Chapter 2
Cells and their Basic Building Blocks
2.1 Chapter Overview
The chemical composition of a typical bacterium and animal (mammalian) cell is shown in Table 2.1.
Table 2.1 Approximate chemical composition of a typical bacterium and mammalian cell. (Adapted from Alberts et al. [1])
| Chemical component | Percentage of total cell weight | |
| Bacterium | Animal cell | |
| Water | 70 | 70 |
| Inorganic ions (e.g. Na+, K+, Mg2+, Ca2+, Cl2−) | 1 | 1 |
| Amino acids, nucleotides, and other small molecules | 1 | 1 |
| Metabolites (e.g. glucose, fatty acids) | 2 | 2 |
| Macromolecules (proteins, nucleic acids, polysaccharides) | 24 | 21 |
| Lipids | 2 | 5 |
Leaving aside the water content of a cell, macromolecules such as proteins, nucleic acids (DNA, RNA), and polysaccharides make up a large percentage of a cell's mass. The building blocks for these macromolecules are small organic molecules, namely fatty acids, sugars, amino acids and nucleotides. This chapter describes the chemical structures and functions of these molecular building blocks, and the biological importance of the macromolecules and macrostructures they combine to form. A summary description is then given of how these macromolecules and microstructures interact and function in different types of cell.
After reading this chapter a basic understanding should be obtained of:
2.2 Lipids and Biomembranes
Cells of higher organisms are separated, but not isolated, from their surroundings by their cytoplasmic membrane, which also serves to act as anchors for proteins that transport or pump specific chemicals into or out of a cell. Membranes also define the boundaries of intracellular organelles and the nucleus in eukaryotic cells. The main structural components of biological membranes are lipids, which exist as derivatives of fatty acids. The term ‘lipid’ covers a wide range of molecules, including oils, waxes, sterols, certain (fat-soluble) vitamins and fats. The one property they all share in common is that they are hydrophobic. When placed in water individual lipid molecules will adopt a configuration that leads to minimum contact with water molecules, and will cluster into a group with other lipid molecules. This is exemplified by the formation of oil droplets in water, and how lipids in an aqueous medium segregate into a separate nonaqueous phase.
2.2.1 Fatty Acids
Fatty acid molecules contain a hydrocarbon chain, commonly consisting of 16 or 18 carbon atoms. An acidic carboxyl group (COOH) is attached to one end of this chain. Stearic acid CH3(CH2)16COOH and arachidic acid CH3(CH2)18COOH are examples, whose general chemical structure is shown below:
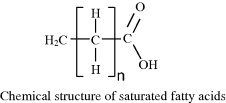
Stearic (n = 16) and arachidic (n = 18) acid are examples of fatty acids with no double (C=C) bonds in their hydrocarbon chain, and are termed as being saturated. If the hydrocarbon chain contains one or more double C=C bonds the fatty acid is termed unsaturated – an example of which is oleic acid CH3(CH2)7CH(CH2)7COOH:
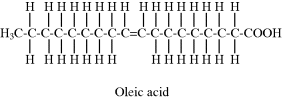
The two hydrogen atoms attached to the carbons in the C=C double bond of oleic acid lie on the same side of the bond, and this configuration is known as the cis form. This cis configuration introduces a bend in the hydrocarbon chain. The other possible configuration, known as the trans form in which the two hydrogen atoms are situated on opposite sides of the C=C double bond, does not result in a bent hydrocarbon chain. The reason why butter and lard are solid at room temperature is because they are composed of saturated fatty acids whose straight hydrocarbon chains can pack closely together. Easily spreadable butter substitutes (e.g. margarine) contain unsaturated fatty acids that are unable to pack closely together because of the ‘kinks’ in their hydrocarbon chains, and have a softer form than butter at room temperature. Plant oils contain polyunsaturated fatty acids (with multiple C=C double bonds) and are liquid at room temperature.
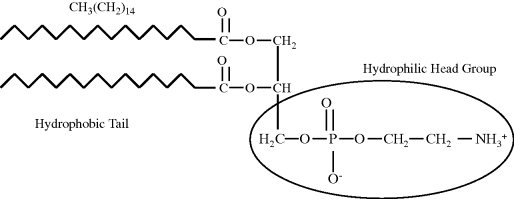
A fatty acid molecule thus has two chemically distinct parts – a long hydrophobic chain that is not very reactive chemically, and a carboxyl group (COOH) which when ionised as COO− is chemically active and hydrophilic. Molecules such as these, which contain both hydrophobic and hydrophilic regions, are termed amphipathic. Fatty acids by themselves will not form a membrane that is capable of acting as a boundary between an aqueous medium and the aqueous cytoplasm of a cell. In aqueous media fatty acids will tend to form clusters, with the hydrocarbon chains packed together inside and the carboxylic acid groups directed outwards towards the surrounding water molecules. To form biomembranes fatty acids need to be converted into a structure that readily form sheets of lipid bilayers. The most common ones adopted in nature are phospholipids composed of two fatty acid side chains attached to a negatively charged (and hence hydrophilic) phosphate group via a glycerol molecule. The two fatty acid ‘tails’ may both be saturated, unsaturated, or adopt one of each form. As shown in Figure 2.1, in some phospholipids the ‘head’ of the molecule may be increased in size with the addition of an amine which can ionise to the hydrophilic form NH3+.
Figure 2.1 The chemical structure of a typical phospholipid (in this case phosphatidylethanolamine) to show its hydrophobic tail and hydrophilic head group.
Phospholipids can spontaneously form sheets of bilayers, two molecules thick, in an aqueous environment. As depicted in Figure 2.2, the hydrocarbon tails keep away from the water by aligning themselves in the middle of the bilayer structure. The close packing of the hydrocarbon tails is stabilised by van der Waals interactions, and the fluidity of the bilayer interior is influenced by the number of C=C double bonds in the hydrocarbon structures of the tails. The polar head groups are stabilised through hydrogen bonding to water molecules, as well as by electrostatic interactions between the phosphate and amine groups. As shown schematically in Figure 2.3 for a fat cell, the outer membrane of a cell is formed by a spherical lipid bilayer structure that encloses the cytoplasm and internal cell structures.
Figure 2.2 Schematic representation of a phospholipid bilayer. The small spheres represent the hydrophilic heads groups, and the lines are the hydrophobic hydrocarbon tails of individual phospholipid molecules.
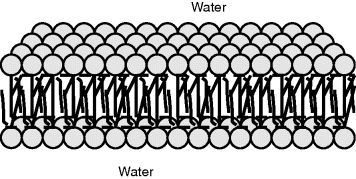
Figure 2.3 Schematic representation of a fat cell (adipocyte).
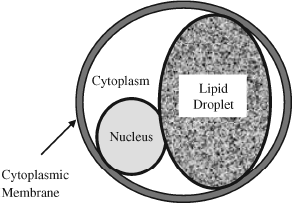
Apart from their importance as precursors to phospholipids, fatty acids are used as a source of energy by tissues. Fat cells, known as adipocytes, contain one large droplet of lipid (see Figure 2.3). When triggered by hormones such as adrenaline these cells release fatty acids into their surrounding environment (normally blood), which are then broken down into smaller molecules identical to those derived from the breakdown of glucose.
2.3 Carbohydrates and Sugars
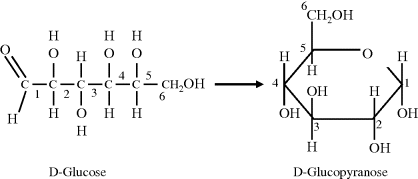
Carbohydrates are composed of carbon atoms and the atoms that form water molecules, namely hydrogen and oxygen. Simple carbohydrates, called mono-saccharides, have the chemical structure (CH2O)n and are often referred to as simple sugars. The number ‘n’ of carbon atoms ranges from 3 to 7 and the corresponding sugar molecules are called trioses, tetroses, pentoses, hexoses and heptoses. We will learn later in this chapter that two pentose sugars (ribose and deoxyribose) are essential components of DNA and RNA. An important hexose is glucose (C6H12O6) because when it is broken down in cells of higher organisms it releases free energy. As shown in Figure 2.4 the linear structure of glucose can form a ring structure arising from the reaction of the aldehyde at the 1 carbon with the hydroxyl group on the 5 carbon, to form glucopyranose. A less common ring structure (glucofuranose) is formed by the reaction of the 1 carbon aldehyde with the hydroxyl on the 4 carbon. The chemical formula of a monosaccharide does not therefore fully describe the molecule. For example, a different sugar is formed if the hydrogen and hydroxyl groups attached to the 2 carbon of the D-glucose molecule switch places. This sugar (mannose) cannot be converted to glucose without breaking and making the relevant covalent bonds. Each of the sugars can also exist in either of two forms that are mirror images of each other, called the D-form and the L-form. The D- or L-form of a molecule signifies the direction, dextro (right) or levro (left), in which the plane of polarisation of light rotates when passing through a solution of the molecules. The most common form of sugars found in, and metabolised by, biological systems is the D-form.
Figure 2.4 The linear and ring form of D-Glucose.
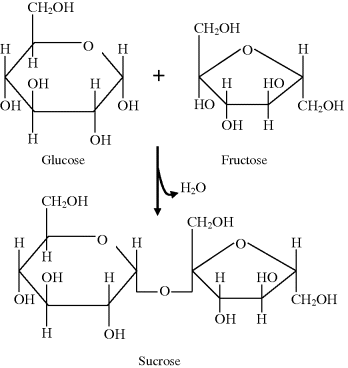
Two monosacharides can be linked by a covalent bond to form a disaccharide, a third can be added to form a trisaccharide, and so on to form a polymeric chain known as an oligosaccharide. If more than around 50 sugar subunits (mers) are joined together the resultant structure is called a polysaccharide. Each covalent bond, known as a glycosidic bond, is formed between an ߝOH group on one sugar and an ߝOH group on another by a condensation reaction that results in a loss of a molecule of water. This reaction is depicted in Figure 2.5 for the formation of one molecule of the disaccharide known as sucrose composed of glucose and a fructose molecule. Polysaccharides, containing hundreds or even thousands of sugar subunits, can act as energy stores in cells – an example being glycogen stored in liver cells. They also form part of the structures of connective tissues, are part of the composition of mucus and slime, and serve to lubricate bone joints. A polysaccharide of glucose, namely cellulose, is the main structural component of plant cell walls and as such is probably the most abundant organic molecule on Earth.
Figure 2.5 Formation of sucrose from the condensation reaction of glucose with fructose.
Proteins and lipids, known as glycoproteins and glycolipids, have oligosaccharide chain attachments and are important building blocks of cell membranes. The attached oligosaccharide chains increase the water solubility of the proteins and lipids and act to orientate them at the interface between the membrane and the surrounding aqueous medium. Oligosaccharides are also responsible for the grouping of human blood cells according to the ABO system, and for the rules that dictate allowed transfusion of blood between donors and recipients.
For example, a group O person can only receive blood from group O individuals, but can donate blood to persons of any blood group. On the other hand, a person with the AB group can receive blood from any donor, but can only be a donor to another AB individual. Such rules are determined by the type of oligosaccharide linked to a protein or lipid in the outer membrane of a person's red blood cells, and how this molecular structure can perform as an antigen. If blood of one group is injected into someone having a different blood group, white blood cells (known as B cells) in the recipient's blood may ‘recognise’ the foreign antigen and then generate antibodies that selectively attach themselves to it. These antibodies, which may already have existed in the recipient's blood serum, are large protein complexes known as immunoglobulins. Very large numbers of different antibodies are normally created in a person's blood from an early age as a result of exposure to antigens present on bacteria or plants, for example. The attachment of antibodies to their specific antigens in turn triggers an immune response that involves specialised cells, known as phagocytes, locating and then ingesting the antibody-coated foreign blood cells. The reason why blood group O individuals can be a donor to all ABO groups is because they do not have either the A- or B-antigen on their red blood cells. However, their blood serum naturally contains both anti-A and anti-B antibodies against the A and B blood group antigens, which does not permit their receiving blood from A, B or AB donors.
