
Top texture: © Laguna Design/Science Source;
Chapter 20: mRNA Stability and Localization
Edited by Ellen Baker

Chapter Opener: © Science Photo Library/Shutterstock, Inc.
20.1 Introduction
RNA is critical at many stages of gene expression. The focus of this chapter is messenger RNA (mRNA), the first RNA to be characterized for its central role as an intermediate in protein synthesis. Many other RNAs play structural or functional roles at other stages of gene expression. The functions of other cellular RNAs are discussed in other chapters: snRNAs and snoRNAs in the chapter titled RNA Splicing and Processing; tRNA and rRNA in the chapter titled Translation; and miRNAs and siRNAs in the chapter titled Regulatory RNA. The subset of RNAs that have retained ancestral catalytic activity are discussed in the chapter titled Catalytic RNA.
Messenger RNA plays the principal role in the expression of protein-coding genes. Each mRNA molecule carries the genetic code for synthesis of a specific polypeptide during the process of translation. An mRNA carries much more information as well: how frequently it will be translated, how long it is likely to survive, and where in the cell it will be translated. This information is carried in the form of RNA cis-elements and associated proteins. Much of this information is located in parts of the mRNA sequence that are not directly involved in encoding protein.
FIGURE 20.1 shows some of the structural features typical of mRNAs in prokaryotes and eukaryotes. Bacterial mRNA termini are not modified after transcription, so they begin with the 5′ triphosphate nucleotide used in initiation of transcription and end with the final nucleotide added by RNA polymerase before termination. The 3′ end of many Escherichia coli mRNAs form a hairpin structure involved in intrinsic (rho-independent) transcription termination (see the chapter titled Prokaryotic Transcription). Eukaryotic mRNAs are cotranscriptionally capped and polyadenylated (see the chapter titled RNA Splicing and Processing). Most of the non-protein-coding regulatory information is carried in the 5′ and 3′ untranslated regions (UTRs) of an mRNA, but some elements are present in the coding region. All mRNAs are linear sequences of nucleotides, but secondary and tertiary structures can be formed by intramolecular base pairing. These structures can be simple, like the stem-loop structures illustrated in Figure 20.1, or more complex, involving branched structures or pairing of nucleotides from distant regions of the molecule. Investigation of the mechanisms by which mRNA regulatory information is deciphered and acted upon by machinery responsible for mRNA degradation, translation, and localization is an important field in molecular biology today.

FIGURE 20.1 Features of prokaryotic and eukaryotic mRNAs. (a) A typical bacterial mRNA. This is a monocistronic mRNA, but bacterial mRNAs may also be polycistronic. Many bacterial mRNAs end in a terminal stem-loop. (b) All eukaryotic mRNAs begin with a cap (m7G), and almost all end with a poly(A) tail. The poly(A) tail is coated with poly(A)-binding proteins (PABPs). Eukaryotic mRNAs may have one or more regions of secondary structure, typically in the 5′ and 3′ UTRs. (c) The major histone mRNAs in mammals have a 3′ terminal stem-loop in place of a poly(A) tail.
20.2 Messenger RNAs Are Unstable Molecules
Messenger RNAs are relatively unstable molecules, unlike DNA, and, to a lesser extent, rRNAs and tRNAs. Although it is true that the phosphodiester bonds connecting ribonucleotides are somewhat weaker than those connecting deoxyribonucleotides due to the presence of the 2′–OH group on the ribose sugar, this is not the primary reason for the instability of mRNA. Rather, cells contain myriad RNA-degrading enzymes, called ribonucleases (RNases), some of which specifically target mRNA molecules.
