part of the antibody variable regions, and that these segments are rearranged differently in the formation of each naïve B cell to produce an extremely diverse primary receptor repertoire. In humans and mice, these rearranged genes are then further acted on after antigen encounter by somatic hypermutation and antigenic selection, resulting in an expanded and exquisitely honed population of antigen-specific B cells. We will describe the process of antibody gene rearrangement in this chapter, but delay a description of somatic hypermutation until Chapter 11, since it occurs following antigenic stimulation.
Breakthrough Experiments Revealed That Multiple Gene Segments Encode the Immunoglobulin Light Chain
From the mid-1970s until the mid-1980s, a small group of brilliant immunologists completed a series of experiments that fundamentally altered the way in which scientists think about genetics. The first breakthrough occurred when Nobumichi Hozumi and Susumu Tonegawa showed that, as Dreyer and Bennett had predicted, multiple gene segments encode the antibody protein chains.
Tonegawa and his colleagues showed that the variable and constant regions of the antibody light-chain gene were encoded in segments located in two distinct places in the germ line. These segments were then brought together by DNA recombination only in mature, antibody-producing cells.
The variable (V) and constant (C) region gene segment families are located kilobases apart in the germ-line DNA. The work of Tonegawa and colleagues demonstrated that these two DNA segments, one containing coding information for the kappa light-chain variable (Vκ) region and the other containing coding information for the Cκ region gene segment families, are stitched together, only in B lymphocytes, to create the complete κ light-chain gene. Each B cell contains only one functional combination of κ gene segments, and different Vκ gene segments are used in each B cell.
How were these ground-breaking discoveries made? Figure 6-3 outlines the experiment. They made use of the ability of the BamH1 restriction endonuclease to cut DNA at a precise nucleotide sequence. (Each restriction endonuclease cuts DNA at a characteristic nucleotide sequence.) Hozumi and Tonegawa showed that in the DNA from non–antibody-producing, embryonic liver cells (used in place of germ-line DNA for technical reasons), there is a BamHI restriction endonuclease site between the DNA that encodes the variable and constant regions of the antibody molecule. We know this because labeled nucleotide probes for the variable and constant regions each recognized a different DNA fragment in BamHI-digested germ-line DNA. However, in an antibody-producing B cell from the same mouse strain, the gene segments encoding the variable and constant regions appeared to be combined into a single fragment (later confirmed by DNA sequencing), thus demonstrating that DNA rearrangement must have occurred during the formation of an antibody light-chain gene. Note that in Figure 6-3 their result is presented as a Southern blot, in which DNA fragments cut by the enzyme are run out on a gel, blotted onto nitrocellulose paper to prevent further nucleotide fragment diffusion, and probed with a labeled short nucleotide sequence. For a more detailed description of their actual experiment, and the original methods they used, see Classic Experiment Box 6-1.

FIGURE 6-3 The κ light-chain gene is formed by DNA recombination between variable and constant region gene segments. DNA from embryonic liver cells was used as a source of germ-line type DNA, and DNA from a B-cell tumor cell line was used as an example of DNA from antibody-producing cells. Each DNA sample was digested with the BamHI restriction endonuclease. The DNA fragments were separated by gel electrophoresis, and the location of fragments containing V and C regions were detected with specific probes for the entire κ light-chain (V and C region) and for the C region alone. In embryonic liver, the DNA sequences encoding the V and C regions, respectively, were located on different restriction endonuclease fragments. In contrast, these two sequences were colocated on a single restriction fragment in the myeloma DNA.
This experiment demonstrated that, in agreement with the Dreyer-Bennett hypothesis, the V and C regions of antibody genes were located in different places in the DNA of antibody-producing, versus non–antibody-producing, cells. However, it yielded no information about the physical relationship of the V and C segments in the animal’s DNA; indeed, the initial experiment did not rule out the possibility that the V and C fragments could be encoded on different chromosomes in the embryonic cells. Subsequent sequencing experiments showed that the segments encoding the V and C segments of the κ light chains are on the same chromosome and that, in non-B cells, the V and C segments are separated by a long noncoding DNA sequence.
The impact of this result on the biological community was profound. For the first time, mammalian DNA was shown to be cut and recombined during the process of cell differentiation. Furthermore, this finding paved the way for the next surprise.
Scientists in Tonegawa’s group sequenced the germ-line DNA segments that hybridized with their probes and analyzed the sequences for open reading frames that encode protein sequences. They found that the (unrearranged) embryonic liver V region segment had a short, protein-coding leader sequence at its 5′ terminus. This is a common feature of membrane proteins; amino-terminal leader sequences (also known as signal peptides) guide the nascent polypeptide chain to the endoplasmic reticulum membrane during translation (Figure 6-4). A 93-base pair (bp) sequence of noncoding DNA separated the leader sequence from a long stretch of DNA that encoded the first 97 amino acids of the V region. But the light-chain V region domain is approximately 110 amino acids long. Where was the coding information for the remaining 13 amino acids of the light-chain variable region?