2.4 Amino Acids, Polypeptides and Proteins
Proteins are the working molecules of cells. They provide a cell with structural rigidity; pumps to drive ions and metabolites across membranes; catalysts for a vast range of biochemical reactions including the functioning of genes; and ‘motors’ to provide motility of objects within cells and of some whole cells. Our present understanding of how proteins are able to perform this amazing range of functions has taken more than 200 years to achieve. Although the importance to biological processes of substances we now call proteins was appreciated before 1800, the word ‘protein’ was first used in the scientific literature by the Dutch chemist G.J. Mulder in 1838 – a term suggested to him by J.J. Berzelius [1, 2]. Using refinements of the chemical analysis procedures of Lavoisier, Liebig, Gay-Lussac, Dalton and others [3], Mulder concluded that proteins were composed of a very large number of carbon, hydrogen, nitrogen and oxygen atoms. Based on the principle that no molecule can contain a nonintegral number of atoms, he obtained the chemical formula C400H620N100O120P1S1 for egg albumin, and exactly the same formula for serum albumin, but with two sulphur atoms instead of one. It is now known that phosphate groups can be linked to the protein structure, so that phosphorus, unlike sulphur, is not considered to be an intrinsic atomic component of a protein molecule. Proteins are thus macromolecules of large molecular weight. At that time it was also found that when proteins are subjected to the hydrolytic action of boiling acid, they decompose into relatively simple crystalline substances – now known as amino acids. Amino acids are the monomers that make up the polymeric chains of proteins. By 1903, 18 of the common 20 amino acids had been isolated and characterised, with the last two, methionine and threonine, being found in 1922 and 1936, respectively [4, 5].
2.4.1 Amino Acids and Peptide Bonds
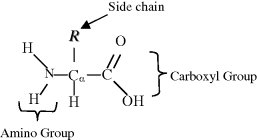
The 20 common amino acids, apart from proline, contain an amino group (ߝNH2) and an acid carboxyl group (ߝCOOH). Proline possesses an imino group (ߝNHߝ) instead of an amino group. In accordance with the description of acids and bases in Chapter 1, at normal physiological pH the acidic carboxyl group is ionised as ߝCOO−, and the basic groups are ionised as ߝNH3+ (or =NH2+ for the case of proline). The basic chemical structure of an amino acid is shown below:
The predominant forms of an amino acid across the pH range are as follows:
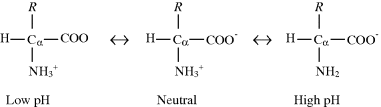
In neutral solutions (pH ≈ 7) amino acids exist predominantly in the dipolar, doubly ionised form, called a zwitterion.
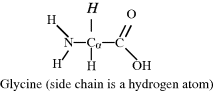
The central carbon atom, called the alpha-carbon (Cα), is bonded to an amino (or imino) group, a carboxyl group and a hydrogen atom. A variable chemical group R, termed the side chain, is also bonded to the Cα carbon, and is what gives an amino acid its special characteristic. Amino acids with a side chain bonded to the Cα carbon are referred to as alpha amino acids, and are by far the most common form found in nature. Glycine has the simplest side chain, namely a single hydrogen atom. This lends to glycine, with its two hydrogen atoms about the Cα carbon atom, the property of symmetry. The remaining amino acids do not possess such symmetry – and so have two mirror-image (sterioisomeric) structures, called the D and L forms, as described for sugars in Chapter 1. Only the L forms of amino acids are found in protein molecules, but D-amino acids form part of bacterial cell walls and occur in some antibiotics.
The chemical structures of the 20 common amino acids are given in Tables 2.2–2.4, and are classified according to whether their side chain is hydrophilic or hydrophobic.
Table 2.2 Amino acids with hydrophobic (nonpolar) side chains R.
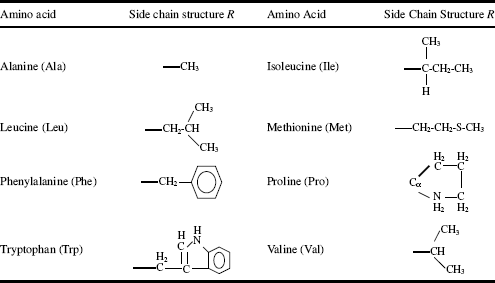
Table 2.3 Amino acids with hydrophilic (uncharged, polar) side chains R.
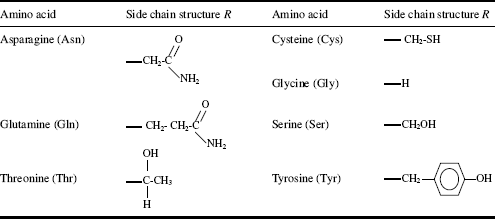
Table 2.4 Amino acids with hydrophilic (charged) side chains R.
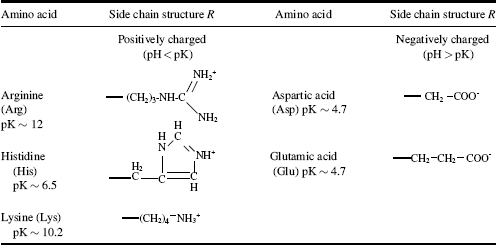
Five of the side chains in the 20 common amino acids are ionisable, and their pK values are given in Table 2.4. The main factors to consider when determining whether a side chain is hydrophobic or hydrophilic are:
- carbon and nonpolar groups do not readily hydrogen-bond to water, and are thus hydrophobic;
- oxygen and nitrogen can hydrogen-bond to water, and are thus hydrophilic;
- ionisable groups (e.g. ߝCOO−, ߝNH3+ or =NH2+) are hydrophilic;
- polar groups are hydrophilic.
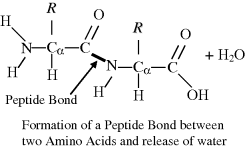
A covalent chemical bond can be formed between two amino acids to form a dipeptide, involving the amino group of one amino acid and the carboxyl group of the other. This bond, known as a peptide bond, results from the elimination of a molecule of water in a so-called condensation reaction:
The carboxyl (C=O) and nitrogen atom forming the peptide bond between the two amino acid residues exhibit a resonating partial double-bond character, as depicted below:
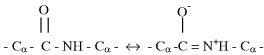
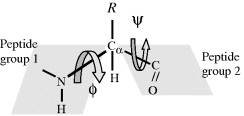
Because of this resonance bonding, the six atoms of the peptide group (the two alpha-carbons, the carbon, oxygen and nitrogen plus hydrogen) all lie in the same plane, as shown in Figure 2.6. Independent rotation of two planar peptide groups about their connecting alpha-carbon is possible. As shown in Figure 2.7, the relative conformation of a pair of planar peptide groups can be defined by two dihedral angles ϕ and ϕ, where ϕ is the angle of rotation about the CαߝC bond, and ϕ is the angle of rotation about the NߝCα bond.
Figure 2.6 The peptide bond has a ‘resonant’ partial double-bonding of the carbon and nitrogen atoms. This results in the peptide group of atoms to lie in the same plane [7].

Figure 2.7 The relative conformation of two adjacent planar peptide groups in a dipeptide is defined by the angles of rotation ϕ and ϕ about the connecting Cα atom.
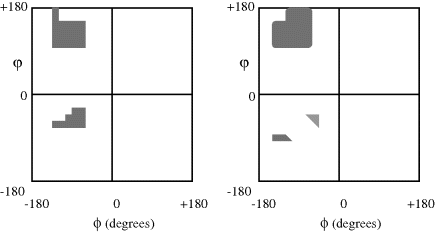
Although the angles ϕ and ϕ shown in Figure 2.7 can theoretically both assume all the values from 0 to 360°, physically realisable conformations are in fact limited by restrictions on the allowed van der Waals contact distances between different atoms in the dipeptide, and especially for those atoms that are located in the side-chain R. Although ϕ = ϕ = 0° is used as a reference point, it is in fact sterically prohibited because this would require the hydrogen atom in the NH group of peptide unit 1 and the oxygen atom in peptide unit 2 attempting to occupy the same space. The situation ϕ = ϕ = 180° corresponds to where the two peptide units are fully extended and lie in the same plane. The allowed values for ϕ and ϕ can be graphically presented in the form of a Ramachandran plot [7], examples of which are shown in Figure 2.8. Glycine, with a side chain R comprising a single hydrogen atom, is far less sterically hindered than the examples shown in Figure 2.8 for valine and isoleucine, and the allowed values for ϕ and ϕ for glycine occupy all four quadrants of the Ramachandron plot and permit a broad range of conformations. With increasing length and complexity of the side chain R, the degree of steric freedom becomes increasingly smaller.
Figure 2.8 Ramachandran plots to show (left) the permissible conformations of valine and isoleucine side groups and (right) for a perfect helix of poly-L-alanine, in terms of the angles ϕ and ϕ shown in Figure 2.7. (Adapted from Ramachandran and Sassiekharan [7, p. 337].)
2.4.2 Polypeptides and Proteins
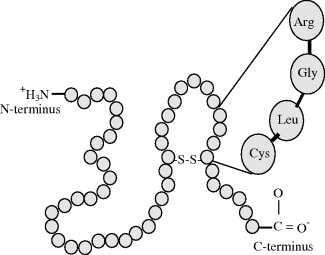
Three amino acids can covalently bond together to form a tripeptide, and when four do so we have a tetrapeptide, and so on to form resulting structures known as oligopeptides. When many amino acid residues bond together to form a long structure, the result is known as a polypeptide chain, as depicted in Figure 2.9. Proteins are formed from one or more polypeptide chains. To be able to perform their biological function (e.g. as an enzyme or a structural element such as a microfilament) proteins fold into one or more specific spatial conformations dictated by the sequence of residues in their polypeptide chains and the corresponding permitted values for the rotational angles ϕ and ϕ. Protein sizes range from a lower limit of around 50 to several thousand amino acid residues. An average protein contains around 300 residues. Very large aggregates can be formed from protein subunits, for example many thousand actin molecules assemble into a microfilament.
Figure 2.9 A polypeptide is formed of a chain of amino acid residues. It has a defined chemical orientation, with an N-terminus (bearing a free amino group) and a C-terminus (bearing a free carboxyl group). A disulphide bond linking two cysteine residues is shown.
There are four distinct categories of a protein's structure:
- Primary structure: This is defined by the amino acid sequence of the polypeptide chains. A specific gene in a cell determines the primary structure of a protein. As will be described later in this chapter, a specific sequence of nucleotides in DNA is transcribed into mRNA, which is then read by structures called ribosomes, in a process called translation. The sequence of a protein is unique to that protein, and defines its structure and function. The primary structure is held together by the covalent peptide bonds made during the process of protein biosynthesis or translation by ribosomes. These peptide bonds provide rigidity to the protein. The primary structure can also be defined by the covalent bonding of sulphur atoms between two cysteine residues in the same or different polypeptide chains. These bonds are termed disulphide bridges, and an example is given in Figure 2.9.
- Secondary structure: This refers to the arrangement of parts of a polypeptide chain into highly regular substructures, the most prominent of which are the alpha helix and the beta-pleated sheet structures shown in Figure 2.10. Hydrogen bonds are responsible for stabilising these two structures. The conformations of the amino acid residues in the alpha helix correspond to values for ϕ of −45° to −50° and ϕ = −60°, and each turn of the helix includes 3.6 residues. Each residue participates in a hydrogen bond, so that each successive helix turn is held in place to an adjacent helix turn by three to four hydrogen bonds. The residues in a beta-pleated sheet structure have conformations with ϕ = −135° and ϕ = +135°, and is also held together by hydrogen bonds. However, because water-amide hydrogen bonds are generally stronger than amide-amide hydrogen bonds, these secondary structures are stable only when the local concentration of water is sufficiently low, as for example in the fully folded protein state.
- Tertiary structure: This is the 3D structure of a single protein molecule, involving the spatial arrangement of the secondary structures, including the folding of parts of the polypeptide chain between α-helices and β-sheets. As depicted in Figure 2.11, it describes the completely folded and compacted polypeptide chain. Several polypeptide chains can be combined into a single protein molecule through ionic interactions (salt bridges) between oppositely charged ionised side-chains, hydrogen bonds, hydrophobic ‘bonding’ interactions, disulphide bridges and intermolecular van der Waals forces between nonpolar groups. As a general rule, the hydrophilic (charged and polar) amino acid residues are located on the outside of a folded protein, with the hydrophobic residues buried inside the polypeptide structure.
- Quaternary structure: The forming of a complex of several protein molecules, or protein subunits, that function as part of a larger assembly or protein complex is referred to as a quaternary structure, an example of which is depicted in Figure 2.11. A protein may shift between several, reversible, similar structures in performing its biological function, either as an enzyme controlling chemical reactions or as a structural element.
Figure 2.10 The α-helix (left) structure is held together by hydrogen bonds (NH O) that are formed nearly parallel to the long axis of the helix. The β-sheet (right) is held together by hydrogen bonds between adjacent sections of a polypeptide chain.
O) that are formed nearly parallel to the long axis of the helix. The β-sheet (right) is held together by hydrogen bonds between adjacent sections of a polypeptide chain.
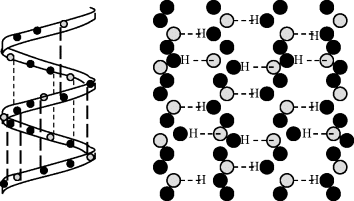
Figure 2.11 The tertiary structure (left) describes the links between α-helices and β-sheets and all of the noncovalent interactions that stabilise the correct folding of a single polypeptide chain in a protein. The quaternary structure (right) refers to the noncovalent interactions that hold together several polypeptide chains into a single protein molecule, as for example chymotrypsin, haemoglobin and RNA polymerase.