Ribonucleases are enzymes that cleave the phosphodiester linkage connecting RNA ribonucleotides. They are diverse molecules because many different protein domains have evolved to have ribonuclease activity. The rare examples of known ribozymes (catalytic RNAs) include multiple ribonucleases, indicating the ancient origins of this important activity (see the chapter titled Catalytic RNA). Ribonucleases, often just called nucleases when the RNA nature of the substrate is obvious, have many roles in a cell, including participation in DNA replication, DNA repair, processing of new transcripts (including pre-mRNAs, tRNAs, rRNAs, snRNAs, and miRNAs), and the degradation of mRNA. Ribonucleases are either endoribonucleases or exoribonucleases, as depicted in FIGURE 20.2 (and as discussed in the chapter titled Methods in Molecular Biology and Genetic Engineering). Endonucleases cleave an RNA molecule at an internal site and may have a requirement or preference for a certain structure or sequence. Exonucleases remove nucleotides from an RNA terminus and have a defined polarity of attack—either 5′ to 3′ or 3′ to 5′. Some exonucleases are processive, remaining engaged with the substrate while sequentially removing nucleotides, whereas others are distributive, catalyzing the removal of only one or a few nucleotides before dissociating from the substrate.

FIGURE 20.2 Types of ribonucleases. Exonucleases are unidirectional. They can digest RNA either from the 5′ end or from the 3′ end, liberating individual ribonucleotides. Endonucleases cleave RNA at internal phosphosphodiester linkages. An endonuclease usually targets specific sequences and/or secondary structures.
Most mRNAs decay stochastically (like the decay of radioactive isotopes), and as a result mRNA stability is usually expressed as a half-life (t½). The term mRNA decay is often used interchangeably with mRNA degradation. mRNA-specific stability information is encoded in cis-sequences (see the section in this chapter titled mRNA-Specific Half-Lives Are Controlled by Sequences or Structures Within the mRNA) and is therefore characteristic of each mRNA. Different mRNAs can exhibit remarkably different stabilities, varying by 100-fold or more. In E. coli the typical mRNA half-life is about 3 minutes, but half-lives of individual mRNAs may be as short as 20 seconds or as long as 90 minutes. In budding yeast, mRNA half-lives range from 3 to 100 minutes, whereas in metazoans half-lives range from minutes to hours, and in rare cases, even days. Abnormal mRNAs can be targeted for very rapid destruction (see the sections in this chapter titled Newly Synthesized RNAs Are Checked for Defects via a Nuclear Surveillance System and Quality Control of mRNA Translation Is Performed by Cytoplasmic Surveillance Systems). Half-life values are generally determined by some version of the method illustrated in FIGURE 20.3.
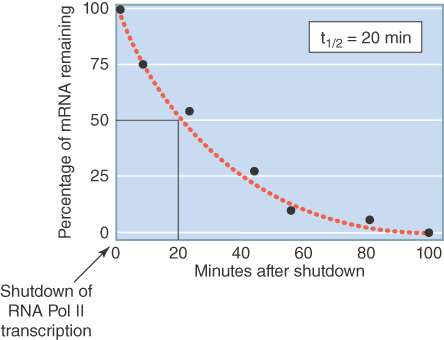
FIGURE 20.3 Method for determining mRNA half-lives. RNA polymerase II transcription is shut down, either by a drug or a temperature shift in strains with a temperature-sensitive mutation in a Pol II gene. The levels of specific mRNAs are determined by northern blot or RT-PCR at various times following shutdown. RNA degradation, once initiated, is usually so rapid that intermediates in the process are not detectible. The half-life is the time required for the mRNA to fall to one-half of its initial value.
The abundance of specific mRNAs in a cell is a consequence of their combined rates of synthesis (transcription and processing) and degradation. mRNA levels reach a steady state when these parameters remain constant. The spectrum of proteins synthesized by a cell is largely a reflection of the abundance of their mRNA templates (although differences in translational efficiency play a role). The importance of mRNA decay is highlighted by large-scale studies that have examined the relative contributions of decay rate and transcription rate to differential mRNA abundance. Decay rate predominates. The great advantage of unstable mRNAs is the ability to rapidly change the output of translation through changes in mRNA synthesis. Clearly this advantage is important enough to compensate for the seeming wastefulness of making and destroying mRNAs so quickly. Abnormal control of mRNA stability has been implicated in disease states, including cancer, chronic inflammatory responses, and coronary disease.