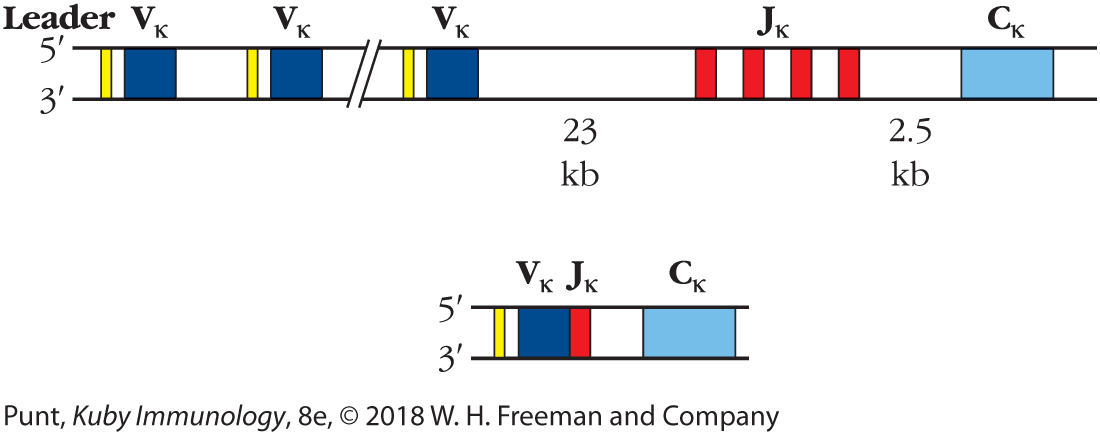
FIGURE 6-4 The antibody κ light-chain locus is composed of three families of DNA segments. In germ-line DNA a large family of Vκ segments is found upstream of a smaller group of Jκ segments. A single Cκ segment is located 2.5 kb downstream from the 3′-most Jκ segment. During B-cell development, DNA recombination events join one Vκ segment and one Jκ segment in each B cell.
A band represents the gene locus. After a short leader sequence, there is a V kappa sequence, and then another leader sequence and a V kappa sequence. Further down there is another leader sequence and V kappa sequence. Next there are four successive J kappa segments 23 kilobases downstream from the V kappa segments. A C kappa segment is located 2.5 kilobases downstream from there. A shorter band shows the B-cell development in which after a leader sequence, one V kappa segment is next to a J Kappa segment, followed by the C Kappa segment downstream.
Sequencing of the light-chain constant region fragment from embryonic DNA provided the answer to this question. Upstream (toward the 5′ end) from the constant region coding sequence, and separated from it by a noncoding DNA segment of 2.5 kilobases (kb), were the 39 bp encoding the remaining 13 amino acids of the V region. This additional light-chain coding segment was named the joining (J) gene segment (shown in red in Figure 6-4), as it was the site of joining of the V-gene segment with the fragment containing the constant region during rearrangement.
Further sequencing of mouse and human light-chain variable and constant region genes confirmed this second, astonishing finding from Tonegawa’s group. Not only had they proved that the variable and constant regions of antibody light chains are encoded by separate DNA segments, as Dreyer and Bennett had predicted; they had also shown, completely unexpectedly, that the light-chain variable regions themselves are encoded by two separate gene segments, the V and J segments, and that they are made contiguous only after the gene rearrangements that occur during the development of B cells. Analogous experiments performed later demonstrated that the lambda light-chain gene segments were similarly, although not identically, arranged (see below).
If the kappa light-chain variable region is encoded in two segments that are recombined in each antibody-producing cell to create a new combination of sequences, what about the heavy chain? By cloning and sequencing immunoglobulin (Ig) heavy-chain genes, Lee Hood’s group showed that the heavy-chain variable region was encoded in not two, but three separate gene segments in the germ line, and that these segments were recombined during B-cell development to create the contiguous coding information for the heavy-chain variable region (Figure 6-5).