Hair is primarily composed of keratin – a fibrous structural protein that is also a key structural component of skin and finger nails. Disulphide bonds between cysteine residues in the keratin structure give hair its elasticity. A strand of straight hair can be transformed into curly hair (or curly transformed to straight) by wrapping it around curved (or straight) rods and breaking the disulphide bonds using a reducing solution of sodium or ammonium thioglycolate at a pH of 8 ~ 10, together with applied heat (the earliest methods used a mixture of cow urine and water!). After 15 ~ 30 minutes an oxidising lotion (e.g. hydrogen peroxide) and an alkali neutraliser is applied to bring down the pH and to close the disulphide bonds again to reform the hair into the shape of the rod.
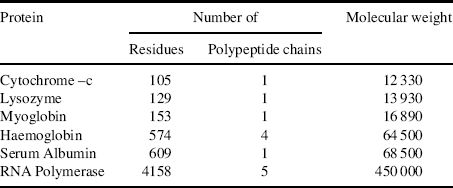
The numbers of constituent amino acid residues and polypeptide chains for several proteins are presented in Table 2.5.
Table 2.5 The composition and molecular weight of some proteins.
Some proteins, known as conjugated proteins, contain chemical components that are not amino acid residues. These components, known as prosthetic groups, are held in place in the protein structure through interactions with peptide unit side chains. Metalloproteins contain complexes that incorporate metals such as zinc, calcium and copper, and haemoglobin contains an iron complex known as a haem or porphyrin group. Casein found in milk contains a phosphate group, and is an example of a phosphoprotein. The lipoproteins found in blood contain lipids, and glycoproteins contain carbohydrates, of which the immunoglobulin antibody is a good example.
For specified conditions and physiological state of a cell, its complete set of proteins is known as its proteome, whose study and application is given the name proteomics (rather like genomics for the study of genes). Proteomics includes determinations of: the dynamics and structure of proteins; their abundance (expression) and localisation in a cell; chemical modifications of proteins after their translation from mRNA, and the extent and types of protein-protein interactions. An important post-translational modification is the phosphorylation (addition of a phosphate) of many enzymes and structural proteins involved in cell signalling processes. Cell signalling and cell-cell communication, to be described further in Section 2.6.11, governs basic cellular activities and coordinates cell actions. The ability of cells to perceive and correctly respond to their microenvironment is the basis of their development, tissue repair, immunity, and the regulation of their internal environment to maintain a stable and constant physiological state (homeostasis). Errors in cell signalling can result in various diseases, including cancer and diabetes, and by understanding this diseases may be treated effectively. Proteomics also offers the promise to identify targets for new drugs. For example, if a certain enzyme is known to be implicated in a disease, its three-dimensional structure can provide the information to design a drug molecule that fits into the active site of the enzyme and blocks its activity. As knowledge of the genetic differences between individuals is gained, the development of personalised drugs will become possible.
A protein's function is determined by its three-dimensional structure, which in turn is determined by the linear sequence(s) of the amino acids in the polypeptide chain(s) of which it is composed. Instructions for the assembly of the amino acid sequence are coded by the linear sequence of nucleotides of the nucleic acid DNA.
2.5 Nucleotides, Nucleic Acids, DNA, RNA and Genes
Nucleic acids are responsible for storing the information and instructing the cell about the proteins it should synthesise. The chemical nature of this genetic process was discovered in 1944, and the description of the structure of the DNA double helix was given by Watson and Crick in 1953 [8]. The DNA double helix has assumed the symbol for the discipline of molecular biology, whose primary function is elucidation of the nature and methods of replication and expression of genetic information. The exciting way in which molecular biology evolved has been described by Judson in his colourful book The Eighth Day of Creation, [9]. What follows here is a basic description of the molecular actors and actions of this story.
The two information-storing molecules in cells are deoxyribonucleic acid (DNA) and ribonucleic acid (RNA). Proteins are polymers constructed from 20 different monomers (the amino acids) but DNA and RNA are comprised of just four monomers – called nucleotides. A nucleotide is composed of a phosphate group (P) and a ‘base’ linked together by a five-carbon sugar molecule (pentose):
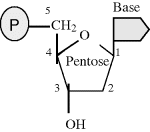
The bases found in DNA are adenine, guanine, cytosine and thymine, often abbreviated as A, G, C and T. In RNA the thymine base (T) is replaced by uracil (U).
In DNA the pentose sugar molecule is deoxyribose, whereas in RNA it is ribose.
2.5.1 DNA
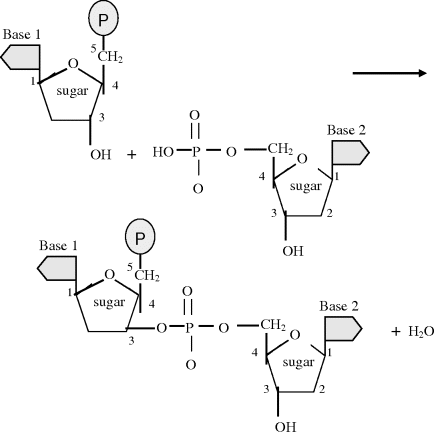
Nucleic acids consist of polymer chains of nucleotides, formed in a condensation reaction to create a phosphodiester bond, in which, as shown in Figure 2.12, a water molecule is released (as with the production of a glycosidic bond between sugars or a peptide bond between amino acids). Two nucleotides joined by such a bond forms a dinucleotide, and a trinucleotide represents a single strand of DNA containing three nucleotides. Additional nucleotides can be added to produce a long DNA single strand having a defined chemical orientation. One end (the so-called 3′ end) of a DNA strand has a free hydroxyl group (attached to carbon 3 of the sugar), whilst the other end (the 5′ end) has a phosphate group. This orientation has important implications regarding the properties of DNA.
Figure 2.12 The condensation reaction that links two nucleotides with a phosphodiester bond.
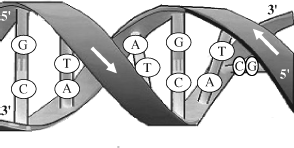
At the beginning of Chapter 1, the ability to self-produce was given as a defining characteristic of a living system. The metaphor has even been proposed that living things are ‘chemical machines, whose object is to make two where there was one before’ [10]. The entity in a cell that is responsible for this ‘secret of life’ is DNA. The biologically native state of DNA is a double helix composed of two intertwined single strands of DNA, as schematically shown in Figure 2.13.
Figure 2.13 A schematic of the DNA double helix.
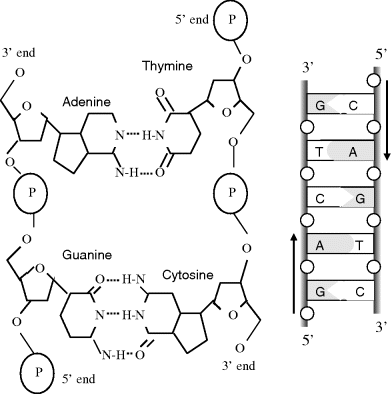
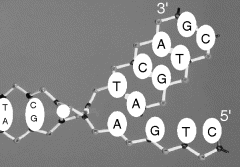
As indicated in Figure 2.13, the two single strands of DNA in the double helix structure proceed from carbon 5′ to 3′, but are directed in opposing directions. The two DNA strands are held together by hydrogen bonds linking their bases, and only if the two strands of the helix are antiparallel can the members of each base pair fit together within the double helix. The two DNA strands can, in theory, form either a right-handed or left-handed helix, but the structure of the sugar-phosphate backbone is such that the right-handed helix is the more favourable geometry. As first proposed by Watson and Crick [8] (with their famous understatement: ‘It has not escaped our notice that the specific pairing we have postulated immediately suggests a possible copying mechanism for the genetic material.’) the size, shape and chemical composition of the bases dictates that base A is always paired with T, and G is paired with C. As shown in Figure 2.14 the A-T pair is held together by two hydrogen bonds and the G-C pair by three. To break the G-C pair thus requires more energy (87.9 kJ/mol) than that required to break the A-T pair (50.6 kJ/mol) [11]. This difference is reflected in the finer details of how the DNA polymer is copied. X-ray studies have determined that the stacked bases are regularly spaced 0.34 nm apart along the helix, and that the length of one complete helix turn is 3.4 nm (to give ~10 pairs of bases per turn). The hydrogen bonds between the bases gives the double helix considerable stability and rigidity, but also allows the double helix a good degree of flexibility, enabling long DNA chains to coil up to form supercoils or condensed structures of very large molecular weight. The polypeptide alpha-helix shown in Figure 2.10 is far less flexible, because the hydrogen bonds hold together adjacent parts of the helix.
Figure 2.14 A schematic of the double-stranded DNA, showing the base-pair complementarity of the A-T and G-C pairings.
From Table 2.5 we note that the number of amino acid residues in proteins ranges from hundreds to several thousands – whereas DNA molecules are typically very much larger. For example, the DNA molecule in the single chromosome of an E. coli bacterium comprises just under five million base pairs (this number of base-pairs defines the genome size of E. coli). Thus, if fully stretched out its DNA would have a length of ~1.5 mm – some three orders of magnitude longer than the E. coli bacterium itself! So how does the chromosome package itself inside this bacterium? The solution lies in the flexibility of a DNA double helix that allows it to coil and fold into a superhelix. This can be simulated by continuously twisting an elastic band and slowly bringing the ends together, so that the twisted band first forms small coils that then proceed to curl into a tight knot. Human cells contain 46 chromosomes, containing a total of 3.2 × 109 base-pairs. If the DNA from all 46 chromosomes of a single human cell were to be connected and straightened out, its total length would be ~2 m!
To assist in the packaging of this DNA into the nucleus of a human cell it is wrapped around protein molecules, called histones, to form structural units called nucleosomes that are spaced at regular intervals along the main DNA chain, rather like beads on a string. Arrays of nucleosomes form chromatin fibres that are then further packaged into chromosomes. This form of DNA packaging occurs in cells with a nucleus (eukaryotic cells) but not in prokaryotic cells such as bacteria, where typically the total DNA forms a large circular molecule. A gene corresponds to a stretch of DNA that contains the sequential information for the production of proteins or RNA chains that have functional roles in the cell. Some stretches of DNA do not encode for proteins or RNA, and at present a quite large percentage of this so-called ‘junk’ DNA has no known biological function. The entirety of the genes and noncoding sequences of DNA in a cell is called its genome. Many types of virus do not possess DNA, and instead their genome consists of the coding information contained in another polynucleic acid called RNA.
At each division of a cell in an organism an exact copy must be made of its genome. How is this accomplished? In Figure 2.14 we see that one strand, say S1, of the DNA double helix is composed of a sequence of nucleotides that is exactly mirrored by its complimentary bases in the other strand S2. Thus, strand S1 can serve as a template for making a new S2 strand, and strand S2 can serve the same purpose for S1. This replication process can be observed, using an electron microscope, in the form of Y-shaped structural forks moving along a DNA double helix (Figure 2.15).
Figure 2.15 A schematic representation (as viewed with an electron microscope) of a portion of a DNA double helix in the process of being replicated. The arrows indicate the locations of four Y-shaped forks and their directions of propagation (Derived from Kornberg & Baker [12].)

The forks shown in Figure 2.15 are produced by initiator proteins that bind to the DNA with the result that strand S1 separates away from strand S2. This requires the breaking of the hydrogen bonds that hold together the base pairs, and tends to happen at an A-T base pair because the energy required is about half of that required to break the G-C base pair. A group of proteins are then attracted to the exposed fork, an important member of which is the enzyme DNA polymerase that places a new complimentary DNA strand onto each separated S1 and S2 to produce two new DNA double-helices. DNA polymerase can synthesise a new DNA strand continuously in only one direction, because it adds new nucleotides to the 3′ end of a strand – not to the 5′ end, as depicted in Figure 2.16. This limitation is overcome through a discontinuous process whereby the polymerase ‘backstitches’ short sections of DNA onto the 5′ end strand with the assistance of another enzyme called DNA ligase. The new nucleotides enter these processes as nucleoside triphosphates, the breaking of whose bonds provides the free energy required for the polymerisation reaction. DNA polymerase also serves as its own ‘proofreader’ to ensure that the correct complimentary base has been inserted. In this way less than one error is typically made for every ten million new base pairs formed. A variety of DNA repair enzymes also continuously scan for and correct replication mistakes, or replace damaged nucleotides, using the uncorrupted DNA strand as the template.
Figure 2.16 At a replication fork DNA polymerase continuously synthesises a new DNA strand by adding new nucleotides to the separated strand with the 3′ end. This produces one copy of the original DNA double helix. Complimentary bases are discontinuously ‘backstitched’ onto the other strand, to form another copy of the original DNA molecule.