20.3 Eukaryotic mRNAs Exist in the Form of mRNPs from Their Birth to Their Death
From the time pre-mRNAs are transcribed in the nucleus until their cytoplasmic destruction, eukaryotic mRNAs are associated with a changing repertoire of proteins. RNA–protein complexes are called ribonucleoprotein particles (RNPs). Many of the pre-mRNA–binding proteins are involved in splicing and processing reactions (see the chapter titled RNA Splicing and Processing), and others are involved in quality control (discussed in the section in this chapter titled Newly Synthesized RNAs Are Checked for Defects via a Nuclear Surveillance System). The nuclear maturation of an mRNA comprises multiple remodeling steps involving both the RNA sequence and its complement of proteins. The mature mRNA product is export competent only when fully processed and associated with the correct protein complexes, including TREX (for transcription export), which mediates its association with the nuclear pore export receptor. Mature mRNAs retain multiple binding sites (cis-elements) for different regulatory proteins, most often within their 5′ or 3′ UTRs.
Many nuclear proteins are shed before or during mRNA export to the cytoplasm, whereas others accompany the mRNA and have cytoplasmic roles. For example, once in the cytoplasm the nuclear cap-binding complex participates in the new mRNA’s first translation event, the so-called pioneering round of translation. This first translation initiation is critical for a new mRNA; if it is found to be a defective template it will be rapidly destroyed by a surveillance system (see the section in this chapter titled Quality Control of mRNA Translation Is Performed by Cytoplasmic Surveillance Systems). An mRNA that passes its translation test will spend the rest of its existence associated with a variety of proteins that control its translation, its stability, and sometimes its cellular location. The “nuclear history” of an mRNA is critical in determining its fate in the cytoplasm.
A large number of different RNA-binding proteins (RBPs) are known, and many more are predicted based on genome analysis. The Saccharomyces cerevisiae genome encodes nearly 600 different proteins predicted to bind to RNA, about one-tenth of the total gene number for this organism. Based on similar proportions, the human genome would be expected to contain more than 2,000 such proteins. These estimates are based on the presence of characterized RNA-binding domains, and it is likely that additional RNA-binding domains remain to be found. The RNA targets and functions of the great majority of these RBPs are unknown, although it is considered likely that a large fraction of them interact with pre-mRNA or mRNA. This kind of analysis does not include the many proteins that do not bind RNA directly, but participate in RNA-binding complexes.
An important insight into why the number of different mRNA-binding proteins is so large has come from the finding that mRNAs are associated with distinct, but overlapping, sets of RBPs. Studies that have matched specific RBPs with their target mRNAs have revealed that those mRNAs encode proteins with shared features such as involvement in similar cellular processes or location. Thus, the repertoire of bound proteins catalogues the mRNA. For example, hundreds of yeast mRNAs are bound by one or more of six related Puf proteins. Puf1 and Puf2 bind mostly mRNAs encoding membrane proteins, whereas Puf3 binds mostly mRNAs encoding mitochondrial proteins, and so on. A current model, illustrated in FIGURE 20.4, proposes that the coordinate control of posttranscriptional processes of mRNAs is mediated by the combinatorial action of multiple RBPs, much like the coordinate control of gene transcription is mediated by the right combinations of transcription factors (see the chapter titled Eukaryotic Transcription Regulation). The set of mRNAs that share a particular type of RBP is called an RNA regulon.

FIGURE 20.4 The concept of an RNA regulon. Eukaryotic mRNAs are bound by a variety of proteins that control their translation, localization, and stability. The subset of mRNAs that have a binding protein in common are considered part of the same regulon. In the diagram, mRNAs a and d are part of regulon 1; mRNAs a, c, and e are part of regulon 2; and so on.
20.4 Prokaryotic mRNA Degradation Involves Multiple Enzymes
Our understanding of prokaryotic mRNA degradation comes mostly from studies of E. coli. So far, the general principles apply to the other bacterial species studied. In prokaryotes, mRNA degradation occurs during the process of coupled transcription/translation. Prokaryotic ribosomes begin translation even before transcription is completed, attaching to the mRNA at an initiation site near the 5′ end and proceeding toward the 3′ end. Multiple ribosomes can initiate translation on the same mRNA sequentially, forming a polyribosome (or polysome): one mRNA with multiple ribosomes.