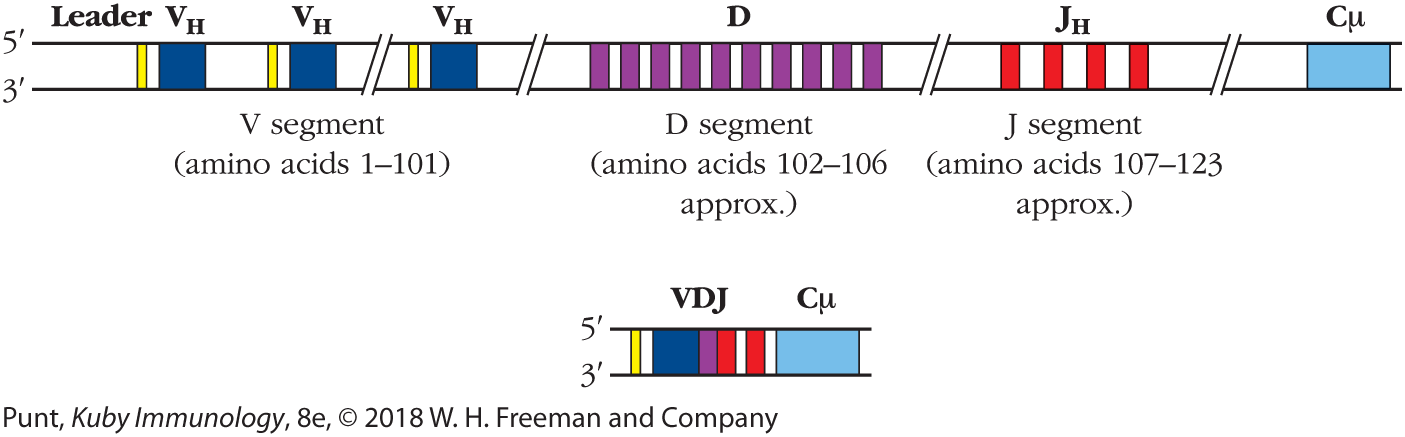
FIGURE 6-5 Variable region of antibody heavy chains is encoded in three segments—V, D, and J. During B-cell development, DNA recombination events bring together three DNA segments—a VH, a D, and a JH segment—to form the variable region of the Ig heavy chain.
A band represents the gene locus. After a short leader sequence, there is a V kappa sequence, and then another leader sequence and a V kappa sequence. Further down there is another leader sequence and V kappa sequence. These together constitute the V segment, amino acids 1 to 101. Next is the D segment, from amino acids approximately 102 to 106. Next is the J segment, comprised of four short J H bands, from amino acids approximately 107 to 123. A C mu segment is at the end. A shorter band shows the V, D, and J segments all close together, with the C mu segment at the end.
Specifically, the heavy-chain variable region was found to be encoded by a germ-line heavy-chain variable (VH) region gene fragment that encodes amino acid residues 1–101 of the antibody heavy chain and a second fragment that included a heavy-chain joining (JH) region gene segment that determined the sequence of amino acid residues 107–123. The DNA sequence necessary to encode residues 102–106 (approximately) of the heavy chain was discovered last. It was located 5′ of the J region in mouse embryonic DNA (see Figure 6-5). One of the features of the D region is that it is quite variable in its length. The importance of the contribution made by this sequence, the D gene segment, to the diversity of antibody specificities is denoted by its name: the diversity (D) region. (Because there is no D region in the light chain, immunologists usually drop the subscript denoting the heavy chain.)
Significantly, DNA in the D gene segment encodes most of the amino acid residues shown to be part of the third complementarity-determining region of the antibody heavy chain, CDR3 (see Figure 3-11a). This region provides amino acid residues that contact antigen for the binding of most antigens. In the light chain, the CDR3 portion of the antibody sequence is located at the V-J junction.
Thus, the variable region of the heavy chain of the antibody molecule is encoded by three discrete gene segments and the variable region of the light chain by two segments. These segments in the germ-line genome are brought together by a process of DNA recombination that occurs only in cells of the B-lymphocyte lineage to create the complete and contiguous coding information for the variable regions of heavy and light chains.
Key to understanding the significance of these findings is the fact that multiple copies of each of these separate gene segments exist in the germ line. Thus, in creating a gene for the variable region of a mouse immunoglobulin heavy chain, a lymphoid-specific recombinase enzyme may select one of 100 different VH gene segments; one of 12–14 D region gene segments, and one of four JH gene segments. These segments are selected at random by the recombinase as B cells develop in the bone marrow, and different combinations of V, D, and J segments are ligated (i.e., joined) in each B cell to form the complete coding sequence for a VH region. This leads to considerable recombinatorial diversity in antibody variable region genes. We will describe the immunoglobulin gene loci in more detail below. In addition, we will describe the mechanisms responsible for Ig gene rearrangements and the generation of further genetic diversity at the junctions between the V, D, and J segment by additional processes unique to the immune system.
Key Concepts:
- Tonegawa’s key experiment showed that two gene segments encoding V and C light-chain regions, which are separate in the germ line, are brought together by recombination at the level of DNA in B cells, to form an intact light-chain gene. Light-chain V region diversity is achieved in part by combinatorial association of multiple gene segments termed V (variable) and J (joining) segments, that together encode the variable region of the light chain of the Ig molecule. The region of greatest diversity in the light chain is encoded at the V-J gene junction.
- Recombination between three gene segments—the VH, D (diversity), and JH segments—is required to encode the variable region of the heavy chain of the Ig molecule. The D region and V-D and D-J junctions encode the regions of greatest sequence diversity in the antibody heavy chain, corresponding to CDR3 regions of the antibody heavy chains.