The research that led to the understanding of DNA replication, and to the discovery of DNA polymerase, is described in a book by Arthur Kornberg who in 1959 was awarded the Nobel Prize in Physiology and Medicine for this work [13].
2.5.2 Ribonucleic Acid (RNA)
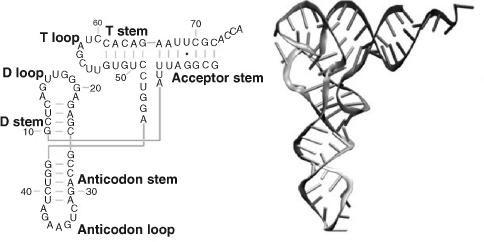
RNA is very similar to DNA but differs in a few important structural details. In a cell, whereas DNA takes the form of a double-stranded helix, RNA is single-stranded. This means that a RNA chain is much more flexible than DNA and can fold up into a variety of three-dimensional shapes containing sections of single strand loops and double helices wherever parallel strands are able to form complimentary nucleotide base pairs. An example of this can be found in the transfer RNA molecule shown in Figure 2.17. Some of the shapes that RNA molecules can adopt enable them to perform catalytic functions. Another difference is that the RNA nucleotides contain ribose (DNA contains deoxyribose – a type of ribose that lacks one oxygen atom) and has the base uracil rather than thymine present in DNA. Examples of complimentary A-U pairing in a RNA molecule can be seen in Figure 2.17.
Figure 2.17 The nucleotide sequence and 3-Dimensional structure of a transfer RNA (tRNA) molecule. (Reproduced with permission from Flores, S.C., and Altman, R.B., RNA, 16: 1769–1778, 2010.)
Different types of RNA are central to the synthesis of proteins and are transcribed from DNA by enzymes called RNA polymerases. These enzymes bind to the DNA in the nucleus of eukaryotic cells, separate the two strands of the nuclear DNA, and pair ribonucleotide bases to the template DNA strand according to the Watson-Crick base-pairing interactions shown in Figure 2.14 (with uracil replacing thymine). Thus, referring to the replication of a DNA molecule depicted in Figure 2.16, the action of RNA polymerase is to produce a strand of RNA with a nucleotide sequence CUGA (rather than the sequence CTGA of a DNA strand if DNA polymerase had been in action). Many RNA polymerases can act on a single strand of DNA at the same time to speed up the transcription process. Roger Kornberg, the son of Arthur Kornberg mentioned for his work leading to the discovery of DNA polymerase, was awarded a Nobel Prise in 2006 for his detailed molecular images of RNA polymerase during various stages of the transcription process.
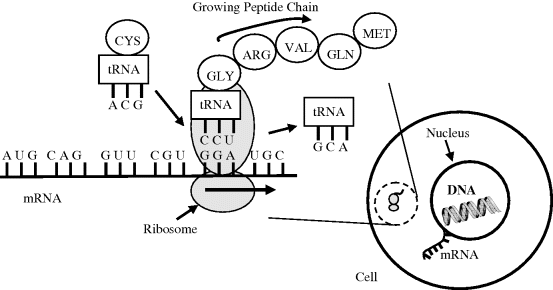
A type of RNA called messenger RNA (mRNA) carries coding information, obtained from the DNA template, in the form of tri-nucleotide units called codons that each code for a single amino acid. Strands of mRNA then interact in the cytoplasm with protein structures called ribosomes (in recent years ribosomes have become important targets in the search for new antibiotics to fight the emergence of drug resistant bacteria). In eukaryotic cells the mRNA is formed inside the nucleus, and has to pass through pores in the nuclear membrane to locate organelles known as ribosomes in the cytoplasm. Ribosomes consist of proteins and ribosomal RNA polymers, which together act as a molecular ‘machine’ to read mRNA and to translate the information it carries into the production of amino acid chains that form proteins. Different types of transfer RNA (tRNA) molecules mediate this process by transferring a specific amino acid to the growing peptide chain. The different tRNA molecules can be attached to only one type of amino acid, and each one contains a three base anticodon that can base pair to the corresponding codon on the mRNA chain. The ‘anticodon’ arm whose loop contains the anticodon is shown in Figure 2.17, together with the 7 base-pair stem that attaches to an amino acid. This process is shown schematically in Figure 2.18.
Figure 2.18 Translation of mRNA (from right to left) into a peptide chain. The ribosome begins at the start triplet codon (AUG) at the 3′ end of the mRNA, which also codes for methionine. The triplet codons (CAGGUUCGUGGA) that follow produce glutamine, valine, arginine and glycine in the growing peptide chain. A transfer RNA molecule, with its anticodon ACG, brings cystine towards the codon UGC site on the mRNA.
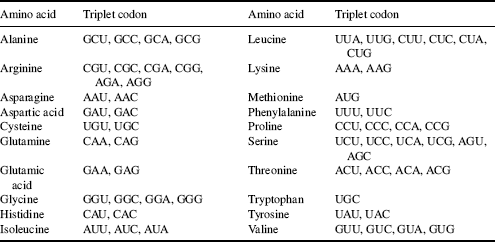
There are 43 = 64 different codon combinations possible with a triplet codon of three nucleotides, and all 64 codons are assigned for either amino acids or start and stop signals during translation of the mRNA code into a polypeptide sequence. Because there are only 20 common amino acids, there is some redundancy in the assignment of the mRNA triplet codons, as shown in Table 2.6. The codons UAA, UGA and UAG are used as instructions to stop the translation process.
Table 2.6 The synthesis (translation) of the common 20 amino acids by ribosomes employs the triplet codons (three nucleotide sequences) carried by mRNA listed in this table.
The translation example shown in Figure 2.18 shows a strand of mRNA with the initial codon AUG that serves as an initiation site, where translation into a polypeptide chain begins, and also as the code to produce methionine. The mRNA sequence, CAGGUUCGUGGAUGC, that follows is translated into a chain of five amino acids comprising glutamine-valine-arginine-glycine-cystine. In Figure 2.18 the tRNA molecule that added arginine to the peptide chain is shown leaving the ribosome, whilst a tRNA with its anticodon ACG carries a cystine molecule to the site on the ribosome where the translation process takes place.
The fact that RNA is able to both store information and catalyse chemical reactions has led some biologists to the conclusion that RNA predates DNA in the evolution of living systems [14]. The reign of the RNA world on Earth may have lasted between 3.6 and 4.2 billion years ago. This would have been based on RNA genomes that are copied and maintained through the catalytic function of RNA. Remnants of this may still exist in some microenvironments that have survived to this day, and the construction of artificial RNA-based life from synthetic oligonucleotides may be possible. Until relatively recently, a difficulty with this concept has been the fact that the single-stranded RNA normally expands into a chain one nucleotide at a time, and that in the primordial world RNA did not have enzymes to catalyse this reaction. In 2009, however, it was demonstrated that under favourable conditions of temperature and pH in water small fragments of RNA can fuse into larger lengths of 120 nucleotides and more [15]. This enzyme- and template-independent synthesis of long oligomers of RNA in water, from chemicals that would have existed in prebiotic times, certainly approaches the concept of spontaneous generation of (pre)genetic information.
2.5.3 Chromosomes
Earlier we deduced that, if stretched out and joined end to end, the total amount of DNA in each human cell (apart from red blood cells which do not have a nucleus) would have a length of about 2 m. This total DNA, the human genome, contains approximately 3 × 109 nucleotides and is distributed as long lengths of DNA in chromosomes. Apart from the germ cells (eggs and sperm) a typical human cell contains two copies of 22 of these chromosomes, numbered from 1 to 22 in order of diminishing physical size. Females have two X chromosomes and males one X and one Y chromosome to give a total of 46 chromosomes. The X chromosome is inherited from the mother and the Y chromosome from the father. The 22 chromosomes, plus the X and Y chromosome, can be distinguished from one another by staining with dyes that distinguish between DNA that is rich in either A-T or G-C nucleotide base pairs. Each chromosome type can be identified by the distinctive patterns of coloured bands along them, and chromosomal abnormalities can also be detected.
2.5.4 Central Dogma of Molecular Biology (DNA Makes RNA Makes Protein)
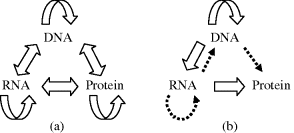
This dogma was first enunciated by Francis Crick [16] and states that the sequential structural information stored in a protein cannot be transferred to another protein or to a nucleic acid. (Crick used the word ‘dogma’ by way of a catch phrase without realising its implied interpretation – in fact he wished his concept to be considered as an hypothesis [Ref. [9], Chapter 6]) In living systems there are three major classes of linear biopolymer, namely DNA, RNA and proteins, whose monomer sequences encode information. There are 9 conceivable direct transfers of information possible between these three classes, as depicted in Figure 2.19a. The transfer of information is assumed to be an error-free transfer in which the molecular sequence of one biopolymer is used as a template to construct another biopolymer with a molecular sequence that depends entirely on that template. Transfers that can occur in all cells, known as general transfers, are the three cases of DNA → DNA (DNA replication), DNA → RNA (transcription), and RNA → protein (translation). Special transfers are ones which do not occur in most cells but may occur in special circumstances, such as in virus-infected cells, and are the three cases of RNA → RNA, RNA → DNA, and DNA → Protein. A known example of the RNA-DNA transfer takes place in retroviruses, where DNA is synthesised using RNA as a template. An enzyme known as reverse transcriptase carries out this process. The human immunodeficiency virus (HIV) is a retrovirus and is the cause of AIDS.
Figure 2.19 (a) The 9 conceivably possible direct transfers of information between DNA, RNA and proteins. (b) The central dogma of molecular biology states that only the transfers represented by the block arrows are possible. The dotted arrows indicate special transfers under specific conditions, such as those involving retroviruses or artificially in a test-tube.
After HIV has bound to a target cell, normally one of the vital blood cells of the immune system, the RNA content of the virus and various enzymes including reverse transcriptase and ribonuclease, and protease, are injected into the cell. The single strand of viral RNA genome is then transcribed into double strand DNA and integrated into a chromosome of the host cell, which can lead to possible reproduction of the virus [17].
The three special transfer possibilities are included in Figure 2.19b as dotted arrows. Finally, the three remaining transfer possibilities, known as unknown transfers, are considered never to occur and are not included in Figure 2.19b.
The block arrows in Figure 2.19b therefore summarise the normal flow of biological information, namely that DNA information can be transcribed into messenger RNA, and then translated into the synthesis of proteins using the information in mRNA as a template. The master template stored in DNA can itself be replicated, so that the cycle DNA-makes-RNA-makes-protein can be repeated in a new generation of cells and biological organisms. The information stored in a protein cannot be used to make new DNA, RNA or another protein.
2.6 Cells and Pathogenic Bioparticles
We complete this chapter with a summary description of different types of cells, including blood cells and bacteria, along with viruses and prions as examples of pathogenic bioparticles.
The cell is the structural and functional unit of all known living organisms. It is the smallest unit of an organism that is classified as living, and is often called the building block of life. Organisms, such as most bacteria, consist of a single cell but other organisms such as animals are composed many cells, they are multicellular. Humans, for example, comprise around 1014 cells of typical diameter 10 μm, and each of mass around 1 nanogram. Cells are limited in size by the ratio between their outer surface area and their volume. A small cell has more surface area through which to exchange nutrients, gases and other chemicals between the external and internal cell media than a large cell for a given volume of nucleus. There is also a limit to the amount of biochemical processes that a nucleus can control in a cytoplasm.
2.6.1 Prokaryotic and Eukaryotic Cells
The names for these two basic cell types are derived from the Greek word Karyose, meaning ‘kernel’, which in biology is used to refer to the nucleus of a cell. Pro means before, and eu means true or good. Thus, Prokaryotic means before a nucleus, and eukaryotic means possessing a true nucleus.
These two cell types perform many similar biological functions. Both are enclosed by plasma membranes, filled with cytoplasm, and loaded with ribosomes. Both have DNA which carries the archived instructions for operating the cell. The DNA in the two cell types has the same structural form, and the genetic code for a prokaryotic cell is exactly the same genetic code used in eukaryotic cells. Eukaryotic animal cells are generally thought to have descended from prokaryotes that lost their cell walls. The cell wall has pores that allow materials to enter and leave the cell, but they are not very selective about what passes through. The plasma membrane, which lines the inner cell wall surface, provides the final filter between the cell interior and the environment. With only the flexible plasma membrane left to enclose them, the primordial prokaryotes would have been able to expand in size and complexity. Eukaryotic cells are generally ten times larger than prokaryotic cells and have membranes enclosing interior components, the organelles. Like the exterior plasma membrane, these membranes also regulate the flow of materials, allowing the cell to segregate its chemical functions into discrete internal compartments.