E. coli mRNAs are degraded by a combination of endonuclease and 3′ to 5′ exonuclease activities. The major mRNA degradation pathway in E. coli is a multistage process illustrated in FIGURE 20.5. The initiating step is removal of pyrophosphate from the 5′ terminus, leaving a single phosphate. The monophosphorylated form stimulates the catalytic activity of an endonuclease (RNase E), which makes an initial cut near the 5′ end of the mRNA. This cleavage leaves a 3′–OH on the upstream fragment and a 5′–monophosphate on the downstream fragment. It functionally destroys a monocistronic mRNA, because ribosomes can no longer initiate translation. The upstream fragment is then degraded by a 3′ to 5′ exonuclease (polynucleotide phosphorylase, or PNPase). This two-step ribonuclease cycle is repeated along the length of the mRNA in a 5′ to 3′ direction as more RNA gets exposed following passage of previously initiated ribosomes. This process proceeds very rapidly as the short fragments generated by RNase E can be detected only in mutant cells in which exonuclease activity is impaired.

FIGURE 20.5 Degradation of bacterial mRNAs. Bacterial mRNA degradation is initiated by cleavage of the triphosphate 5′ terminus to yield a monophosphate. mRNAs are then degraded in a two-step cycle: an endonucleolytic cleavage, followed by 3′ to 5′ exonuclease digestion of the released fragment. The endonucleolytic cleavages occur in a 5′ to 3′ direction on the mRNA, following the passage of the last ribosome.
PNPase, as well as the other known 3′ to 5′ exonucleases in E. coli, are unable to progress through double-stranded regions. Thus, the stem-loop structure at the 3′ end of many bacterial mRNAs protects the mRNA from direct 3′ attack. Some internal fragments generated by RNase E cleavage also have regions of secondary structure that would impede exonuclease digestion. PNPase is, however, able to digest through double-stranded regions if there is a stretch of single-stranded RNA at least 7 to 10 nucleotides long located 3′ to the stem-loop. The single-stranded sequence seems to serve as a necessary staging platform for the enzyme. Rho-independent termination leaves a single-stranded region that is too short to serve as a platform. To solve this problem a bacterial poly (A) polymerase (PAP) adds 10 to 40 nucleotide poly(A) tails to 3′ termini, making them susceptible to 3′ to 5′ degradation. RNA fragments terminating in particularly stable secondary structures may require repeated polyadenylation and exonuclease digestion steps. It is not known whether polyadenylation is ever the initiating step for degradation of mRNA, or whether it is used only to help degrade fragments, including the 3′ terminal one. Some experiments indicate that RNase E cleavage of an mRNA may be required to activate the PAP. This would explain why intact mRNAs do not seem to be degraded from the 3′ end.
RNase E and PNPase, along with a helicase and another accessory enzyme, form a multiprotein complex called the degradosome. RNase E plays dual roles in the complex. Its N-terminal domain provides the endonuclease activity, whereas its C-terminal domain provides a scaffold that holds together the other components. Although RNase E and PNPase are the principal endo- and exonucleases active in mRNA degradation, others also exist, probably with more restricted roles. The role of other nucleases in mRNA degradation has been addressed by evaluating the phenotypes of mutants in each of the enzymes. For example, the inactivation of RNase E slows mRNA degradation without completely blocking it. Mutations that inactivate PNPase or either of the other two known 3′ to 5′ exonucleases have essentially no effect on overall mRNA stability. This reveals that any pair of the exonucleases can carry out apparently normal mRNA degradation. However, only two of the three exonucleases (PNPase and RNase R) can digest fragments with stable secondary structures. This was demonstrated in double-mutant studies, in which both PNPase and RNase R are inactivated. mRNA fragments that contain secondary structures accumulated in these mutants.