Other important differences between prokaryotic and eukaryotic cells include:
- Eukaryotic cells have a true nucleus, representing the largest organelle in a cell, bound by a double membrane. Prokaryotic cells have no nucleus. The purpose of the nucleus is to sequester the DNA-related functions of the big eukaryotic cell into a smaller chamber, for the purpose of increased efficiency. This function is unnecessary for the prokaryotic cell, because it's much smaller size means that all materials within the cell are relatively close together.
- Eukaryotic DNA is linear; prokaryotic DNA is circular and has no ends.
- Eukaryotic DNA is complexed with proteins called histones, and is organised into chromosomes. Prokaryotic DNA is naked, meaning that it has no histones associated with it, and it is not formed into chromosomes. A eukaryotic cell contains a number of chromosomes; a prokaryotic cell contains only one circular DNA molecule and a varied assortment of much smaller circlets of DNA called plasmids. The smaller and simpler prokaryotic cell requires far fewer genes to operate than the eukaryotic cell.
- Both cell types have a large number of ribosomes, but the ribosomes of the eukaryotic cells are larger and more complex than those of the prokaryotic cell. Ribosomes are composed of a special class of RNA molecules (ribosomal RNA, or rRNA) and a specific collection of different proteins. A eukaryotic ribosome is composed of five kinds of rRNA and about eighty kinds of proteins. Prokaryotic ribosomes are composed of only three kinds of rRNA and about 50 kinds of protein.
- The cytoplasm of eukaryotic cells takes the form of a gel-like material filled with a complex collection of organelles, many of them enclosed in their own membranes. A prokaryotic cell contains no membrane-bound organelles. This is a significant difference and the source of the vast majority of the greater complexity of the eukaryotic cell. There is much more space within a eukaryotic cell than within a prokaryotic cell, and many of the organelles structures, like the nucleus, increase the efficiency of bioreactions by confining them within small volumes. If the organelles are removed, the soluble part of the cytoplasm that remains is called the cytosol, consisting mainly of water and dissolved substances such as mineral salts and amino acids.
A summary of the main characteristics that distinguish prokaryotic from eukaryotic cells is given in Table 2.7.
Table 2.7 The characteristic differences between prokaryotic and eukaryotic cells.
| Feature | Prokaryote | Eukaryote |
| Size | Small: 0.5 ~ 5 μm | 5 ≥ 50 μm |
| Genetic material | Circular DNA (in cytoplasm) | DNA in form of linear chromosomes (in nucleus) |
| Organelles | Few present | Many organelles |
| Cell walls and other structures | Rigid, formed from glycoproteins. (Bacteria also contain flagellum, plasmid and capsule) | Fungi: Rigid, formed from polysaccharides (chitin). |
| Plant: Rigid, formed from polysaccharides (e.g. cellulose). | ||
| Animals: No cell wall |
2.6.2 The Plasma Membrane
All living cells have a plasma membrane that encloses their contents and serves as a semiporous barrier to the outside environment. The membrane acts as a boundary, holding the cell constituents together and keeping other substances from entering. The plasma membrane is permeable to specific molecules, however, and allows nutrients and other essential elements to enter the cell and waste materials to leave the cell. Small molecules, such as oxygen, carbon dioxide, and water, are able to pass freely across the membrane, but the passage of larger molecules (e.g. amino acids and sugars) is carefully regulated. The biophysical and electrical properties of membranes are discussed further in Chapter 3.
According to the accepted current model, known as the fluid mosaic model, the plasma membrane is composed of a phospholipid bilayer (see Figure 2.2). Individual lipids and proteins can move freely within the bilayer as if it was a fluid, and the membrane-bound proteins form a mosaic pattern when looking at the membrane surfaces. Within the phospholipid bilayer of the plasma membrane, many diverse proteins are embedded, while other proteins simply adhere to the surfaces of the bilayer. Some have carbohydrates attached to their outer surfaces and are referred to as glycoproteins. The positioning of proteins on the plasma membrane is related in part to the organisation of the filaments that comprise the cytoskeleton, which help anchor them in place. The cytoskeleton forms the framework of a cell. It consists of protein microfilaments and larger microtubules that support the cell, to give it its shape and help with the movement of its internal organelles. The arrangement of proteins also involves the hydrophobic and hydrophilic regions found on the surfaces of the proteins. The hydrophobic regions of the protein associate with the hydrophobic interior of the plasma membrane, whereas hydrophilic regions extend past the surface of the membrane into either the cytosol of the cell or the outer environment. Many of the transmembrane protein structures form channels and pumps.
An important membrane ion pump is the Na+-K+ pump. This pump actively transports Na+ ions out of a cell, and K+ ions into a cell, against their electrochemical gradients. This is described in more detail in Chapter 3. The Na+-K+ pump plays a direct role in regulating the osmolarity of the cytosol, along with the action of water channel proteins (aquaporins) that allows water to flow down its activity gradient into or out of a cell. To avoid influencing the ion gradients across the membrane, an aquaporin permits the rapid passage of water molecules but blocks the passage of ions. This is achieved through the special structure of the aquaporin channel, which consists of a narrow pore lined on one side by hydrophilic amino acids and on the other side by hydrophobic amino acids. The water molecules follow the path, one by one in single file, presented by the hydrophilic groups, to which they make transient hydrogen bonds with carbonyl oxygens [18]. The pore diameter is too small to permit the passage of hydrated ions and as explained in the next chapter the energy required to remove the hydration shell around an ion, plus the electrostatic interaction of a bare ion with a hydrophobic surface, presents an insurmountable energy barrier for the passage of an ion. We also learnt in Chapter 1 that protons in solution exist as the hydronium ion (H2O)3H3O+, shown in Figure 1.4, so that along with Na+, K+, Ca2+ and Cl- ions we can also understand why aquaporins do not allow the passage of H+ ions (protons).
2.6.3 The Cell Cycle
A necessary ability of all living cells is to duplicate their genomic DNA and to pass identical copies of it to their daughter cells. All growing cells perform this function in their cell cycle, which consists of two periods, namely the period of cell division and an interphase period of cell growth. The ways in which prokaryotic and eukaryotic cells coordinate their DNA synthesis and cell division are quite different.
As indicated above, the genome of a prokaryotic cell is a single circular molecule of DNA. In rapidly growing prokaryotes, such as bacteria, its DNA is replicated throughout much of the cell cycle process. The circular chromosome of the mother cell is attached to the internal plasma membrane surface to facilitate the DNA-replication process. When this replication is complete, the new chromosome is attached at another site on the membrane. A new membrane and cell form in the region between the points of attachment of the two chromosomes, parts of which invaginate to produce a septum that divides the cell. The two daughter cells separate, each one with its chromosome attached to its inner membrane surface. The cell cycle is complete.
The life-cycle scheme for all growing and dividing eukaryotic cells is shown in Figure 2.20. Nerve cells and striated muscle cells do not divide. These cells do continue to synthesise RNA, proteins and membrane material, but do not replicate their DNA. The red blood cells of mammals also do not divide – they do not possess a nucleus.
Figure 2.20 The cell cycle for dividing mammalian cells. During the interphase period consisting of G1, S and G2, DNA and other cellular materials such as RNA, protein and membrane are synthesised. Cell division occurs during the mitosis (M) period. Terminally differentiated cells, or cells in a culture with depleted nutrients, stop at the G0 state.
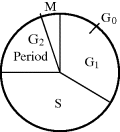
The major part of the cell cycle shown in Figure 2.20 comprises the G1 (first gap), the S (synthetic) phase, and the G2 (second gap) period. These make up the interphase when new DNA, membrane material and other macromolecules are synthesised. The remaining, relatively short, part of the cell cycle is the M (mitosis) period during which time the cell divides (cytokinesis). The replication of DNA and the synthesis of histone proteins take place only during the S phase, during which each DNA double-helix is replicated and quickly combined with histones and other chromosomal proteins. During the two gap periods, G1 and G2, no net synthesis of DNA occurs but damaged DNA can be repaired. During the G2 period a cell should contain two copies of each of the DNA molecules existing in the call at G1. Throughout the interphase (G1, S and G2) there is continuous cell growth and synthesis of macromolecules such as RNA, proteins and membranes. Finally, during the relatively brief mitotic (M) period the cell divides, and identical copies of the DNA are distributed to each of the two daughter cells. Nonreplicating cells such as nerve or striated muscle cells are in the so-called terminally differentiated state and are usually halted in the cell cycle at the G0 stage shown in Figure 2.20. During cell culture growth of mammalian cells the S, G2 and M periods are roughly constant, whereas the G1 period can vary greatly depending on the culture conditions. If the culture medium becomes depleted of the required nutrients or hormones, for example, they can remain at the G0 stage for many hours or days, until stimulated back into G1 by the addition of the missing growth medium components.
2.6.4 Blood Cells
An adult human body contains 5 ~ 6 litres of blood, 55% of which is a liquid called plasma or serum (when clotting factors are removed). The remaining 45% comprises the blood cells. The blood performs important functions. These functions include: carrying oxygen to the tissues and collecting carbon dioxide; conveying nutritive substances (e.g. amino acids, sugars, mineral salts) and collecting waste material to be excreted; transporting hormones, enzymes and vitamins. A very important function is to protect an organism against disease agents, using the immune response provided by lymphocytes, the phagocytic activity of leukocytes, and the bactericidal power of the plasma. These topics are of relevance to bioelectronics for situations where a sensor or other type of electronic device is implanted in the body. Tissue damage will occur on implanting such a device, and its performance can be degraded as a result of the inflammatory and immune responses involved in the healing of this wound. This is described further in Chapter 6, Section 6.12.
The cellular composition of normal human blood is given in Table 2.8. 1 μL of whole blood (equivalent to about one-fortieth of a drop of blood) contains up to six million red blood cells and a much smaller number (~300) of B cells. An increase in the number of lymphocytes is normally an indication of a viral infection, whilst a lower than normal concentration can be associated with an increased rate of infection after surgery or traumatic injury. A reduction of T cells occurs when the human immuno-deficiency virus (HIV) infects and destroys T cells.
Table 2.8 The composition of 1 μL of human blood (equivalent to ~1/40th of a drop of blood).
| Erythrocytes (red cells) | 5 ~ 6 million |
| Platelets | ~ ¼ million |
| Leukocytes (white cells) | ~ 7 thousand |
| Comprising: | |
| 4400 Granulocytes | |
| 400 Monocytes | |
| 2200 Lymphocytes | |
| Comprising: | |
| 1500 T-cells | |
| 400 NK-cells | |
| 300 B-cells |
2.6.4.1 The Plasma
If a sample of blood is centrifuged, the cells sediment to the bottom of the tube to leave ~55% of the sample at the top in the form of a slightly alkaline (pH 7.4) and pale yellow fluid, comprising 90% water and 10% solid matter (9 parts organic and 1 part mineral). The organics include amino acids, glucose, hormones, lipids, proteins and vitamins. The minerals take the form of ions such as Na+, K+, Ca2+, Mg2+ and Cl−.
2.6.4.2 Platelets (Thrombocytes)
The main function of platelets is to prevent the loss of blood in injured tissues, by aggregating and releasing chemicals to promote blood coagulation. Released substances include serotonin which reduces the diameter of damaged blood vessels, and fibrin to trap cells and form a clot. They have a diameter of 2 ~ 3 microns and are not considered to be real cells.
2.6.4.3 Erythrocytes (Red Cells)
These cells are rich in haemoglobin (~250 million per cell), which is a protein able to bind oxygen and thus responsible for providing oxygen to tissues. Where there is a high concentration of oxygen in the body, such as in the alveoli of the lungs, each haemoglobin molecule binds four oxygen molecules to form oxyhaemoglobin. When an erythrocyte reaches tissue with low oxygen concentration the haemoglobin releases these oxygens. Erythrocytes are also partly responsible for recovering carbon dioxide produced as waste, but most CO2 is carried by plasma in the form of soluble carbonates. The mean lifetime of erythrocytes is about 120 days, at which time they are retained by the spleen and then phagocyted (eaten) by macrophages.
In man and in all mammals, erythrocytes are devoid of a nucleus and have the shape of a biconcave lens, which allows more room for haemoglobin and raises the cell surface and cytoplasmic volume ratio. These characteristics maximise the efficiency of oxygen diffusion by these cells. In fishes, amphibians, reptilians and birds, erythrocytes do have a nucleus.
2.6.4.4 Leukocytes (White Cells)
These cells are responsible for the defence of the organism, and are of two types, namely granulocytes and lymphoid cells. The number ratio of white to red blood cells in normal human blood is ~1:700.
Granulocytes contain granules in their cytoplasm, which have different properties including a different affinity towards neutral, acid or basic stains. Granulocytes can thus be distinguished as neutrophils, eosinophil (or acidophils) and basophils;
- Neutrophils: Act to phagocyte bacteria and are present in large numbers in the pus of wounds. They are unable to renew the lysosomes used in digesting the bacteria and die after having phagocyted a few of them.
- Eosinophils: Attack parasites and phagocyte antigen-antibody complexes.
- Basophils: Possess a phagocytory capability, but also secrete anticoagulant and vasodilatory substances such as histamines and serotonin.
Lymphoid Cells consist of two types, namely lymphocytes and monocytes:
- Lymphocytes are cells, which besides being present in blood populate the lymphoid tissues and organs (e.g. thymus, bone marrow, spleen). They are slightly larger than erythrocytes, and have a nucleus that occupies nearly all of the internal cellular volume. They are also the main constituents of the immune system, which is the defence against the attack of pathogenic microorganisms such as viruses, bacteria, fungi and protista (e.g. unicellular organisms). Lymphocytes produce antibodies, which appear on their outer plasma membrane. An antibody is a molecule able to ‘recognise’ and bind itself to molecules called antigens. As for all proteins, these antibodies are coded by genes. On the basis of a recombination mechanism of some of these genes, every lymphocyte produces antibodies of a specific molecular shape. The number of lymphocytes circulating in the blood is so large that they are able to recognise practically all the chemicals existing in the organism, both its own natural and foreign ones. They recognise hundreds of millions of different molecules!
The cells of the immune system, chiefly lymphocytes, cooperate amongst themselves to activate, boost or make more precise the immune response. To attain this scope, there exist different types of lymphocytes, with different functions, namely B and T lymphocytes. When the B cells are activated, they quickly multiply and secrete hosts of antibodies, which on meeting microorganisms with complementary shape (epitopes) bind to them and form complexes to immobilise the microorganisms. Other cells, which are not specific but able to recognise antibodies, phagocyte these complexes. In their turn, the T cells are divided into three categories: Tc (cytotoxic) cells kill infected cells directly by inducing them to undergo apoptosis (programmed cell death); Th (helpers) assist in activating B cells to make antibody responses; Ts (suppressors) suppress the activity of other T cells and are crucial for self tolerance. The immune system also produces memory cells, which are deactivated lymphocytes ready to be reactivated on further encounters with the same antigen.
Another population of lymphocytes in the peripheral blood and lymphoid organs do not have receptors for antigens. These lymphocytes have a nonspecific defence function that is not activated by Th lymphocytes. These cells represent the more ancient component of the immune system and they are characterised by their cytotoxic activity. They are called Natural Killer(NK) cells. Apart from killing viruses, bacteria, infected and neoplastic (abnormal) cells, these lymphocytes also regulate the production of other haematic cells such as erythrocytes and granulocytes.
- Monocytes are the precursors of macrophages. They are larger blood cells, which after attaining maturity in the bone marrow, enter the blood circulation where they stay for 24–36 hours. Then they migrate into the connective tissue, where they become macrophages and move within the tissues. In the presence of an inflammation site, monocytes quickly migrate from blood vessels and start an intense phagocytory activity. Macrophages also cooperate in the immune defence by exposing molecules of digested bodies on their membrane, presenting them to more specialised cells such as B and T lymphocytes.
2.6.5 Bacteria
Bacteria are prokaryotic cells and are about one-tenth the size of eukaryotic cells, typically 0.5 ~ 5.0 μm in length. They display a wide range of morphologies. Most are either spherical (cocci), rod-shaped (bacilli) or spiral-shaped (spirilla). Many bacterial species exist simply as single cells, but others associate in characteristic patterns. For example, Streptococcus form chains, and Staphylococcus group together into clusters. Only a small number of bacterial species cause disease in humans. Some of those that do so can only replicate inside the body and are called obligate pathogens. Other bacteria, called facultative pathogens, replicate in environments such as water or soil and only cause disease on encountering a susceptible host. Opportunistic pathogens are bacteria that are normally harmless but have a latent ability to cause disease in an injured or immuno-compromised host.
A plasma membrane encloses the contents of a bacterial cell and acts as a barrier to hold nutrients, proteins and other essential components of the cytoplasm within the cell. Many important biochemical reactions are driven by concentration gradients of ions and molecules across the membrane. Bacteria lack a nucleus, mitochondria, and the other organelles present in eukaryotic cells. Their genetic material is typically a single circular chromosome located in the cytoplasm in an irregularly shaped body called the nucleoid, which contains the chromosome with associated proteins and RNA. Around the outside of the plasma membrane is the bacterial cell wall, composed of peptidoglycan and made from polysaccharide chains crosslinked by peptides. There are broadly speaking two different types of cell wall in bacteria – Gram-positive and Gram-negative, according to their reaction with the Gram stain. Gram staining involves applying crystal violet to a heat-fixed smear of a bacterial culture, followed by the addition of iodine, rapid decolourisation with alcohol or acetone, and counterstaining. After decolourisation the Gram-negative cell has lost its outer membrane and loses its purple colour, whereas the Gram-positive cell remains purple. The counter-stain, safranin or basic fuchsin, is applied last to give decolourised Gram-negative bacteria a pink or red colour. This staining procedure works because Gram-positive bacteria possess a thick cell wall containing many layers of peptidoglycan and teichoic acids. The crystal violet-iodine complex becomes trapped within the peptidoglycan multilayers. Gram-negative bacteria, on the other hand, have a relatively thin cell wall consisting of a few layers of peptidoglycan surrounded by a second lipid membrane containing lipopolysaccharides and lipoproteins. These differences in structure can produce differences in antibiotic susceptibility. As a general rule Gram-negative bacteria are more pathogenic, because the lipopolysacharide in their outer membrane breaks down into an endotoxin which increases the severity of inflammation. The human body does not contain peptidoglycan, and humans produce an enzyme called lysozyme that attacks the open peptidoglycan layer of Gram-positive bacteria. Gram-positive bacteria are also more susceptible to antibiotics such as penicillin, which inhibit a step in the synthesis of peptidoglycan.
Bacteria are often grown in solid media, such as agar plates, to isolate and identify pure cultures of a bacterial strain. However, liquid growth media are used when measurement of growth or large volumes of cells are required. Growth in stirred liquid media occurs as an even cell suspension, making the cultures easy to divide and transfer, although isolating single bacteria from liquid media is difficult. The use of selective media (media with specific nutrients added or deficient, or with antibiotics added) can help identify specific organisms. Bacterial growth follows three phases. When bacteria first enter a high-nutrient environment that allows growth, the cells need to adapt to their new environment. The first phase of growth is the lag phase, a period of slow growth when the cells are adapting to the high-nutrient environment and preparing for fast growth. The lag phase has high biosynthesis rates, as proteins necessary for rapid growth are produced. The second phase of growth is the logarithmic phase, marked by rapid exponential growth. The rate at which cells grow during this phase is known as the growth rate, and the time it takes the cells to double is known as the doubling time or generation time. During log phase, nutrients are metabolised at maximum speed until one of the nutrients is depleted and starts limiting growth. The final phases are the stationary phase, followed by the death phase, caused by depleted nutrients. The cells reduce their metabolic activity, consume nonessential cellular proteins, and then die.
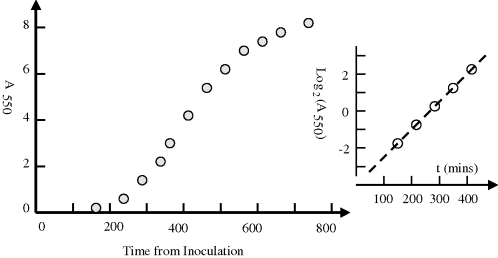
A practical way to monitor bacterial growth and to determine the doubling time is to periodically measure the optical absorbance of a sample of a bacterial suspension. The size of a typical bacterium is such that it will scatter light of wavelengths around 450 ~ 600 nm. An example of an absorbance plot taken over the lag phase, the logarithmic phase and into the stationary phase of a growing culture of Micrococcus luteus is given in Figure 2.21.
Figure 2.21 Growth curve obtained for Micrococcus Luteus obtained by measuring the absorbance at 550 nm of the culture at periodic times after inoculation at time zero (unpublished work). The reciprocal of the Log2(A) plot at the logarithmic growth phase equals the bacteria doubling time (~71 mins).
Exponential growth of the number of bacteria n with time t, commencing with an intitial number n0, can be written as:
where k is the growth constant defined as the number of times per unit time of growing by a factor exponential e. T is the time it takes for the number to double, namely the doubling time. If the absorbance plot is converted to a log2 plot, as indicated in Figure 2.20, a change of 1 absorbance units means the absorbance has doubled. The slope of this log2 plot thus gives the time for the culture to double in number density. The derived doubling time for the M. luteus sample is close to 71 minutes. The ‘textbook’ doubling times for bacteria such as M. luteus and E. coli are often given as 30 and 20 minutes, respectively. However, these values are valid only for optimum conditions of temperature, nutrient concentration and cell density and when no growth suppressing substances are present. Many bacteria produce such substances if their cell density becomes too high. Examples of other typical doubling times are 2 hours and 24 hours, respectively, exhibited by B. subtilis and M. tuberculosis, respectively.
2.6.6 Plant, Fungal and Protozoal Cells
Plant cells deposit an extracellular matrix around themselves, composed of a crosslinked network of a polysaccharide called cellulose, to form a thick cell wall. Secondary cell walls contain additional molecules to add rigidity and further protection. The cellulose gives the primary cell wall high tensile strength, which allows the plant cell to build up a high internal hydrostatic pressure, known as turgor pressure. This turgor pressure may reach 10 or more atmospheres, in order to attain osmotic equilibrium with the external environment. At equilibrium there is no net influx of water into the cell despite the large osmotic difference caused by the higher concentration of solutes inside the cell wall compared to that outside it. The turgor pressure provides rigidity and the main driving force for plant growth.
Fungi include both unicellular yeast cells, such as Saccharomyces cerevisiae, and filamentous, multicellular, moulds such as those found on mouldy fruit or bread. Most pathogenic fungi exhibit dimorphism, which is the ability to grow in either yeast or mould form. The yeast-to-mould or mould-to-yeast transition is frequently associated with infection. For example, some fungi grow as a mould at low temperatures in soil, but then change to a harmful yeast form when inhaled into the lungs.
Protozoan parasites exist as single cells and frequently require more than one host in a complex life-cycle. The most common protozoal disease is malaria, transmitted to humans by the bite of the female of any of 60 species of Anopheles mosquito. The most intensively studied of the malaria-causing parasites, Plasmodium falciparum, exists in eight distinct forms, and requires both human and mosquito hosts to complete its life-cycle. Because fungi and protozoan parasites are eukaryotes, their pathogenic varieties are difficult to kill with drugs without harming the host. The tendency of fungal and parasitic infecting organisms to switch amongst several different forms during their life-cycles also makes them more difficult to treat. A drug that is effective at killing one form is often ineffective at killing another form, which therefore survives the treatment. As a result, antifungal and antiparasitic drugs are often less effective and more toxic than antibiotics.
2.6.7 Viruses
Viruses are not cells. They are bioparticles that can vary from simple helical or icosahedral shapes, to more complex structures. They can reproduce only inside a host cell. They are about 1/100th the size of bacteria, with diameters ranging from around 10 to 300 nm. And so, unlike cells and bacteria which can be viewed using a conventional light microscope, most viruses can only be seen using scanning and transmission electron microscopes. A complete virus particle, known as a virion, consists of nucleic acid (DNA or RNA) surrounded by a protective coat of protein called a capsid, made from proteins encoded by the viral genome. The capsid shape serves as the basis for morphological distinction (helical, icosahedral, envelope, complex).
Some species of virus surround themselves with a modified form of one of the host cell membranes, either the outer membrane of the infected host cell, or internal membranes such as nuclear membrane or endoplasmic reticulum. The virus thus gains an outer lipid bilayer, known as a viral envelope. This membrane is studded with proteins coded for by the viral genome and host genome; the lipid membrane itself and any carbohydrates present originate entirely from the host cell. The influenza virus and HIV use this strategy. Most membrane enveloped viruses are dependent on the envelope for their infectivity. The complex viruses possess a capsid that is neither purely helical, nor purely icosahedral, and may possess extra structures such as protein tails or a complex outer wall. For example, the T4 bacteriophage has a complex structure consisting of an icosahedral head bound to a helical tail with protruding protein tail fibres. This tail structure acts like a molecular syringe, attaching to the bacterial host and then injecting the viral genome into the cell.
All viral genomes encode three types of proteins, namely proteins for replicating the genome, proteins for packaging the genome and delivering it to more host cells, and proteins that modify the structure or function of the host cell to enhance the replication of the virions. Many viral genomes also encode a protein that subverts the host's normal immune defence mechanisms. The design of effective antiviral drugs is difficult because viruses use the host cell's ribosomes to make their proteins and enable viral replication. Antibiotics such as tetracycline specifically poison bacterial ribosomes, but it is not possible to find an equivalent drug to target viral ribosomes without destroying the host cells. In most cases viral infections in animals cause an immune response that eliminates the infecting virus. These immune responses can also be produced by vaccines that give immunity to a viral infection. The best strategy for containing viral diseases is thus to prevent them by vaccinating the potential hosts. Vaccination programs have effectively eliminated the smallpox virus, and the eradication of poliomyelitis is well advanced.