Many questions about mRNA degradation in E. coli remain to be answered. Half-lives for different mRNAs in E. coli can differ more than 100-fold. The basis for these extreme differences in stability is not fully understood but appears to be largely due to two factors. Different mRNAs exhibit a range of susceptibilities to endonuclease cleavage, with some protection being conferred by the secondary structure of the 5′ end region. Some mRNAs are more efficiently translated than others, resulting in a denser packing of protective ribosomes. Whether or not there are additional pathways of mRNA degradation is not known. No 5′ to 3′ exonuclease has been found in E. coli, though one has been identified in Bacillus subtilis and some other bacterial species. So far, the bacterial species found to have the 5′ to 3′ exonuclease RNase J lack the endonuclease RNase E (the major degradative RNase in E. coli). This suggests there is at least one alternative mRNA decay pathway in bacteria. It is likely that the different endonucleases and exonucleases have distinct roles. A genome-wide study using microarrays looked at the steady-state levels of more than 4,000 mRNAs in cells mutant for RNase E or PNPase or other degradosome components. Many mRNA levels increased in the mutants, as expected for a decrease in degradation. Others, however, remained at the same level or even decreased. The half-lives of specific mRNAs can be altered by different cellular physiological states such as starvation or other forms of stress, and mechanisms for these changes remain mostly unknown.
20.5 Most Eukaryotic mRNA Is Degraded via Two Deadenylation-Dependent Pathways
Eukaryotic mRNAs are protected from exonucleases by their modified ends (Figure 20.1). The 7-methyl guanosine cap protects against 5′ attack; the poly(A) tail, in association with bound proteins, protects against 3′ attack. Exceptions are the histone mRNAs in mammals, which terminate in a stem-loop structure rather than a poly(A) tail. A sequence-independent endonuclease attack—the initiating mechanism used by bacteria—is rare or absent in eukaryotes. mRNA decay has been characterized most extensively in budding yeast, although most findings apply to mammalian cells as well.
Degradation of the vast majority of mRNAs is deadenylation dependent; that is, degradation is initiated by breaching their protective poly(A) tail. The newly formed poly(A) tail (which is about 70 to 90 adenylate nucleotides in yeast and about 200 in mammals) is coated with poly(A)-binding proteins (PABPs). The poly(A) tail is subject to gradual shortening upon entry into the cytoplasm, a process catalyzed by specific poly(A) nucleases (also called deadenylases). In both yeast and mammalian cells, the poly(A) tail is initially shortened by the PAN2/3 complex, followed by a more rapid digestion of the remaining 60- to 80-A tail by a second complex, CCR4-NOT, which contains the processive exonuclease CCR4 and at least eight other subunits. Remarkably, similar CCR4-NOT complexes are involved in a variety of other processes in gene expression, including transcriptional activation. It is thought to be a global regulator of gene expression, integrating transcription and mRNA degradation. Other poly(A) nucleases exist in both yeast and mammalian cells, and the reason for this multiplicity is not yet clear.
Two different mRNA degradation pathways are initiated by poly(A) removal, as shown in FIGURE 20.6. In the first pathway (Figure 20.6, left), digestion of the poly(A) tail down to oligo(A) length (10 to 12 As) triggers decapping at the 5′ end of the mRNA. Decapping is catalyzed by a decapping enzyme complex consisting of two proteins in yeast (Dcp1 and Dcp2) and their homologs plus additional proteins in mammals. Decapping yields a 5′ monophosphorylated RNA end (the substrate for the 5′ to 3′ processive exonuclease Xrn1), which rapidly digests the mRNA. In fact, this digestion is so fast that intermediates could not be identified until investigators discovered that a stretch of guanosine nucleotides (poly[G]) could block Xrn1 progression in yeast. As illustrated in FIGURE 20.7, they engineered mRNAs to contain an internal poly(G) tract and found that the oligoadenylated 3′ end of the mRNAs accumulated. This result showed that 5′ to 3′ exonuclease digestion is the primary route of decay and that decapping precedes complete removal of the poly(A) tail.

FIGURE 20.6 The major deadenylation-dependent decay pathways in eukaryotes. Two pathways are initiated by deadenylation. In both, poly(A) is shortened by a poly(A) nuclease until it reaches a length of about 10 A. Then an mRNA may be degraded by the 5′ to 3′ pathway or by the 3′ to 5′ pathway. The 5′ to 3′ pathway involves decapping by Dcp and digestion by the Xrn1 exonuclease. The 3′ to 5′ pathway involves digestion by the exosome complex.

FIGURE 20.7 Use of a poly(G) sequence to determine direction of decay. A poly(G) sequence, engineered into an mRNA, will block the progression of exonucleases in yeast. The 5′ or 3′ mRNA fragment resistant to degradation accumulates in the cell and can be identified by northern blot.
The cap is normally resistant to decapping during active translation because it is bound by the cytoplasmic cap-binding protein, a component of the eukaryotic initiation factor 4F (eIF4F) complex required for translation (described in the chapter titled Translation). Thus, the translation and decapping machineries compete for the cap. How does deadenylation at the 3′ end of the mRNA render the cap susceptible? Translation is known to involve a physical interaction between bound PABP at the 3′ end and the eIF4F complex at the 5′ end. Release of PABP by deadenylation is thought to destabilize the eIF4F–cap interaction, leaving the cap more frequently exposed. The mechanism is not this simple, though, because additional proteins are known to be involved in the decapping event. A complex of seven related proteins, Lsm1–7, binds to the oligo(A) tract after loss of PABP and is required for decapping. Furthermore, a number of decapping enhancers have been discovered. The mechanisms by which these proteins stimulate decapping are not fully understood, although they appear to act either by recruiting/stimulating the decapping machinery or by inhibiting translation.
In the second pathway (Figure 20.6, right), deadenylation to oligo(A) is followed by 3′ to 5′ exonuclease digestion of the body of the mRNA. This degradation step is catalyzed by the exosome, a ring-shaped complex consisting of a nine-subunit core with one or more additional proteins attached to its surface. A recent report showed that the exosome also has endonuclease activity, and the function of this activity in mRNA decay remains unknown. The exosome exists in similar form in archaea and is also analogous to the bacterial degradosome in that its core subunits are structurally related to PNPase. Thus, the exosome is an ancient piece of molecular machinery. The exosome also plays an important role in the nucleus, described in the section in this chapter titled Newly Synthesized RNAs Are Checked for Defects via a Nuclear Surveillance System.
The relative importance of each mechanism is not yet known, although in yeast the deadenylation-dependent decapping pathway seems to predominate. The pathways are at least partially redundant. Hundreds of yeast mRNAs were examined by microarray analysis in cells in which either the 5′ to 3′ or 3′ to 5′ pathway was inactivated. In either case, only a small percentage of transcripts increase in abundance relative to wild-type cells. This finding suggests that few yeast mRNAs have a requirement for one or the other pathway. It has been proposed that these deadenylation-dependent pathways represent the default degradation pathways for all polyadenylated mRNAs, though subsets of mRNAs can be targets for other specialized pathways, described in the next section in this chapter titled Other Degradation Pathways Target Specific mRNAs. Even those mRNAs that are degraded by the default pathways, however, are degraded at different mRNA-specific rates.
20.6 Other Degradation Pathways Target Specific mRNAs
Four other pathways for mRNA degradation have been described. FIGURE 20.8 and TABLE 20.1 summarize these, along with the two major pathways. These pathways are specific for subsets of mRNAs and typically involve regulated degradation events.

FIGURE 20.8 Other decay pathways in eukaryotic cells. The initiating event for each pathway is illustrated. (a) Some mRNAs may be decapped before deadenylation occurs. (b) Histone mRNAs receive a short poly(U) tail to become a decay substrate. (c) Degradation of some mRNAs can be initiated by a sequence-specific endonucleolytic cut. (d) Some mRNAs can be targeted for degradation or translational silencing by complementary guide miRNAs.
TABLE 20.1 Summary of key elements of mRNA decay pathways in eukaryotic cells.