Viral vaccines take the form of being either live or inactivated. Live vaccines are prepared from attenuated strains that are devoid of pathogenecity but are capable of inducing a protective immune response. They multiply in the human host and provide continuous antigenic stimulation over a period of time. For inactivated whole virus vaccines, the original virus is grown by normal virus culture methods, as for example in tissue culture (the polio Salk vaccine), in eggs (influenza) or mouse brain (rabies Semple vaccine). The virus is then inactivated by formalin or B-propiolactone so that the replication function of the virus is destroyed.
Each year the World Health Organization (WHO) recommends specific vaccine viruses for vaccine production, but then individual countries make their own decision for licensing of vaccines in their country. For vaccination against influenza, three or more vaccine viruses are chosen to maximise the likelihood that the main circulating viruses during the upcoming flu season will be well covered by the vaccine. For example, WHO recommended that the Northern Hemisphere's 2010–11 seasonal influenza vaccine should contain the following three vaccine viruses:
- an A/California/7/2009 (H1N1)-like virus,
- an A/Perth/16/2009 (H3N2)-like virus,
- a B/Brisbane/60/2008-like virus.
This recommended composition of the 2010–11 seasonal vaccine for the Northern Hemisphere, including the EU and the United States, was the same composition that was recommended for the Southern Hemisphere's 2010 influenza vaccines. In the nomenclature used, the letter A or B corresponds to the type of human influenza which tend to become pandemic, the numbers correspond to virus strains, and the letters H and N refer to the two viral proteins hemagglutinin and neuraminidase. Neuraminidase promotes influenza virus release from infected cells and facilitates virus spread within the respiratory tract.
2.6.8 Prions
Prions are infectious agents in the form of misfolded proteins that replicate and propagate in the host cell. They cause various neurodegenerative diseases in mammals, a well-known example being bovine spongiform encephalopathy (BSE), otherwise known as mad cow disease. This fatal disease (Creutzfeld-Jacob disease) can be transmitted to humans who eat infected beef, and can also be transmitted from human to human via blood transfusions. The brain tissue develops holes and takes on a spongelike appearance. The DNA code for making prion protein is in a gene that all mammals possess and is mainly active in nerve cells.
The prion has the identical amino acid sequence to the normal form of the protein. The only difference between them is in their folded three-dimensional structure. The misfolded protein can cause a normal folded form to unfold and to aggregate with other prions to produce regular helical structures called amyloid fibres. The prion is thus able to cause the normal protein form to adopt its misfolded prion conformation, causing it to become infectious. This is equivalent to prions being able to replicate themselves in the host cell. If an amyloid fibre is broken into smaller pieces, each piece can initiate the prion polymerisation process in a new cell. The prion can therefore propagate as well as replicate. Furthermore, if consumed by another host organism, the newly formed misfolded prions may transmit the infection to that organism. How polypeptide chains explore their conformation-energy space and fall into a global free energy minimum state corresponding to their correct three-dimensional folded form is poorly understood. The linear order of the peptides is reliably given by the pertinent gene's DNA sequence, but referring to Figures 2.7 and 2.8 we can appreciate that the number of possible polypeptide folding possibilities must be enormous (one of the authors of this book once heard Sydney Brenner remark that there are possibly as many folding possibilities as there are proteins!). The chances of misfolded protein production must be high, so what special circumstances are required to generate the relatively uncommon prion disease? Finding the answers to such questions are currently the objectives of active research.
2.6.9 Cell Culture
This is the process by which cells, mainly mammalian, are grown under controlled ex vivo conditions. Applications of cell culture include the manufacture of viral vaccines and many products of biotechnology. Biological products produced by recombinant DNA (rDNA) technology in animal cell cultures include enzymes, synthetic hormones, antibodies and anticancer agents.
Cells to be cultured can be obtained in several ways. White cells purified from blood are capable of growth in culture (human red blood cells do not possess a nucleus containing the DNA required for cell replication). Mononuclear cells can be released from soft tissues by enzymatic digestion with enzymes, such as collagenase, trypsin, or pronase, which break down the extracellular matrix. Alternatively, in a method known as explant culture, pieces of tissue can be placed in growth media and the cells that grow out are available for culture. Cells that are cultured directly from an organism are known as primary cells. With the exception of some derived from tumors, most primary cell cultures have limited lifespan. After a certain number of population doublings, cells undergo the process of senescence and stop dividing, while generally retaining viability. An established or immortalised cell line has acquired the ability to proliferate indefinitely, either through random mutation or by deliberate genetic modification.
Cells are grown and maintained at an appropriate temperature and gas mixture (typically, 37 °C, 5% CO2 for mammalian cells) in a cell incubator. Culture conditions vary widely for each cell type, and variation of conditions for a particular cell type can result in different phenotypes being expressed. Aside from temperature and gas mixture, the most commonly varied factor in culture systems is the growth medium. Recipes for growth media can vary in pH, glucose concentration, growth factors, and the presence of other nutrients. The growth factors used to supplement media are often derived from animal blood, such as calf serum. The culture medium is also generally supplemented with antibiotics, fungicides, or both to inhibit contamination.
Cells can be grown in suspension or as adherent cultures. Some cells naturally live in suspension, without being attached to a surface, such as cells that exist in the bloodstream. There are also cell lines that have been modified to be able to survive in suspension cultures so that they can be grown to a higher density than adherent conditions would allow. Adherent cells require a surface, such as tissue culture plastic, which may be coated with extracellular matrix components to increase adhesion properties and provide other signals needed for growth and differentiation. Most cells derived from solid tissues are adherent. Another type of adherent culture is organotypic culture which involves growing cells in a three-dimensional environment as opposed to two-dimensional culture dishes. This 3-D culture system is biochemically and physiologically more similar to in vivo tissue, but is technically challenging to maintain because of factors such as limited diffusion of chemicals to and from the innermost cells.
As cells reach confluency, they must be subcultured or passaged. Failure to subculture confluent cells results in reduced mitotic index and eventually cell death. The first step in subculturing monolayers is to detach cells from the surface of the primary culture vessel by trypsinisation or mechanical means. The resultant cell suspension is then subdivided or reseeded into fresh cultures. Secondary cultures are checked for growth, fed periodically, and may be subsequently subcultured to produce tertiary cultures. The time between passaging cells depends on the growth rate and varies with the cell line.
As cells generally continue to divide in culture, they generally grow to fill the available area or volume. This can generate the following issues:
- nutrient depletion in the growth media;
- accumulation of dead (apoptotic/necrotic) cells;
- cell-to-cell contact can stimulate cell cycle arrest, causing cells to stop dividing, and is known as contact inhibition or senescence;
- cell-to-cell contact can stimulate cellular differentiation.
Amongst the common manipulations carried out on cells in culture are media changes, passaging cells, and transfecting cells. These are generally performed using tissue culture methods that rely on sterile techniques, which aim to avoid contamination with bacteria, yeast, or other cell lines. Manipulations are typically carried out in a biosafety hood or laminar flow cabinet to exclude contaminating microorganisms. Antibiotics (e.g. penicillin and streptomycin) and antifungals can also be added to the growth media. As cells undergo metabolic processes, acid is produced and the pH decreases. Often, a pH indicator is added to the medium in order to measure nutrient depletion. Passaging (also known as subculture or splitting cells) involves transferring a small number of cells into a new vessel. Cells can be cultured for a longer time if they are split regularly, as it avoids the senescence associated with prolonged high cell density. Suspension cultures are easily passaged with a small amount of culture containing a few cells diluted in a larger volume of fresh media. For adherent cultures the cells first need to be detached, using a mixture of trypsin and EDTA, for example. A small number of detached cells can then be used to seed a new culture. Another common method for manipulating cells involves the introduction of foreign DNA by transfection. This is often performed to cause cells to express a protein of interest. DNA can also be inserted into cells using viruses.
2.6.10 Tissue Engineering
This is the interdisciplinary field that employs cell biology, engineering, and materials science to repair or replace portions of diseased tissues (e.g. bone, cartilege, blood vessels, bladder, etc.). The term regenerative medicine is also used in this context, but in this case, emphasis is placed on the use of stem cells to produce tissues. There are three main strategies:
Examples include artificial:
- bladders
- bone
- bone marrow
- cartilage (using chondrocytes for knee repair)
- liver (using hepatocytes)
- pancreas (using islet of Langerhans cells (beta-cells) to produce and regulate insulin for control of diabetes)
- skin (using fibroblasts).
Cells are often categorised by their source:
- Autologous cells are obtained from the same individual to whom they will be reimplanted. This reduces problems associated with rejection and pathogen transmission, but is not possible for patients suffering from a genetic disease.
- Allogenic cells come from the body of a donor of the same species.
- Xenogenic cells are those isolated from individuals of another species. In particular, animal cells have been used quite extensively in experiments aimed at the construction of cardiovascular implants.
- Syngenic or isogenic cells are isolated from genetically identical organisms, such as twins, clones, or highly inbred research animal models.
- Primary cells are from an organism.
- Secondary cells are from a cell bank.
Cells are often implanted or seeded into an artificial structure (scaffold) capable of supporting three-dimensional tissue formation. Scaffolds usually serve at least one of the following purposes:
- allow cell attachment and migration;
- deliver and retain cells and biochemical factors;
- enable diffusion of vital cell nutrients and expressed products;
- exert certain mechanical and biological influences to modify the behaviour of the attached cells.
To achieve the goal of tissue reconstruction, scaffolds must meet some specific requirements. A high porosity and an adequate pore size are necessary to facilitate cell seeding and diffusion throughout the whole structure of both cells and nutrients. Biodegradability is often an essential factor since scaffolds should preferably be absorbed by the surrounding tissues without the necessity of surgical removal. The rate at which degradation occurs has to coincide as much as possible with the rate of tissue formation. This means that while cells are fabricating their own natural matrix structure around themselves, the scaffold is able to provide structural integrity within the body and eventually it will break down leaving the newly formed tissue to take over the mechanical load. The ability to be able to inject scaffold material into a tissue is also important for some clinical uses.
Many different materials (natural and synthetic, biodegradable and permanent) have been investigated. Examples of these materials are collagen and some polyesters. A commonly used synthetic material is polylactic acid (PLA) – a polyester which degrades within the human body to form lactic acid, a naturally occurring chemical which is easily released from the body. Similar materials are polyglycolic acid (PGA) and polycaprolactone (PCL). The degradation mechanism for these materials is similar to that of PLA, but PGA exhibits a faster rate and PCL a slower rate of degradation compared to PLA.
Scaffolds may also be constructed from natural materials. Examples include collagen or fibrin, and polysaccharidic materials such as chitosan. Glycosaminoglycans (GAGs) have also proved suitable in terms of cell compatibility, but some issues with potential immunogenicity still remain. Amongst GAGs, hyaluronic acid in combination with cross-linking agents (e.g. glutaraldehyde) has also been used as scaffold material. Chemically functionalised groups incorporated into the scaffolds may also be useful in the delivery of small molecules (drugs) to specific tissues.
2.6.10.1 Stem Cells
Stem cells are undifferentiated cells with the ability to divide in culture and give rise to different forms of specialised cells. According to their source, stem cells are divided into adult and embryonic stem cells – the first class being multipotent and the latter mostly pluripotent. Multipotent progenitor cells have the potential to give rise to a limited number of different cell lineages. An example of a multipotent stem cell is a haematopoietic cell, a blood stem cell that can develop into several types of blood cells but cannot develop into muscle or brain cells, for example. Pluripotency is the ability of the human embryonic stem cell to differentiate or become almost any cell type in the body. Some cells are totipotent in the earliest stages of the embryo. Totipotency is the ability of a single cell to divide and produce all the differentiated cells in an organism, including extra-embryonic tissues such as the placenta. While there is still a large ethical debate related with the use of embryonic stem cells, there is active research and clinical studies directed towards using stem cells to repair diseased or damaged tissues, or to grow new organs.
2.6.11 Cell–Cell Communication
Most cells in multicellular organisms emit and receive chemical signals. They do this to organise themselves into a coordinated system. A cell usually receives a chemical signal by the binding of a signalling molecule onto a receptor protein on the cell membrane surface. This binding activates one or more sequences of intracellular signalling proteins that are directed towards influencing the activity of target effector proteins inside the cell. The types of effector protein are diverse. They can be a component of the cytoskeleton that influences the shape or motility of a cell, or an enzyme involved in a metabolic pathway, the regulation of gene expression, or operation of an ion channel, for example. The different ways an extracellular signal protein can activate signalling pathways within a cell are depicted in Figure 2.22.
Figure 2.22 This binding of a signalling protein to a membrane receptor protein can activate a sequence of intracellular signalling proteins that are designed to influence the activity of a target effector protein inside the cell. Depending on the type of effector protein, this can result in a change of the shape or motility of a cell, alter cell metabolism or the regulation of gene expression, for example.
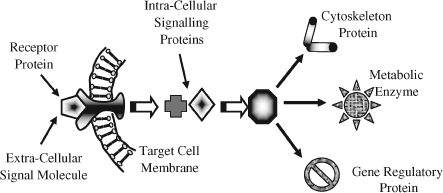
The chemical signals secreted from a signalling cell and communicated to another cell are also diverse, and include peptides, amino acids, proteins, nucleotides, proteins, steroids, or hormones, as well as dissolved gases. Some signalling molecules, such as proteins and carbohydrates, are synthesised in the endoplasmic reticulum of the signalling cell and then processed further by its Golgi complex to enable its release by exocytosis into the extracellular medium. Other signalling molecules are small enough to diffuse through the membrane of the signalling cell, to be either released into the extracellular medium or to remain attached to the outer surface of the signalling cell. Signalling molecules that remain attached to the outer surface of the signalling cell can only perform their function through contact with the receiving cell. Most of the signal molecules released into the extracellular medium are hydrophilic and, as shown in Figure 2.23, bind to cell-surface receptor proteins that span across the membrane of the receiving cell. As is also shown in Figure 2.23, small hydrophobic signal molecules have to be transported by proteins to the target cell, where they can then diffuse across the cell membrane to interact directly with an internal effector protein.
Figure 2.23 Hydrophilic signal molecules released into the extracellular medium can bind directly to cell-surface receptor proteins, but small hydrophobic signal molecules are transported by proteins to the target cell, where they can then diffuse across the cell membrane to interact directly with an internal effector protein.
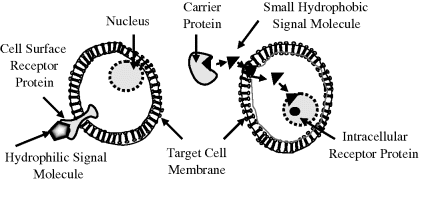
The concentration of molecular signals in the extracellular medium is very low, typically around 0.1 nM or smaller. To compensate for this low concentration the binding of a signal molecule to its receptor on the target cell is very specific, and is usually characterised by a high affinity constant value exceeding 108 litres/mole. (The affinity constant is also known as the association or equilibrium constant Keq described in Chapter 1).
There are four main types of signalling processes between cells:
Figure 2.24 Left: Contact-dependent signalling between cells can only occur if there is direct membrane-membrane contact between the cells. This type of signalling is important in the early stages of an organism's development, and in immune responses. Right: When an action potential reaches the synapse at the end of a neuron's axon, chemical signals called neurotransmitters are secreted and diffuse quickly to their target cell across a narrow synaptic gap. Synaptic signalling is efficient in terms of the speed and repetition rates at which signals can be transmitted between cells.
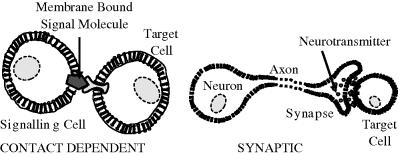
Figure 2.25 In paracrine signalling the secreted signal molecules only affect target cells in close proximity to the signalling cell. This is often used as developmental cues made by embryonic cells to each other. Right: An example of endocrine signalling is the secretion of hormones from an endocrine cell into the bloodstream, which carries them to target cells over the entire body.
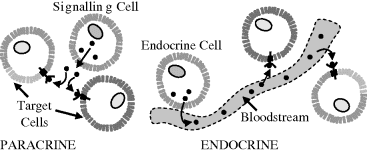
2.6.11.1 Gap Junctions
Gap junctions are water-filled channels (diameter ≤ 1.5 nm) formed by proteins that directly connect the cytoplasms of adjacent cells. They commonly occur in connective tissues, epithelial (skin) and liver cells, as well as for some types of neurons, for example. Gap junctions allow cell neighbours to communicate both chemically and electrically with each other. This is required in the liver, to coordinate the response to nerve signals that connect directly to only a subpopulation of the liver cells. Small molecules (≤1000 daltons) such as inorganic ions, sugars, amino acids, nucleotides, vitamins (but not proteins and nucleic acids) can pass readily through gap junctions. Microelectrodes inserted into cells connected by gap junctions reveal that they are electrically connected. Defective gap junctions can lead to such health problems as congenital deafness and peripheral nerve dysfunction.
2.7 Summary of Key Concepts
Fatty acids in the form of phospholipids can spontaneously form bilayer structures that are used to construct biomembranes, such as the cytoplasmic membrane that separates a cell from its external environment or the membranes of organelles inside a cell. Carbohydrates in the form of sugars are also used as a source of energy by cells and tissues. The importance of amino acids to the cell comes from their role in making proteins, which are polymers of amino acids covalently linked together by peptide bonds into a long polypeptide chain. Polypeptide chains fold into a three-dimensional structure to form proteins. A protein's function is determined by its three-dimensional structure, which in turn is determined by the linear sequence of the amino acids in its polypeptide chain. Instructions for the assembly of the amino acid sequence are coded by the linear sequence of nucleotides of the nucleic acid DNA. DNA in cells of higher organisms takes the form a double helix, in which each polynucleotide strand can act as a template for the synthesis of the other strand. The process of DNA replication results in a copying of the genetic information from cell to daughter cell and from parents to their offspring.
Selected portions of the DNA are transcribed into several types of RNA by enzymes called RNA polymerases. This transcription process produces single strands of RNA complementary to one strand of DNA – with uracil (U) replacing the thymine (T) of DNA. Some types of RNA can fold up into structures that can act as catalysts. Single strands of messenger RNA (mRNA) contain coding information, obtained from the DNA template, in the form of a sequence of tri-nucleotide units called codons. Each codon specifies a single amino acid to be translated into a polypeptide chain by the interaction of the mRNA strand with a ribosome in the cytoplasm. Transfer RNA (tRNA) mediates this process and provides the corresponding amino acid. The central dogma of molecular biology, that DNA makes RNA makes Protein, implies that the sequential structural information stored in a protein by its amino acid sequence cannot be transferred to another protein or to a nucleic acid. The normal flow of biological information is that DNA information can itself be replicated, and then transcribed into messenger RNA from whence it is translated into amino acids to form proteins.
The cell is the structural and functional unit of all known living organisms. There are two basic types of cell, those that do not possess a nucleus (prokaryotes) and those that do have a nucleus (eukaryotes). The nucleus serves to concentrate the DNA-related functions so as to increase their efficiency, which is not necessary for the much smaller prokaryotic cell because the internal reacting materials are relatively close together and many of its biochemical functions are less complicated. Prokaryotic and eukaryotic cells differ significantly in their cell division schemes. For example, the condensation and decondensation of the DNA that occurs in eukaryotic cells during mitosis does not occur at all in prokaryotes. Both cell types possess a plasma membrane that encloses their contents and serves as a semi-porous barrier to the outside environment. Small molecules, such as oxygen, carbon dioxide, and water, are able to pass freely across this membrane, but the passage of larger molecules (e.g. amino acids and sugars) is carefully regulated. Unlike most prokaryotes, eukaryotic cells also contain extensive internal membranes, which enclose subcellular structures called organelles.
Blood and its cellular components perform important functions, such as the transport of oxygen and nutrients to tissues and providing protection against disease agents, through the immune response provided by lymphocytes and the phagocytic activity of leukocytes, for example. Most cells in multicellular organisms emit and receive chemical signals, so as to organise themselves into a coordinated system. The binding of a signalling molecule by the target cell can activate intracellular signalling proteins, that in turn can influence target effector proteins inside the cell, to influence its shape, motility, metabolism or gene expression, for example.
1. Alberts, B., Johnson, A. Lewis, J. et al. (2007) Chapter 2 (Tables 2.3 & 2.4), in Molecular Biology of the Cell, 5th edn, Garland Science.
2. Mulder, G.J. (1838) Zusammensetzung von Fibrin, Albumin, Leimzucker, Leucin u.s.w. Annalen der Pharmacie, 28, 73–82.
3. Vickery, H.B. (1950) The origin of the word protein. Yale Journal of Biology and Medicine, 22, 387–393.
4. Holmes, F.L. (1963) Elementary analysis and the origins of physiological chemistry. ISIS, 54, 50–81.
5. Vickery, H.B. and Schmidt, C.L.A. (1931) The history of the discovery of the amino acids. Chemical Reviews, 9, 169–318.
6. Vickery, H.B. (1972) The history of the discovery of the amino acids II. A review of amino acids described since 1931 as components of native proteins. Advances in Protein Chemistry, 26, 81–171.
7. Ramachandran, G.N. and Sassiekharan, V. (1968) Conformation of polypeptides and proteins. Advances in Protein Chemistry, 23, 283–437.
8. Watson, J.D. and Crick, F.H.C. (1953) A structure for deoxyribose nucleic acid. Nature, 171, 737–738.
9. Judson, H.F. (1979) The Eighth Day of Creation, Simon & Schuster, New York.
10. Harold, F.M. (2001) The Way of the Cell, Oxford University Press.
11. Guerra, C.F., Bickelhaupt, F.M., Snijders, J.G. and Baerends, E.J. (2000) Hydrogen bonding in DNA base Pairs: reconciliation of theory and experiment. Journal of the American Chemical Society, 122, 4117–4128.
12. Kornberg, A. and Baker, T.A. (2005) DNA Replication, 2nd edn, University Science Books, Sausalito, Ca.
13. Kornberg, A. (1989) For the Love of Enzymes: The Odyssey of a Biochemist, Harvard University Press.
14. Joyce, G.F. (2002) The antiquity of RNA-based evolution. Nature, 418, 214–221.
15. Costanzo, G., Pino, S., Ciciriello, F. and Di Mauro, E. (2009) Generation of long RNA chains in water. Journal of Biological Chemistry, 284 (48), 33206–33216.
16. Crick, F. (1970) Central dogma of molecular biology. Nature, 227, 561–563.
17. Greene, W.C. (1993) AIDS and the immune system. Scientific American, 269 (3), 99–105.
18. Lee, K.L., Kozono, D. Remis, J. et al. (2005) Structural basis for conductance by the archaeal aquaporin AqpM at 1.68 Å. Proceedings of the National Academy of Sciences of the United States of America, 102 (52), 18932–18937.
Alberts, B., Johnson, A. Lewis, J. et al. (2007) Chapter 2, in Molecular Biology of the Cell, 5th edn, Garland Science.
Bauer, W.R., Crick, F.H.C. and White, J.H. (1980) Supercoiled DNA. Scientific American, 243 (1), 118–133.
Becker, W.M., Reece, J.M. and Peonie, M.F. (1995) The World of the Cell, 3rd edn, Benjamin/Cummings Publ. Co., Redwood City, CA.
Capra, J.D. and Edmonson, A.B. (1977) The antibody combining site. Scientific American, 236 (1), 50–59.
Crick, F.H.C. (1966) The genetic code III. Scientific American, 215 (4), 55–62.
Dickerson, R.E. (1983) The DNA helix and how it is read. Scientific American, 249 (6), 94–111.
Crick, F. (1988) What Mad Pursuit: A Personal View of Scientific Discovery, Basic Books Inc., New York.
Doolittle, R. (1985) Proteins. Scientific American, 253 (4), 88–99.
Judson, H.F. (1979) The Eighth Day of Creation, Simon & Schuster, New York (reprinted, Cold Spring Harbor Laboratory Press, New York, 1996).
Lake, J.A. (1981) The ribosome. Scientific American, 245 (2), 84–97.
Lodish, H.F. and Rothman, J.E. (1978) The assembly of cell membranes. Scientific American, 240 (1), 48–63.
Nelson, D.L. and Cox, M.M. (2009) Chapters 2, 5 and 6, in Lehninger Principles of Biochemistry, 5th edn, W.H. Freeman.
Perutz, M.F. (1964) The hemoglobin molecule. Scientific American, 211 (5), 64–76.
Perutz, M.F. (1978) Hemoglobin structure and respiratory transport. Scientific American, 239 (6), 92–125.
Rich, A. and Kim, S.H. (1977) The three-dimensional structure of transfer RNA. Scientific American, 238 (1), 51–62.
Sharon, N. (1980) Carbohydrates. Scientific American, 243 (5), 90–116.
Tanford, C. and Reynolds, J. (2001) Nature's Robots: A History of Proteins, Oxford University Press.
Watson, J.D. (1968) The Double Helix, Simon & Schuster, New York, (Touchstone Edition, 2001).