| Pathway | Initiating Event | Secondary Step(s) | Substrates |
|---|---|---|---|
| Deadenylation-dependent 5′ to 3′ digestion | Deadenylation to oligo(A) | Oligo(A) binding by Lsm complexDecapping and 5′ to 3′ exonuclease digestion by XRN1 | Probably most polyadenylated mRNAs |
| Deadenylation-dependent 3′ to 5′ digestion | Deadenylation to oligo(A) | 3′ to 5′ exonuclease digestion | Probably most polyadenylated mRNAs |
| Deadenylation-independent decapping | Decapping | 5′ to 3′ exonuclease digestion | Few specific mRNAs |
| Endonucleolytic pathway | Endonuclease cleavage | 5′ to 3′ and 3′ to 5′ exonuclease digestion | Few specific mRNAs |
| Histone mRNA pathway | Oligouridylation | Oligo(U) binding by Lsm complex Decapping and 5′ to 3′ exonuclease digestion by XRN13′ to 5′ digestion by exosome | Histone mRNAs in mammals |
| miRNA pathway | Base pairing with miRNA in RISC | Endonucleolytic cleavage or translational repression | Many mRNAs (extent unknown) |
One pathway involves deadenylation-independent decapping; that is, decapping proceeds in the presence of a still long poly(A) tail. Decapping is then followed by Xrn1 digestion. Bypassing the deadenylation step requires a mechanism to recruit the decapping machinery and inhibit eIF4F binding without the help of the Lsm1–7 complex. One of the mRNAs degraded by this pathway is RPS28B mRNA, which encodes the ribosomal protein S28 and has an interesting autoregulation mechanism. A stem-loop in its 3′ UTR is involved in recruiting a known decapping enhancer. The recruitment occurs only when the stem-loop is bound by S28 protein. Thus, an excess of free S28 in the cell will cause the accelerated decay of its mRNA.
A second specialized pathway is used to degrade the cell cycle–regulated histone mRNAs in mammalian cells. These mRNAs are responsible for synthesis of the huge number of histone proteins needed during DNA replication. They accumulate only during S-phase and are rapidly degraded at its end. The nonpolyadenylated histone mRNAs terminate in a stem-loop structure similar to that of many bacterial mRNAs. Their mode of degradation has striking similarities to bacterial mRNA decay. A polymerase, structurally similar to the bacterial poly(A) polymerase, adds a short poly(U) tail instead of a poly(A) tail. This short tail serves as a platform for the Lsm1–7 complex and/or the exosome, activating the standard decay pathways. This mode of degradation provides an important evolutionary link between mRNA decay systems in prokaryotes and eukaryotes.
A third pathway is initiated by sequence- or structure-specific endonucleotic cleavage. The cleavage is followed by 5′ to 3′ and 3′ to 5′ digestion of the fragments, and a scavenging decapping enzyme, different from the Dcp complex, can remove the cap. Several endonucleases that cleave specific target sites in mRNAs have been identified. One interesting case is the targeted cleavage of yeast CLB2 (cyclin B2) mRNA, which occurs only at the end of mitosis. The endonuclease that catalyzes the cleavage, RNase MRP, is restricted to the nucleolus and mitochondria for most of the cell cycle, where it is involved in RNA processing but is transported to the cytoplasm in late mitosis.
The fourth, and most important, pathway is the microRNA (miRNA) pathway. This pathway usually leads directly to endonucleolytic cleavage of mRNA in plants; in animal cells it directs targeted deadenylation-dependent degradation and, more commonly, translational repression. MicroRNAs are short RNAs (about 22 nucleotides) derived from transcribed miRNA genes and are generated by cleavage from longer precursor RNAs. In all cases, an mRNA is targeted for silencing by the base pairing of the short complementary miRNAs presented in the context of a protein complex called RISC (RNA-induced silencing complex). Thus, the silencing of target mRNAs is controlled by regulated transcription of the miRNA genes. The details of this mechanism are described in the Regulatory RNA chapter.
The significance of the microRNA pathway to total mRNA decay is substantial. At least 1,000 miRNAs are predicted to function in humans. By identification of conserved complementary target sites in the vertebrate transcriptome, it has been estimated that 50% of all mRNAs could be regulated by miRNAs. Potentially regulated mRNAs often contain multiple target sites in their 3′ UTRs. Mutation of miRNA target sites is likely to explain many genetic disease alleles, and dysregulation of miRNA has already been associated with hundreds of diseases.
An integrated model of mRNA degradation has been proposed. This model suggests that the deadenylation-dependent decay pathways represent the default systems for degrading all polyadenylated mRNAs. The rate of deadenylation and/or other steps in degradation by these pathways can be controlled by cis-acting elements in each mRNA and trans-acting factors present in the cell. Superimposed on the default system are the mRNA decay pathways described earlier for targeting specific mRNAs.
20.7 mRNA-Specific Half-Lives Are Controlled by Sequences or Structures Within the mRNA
What accounts for the large range of half-lives of different mRNAs in the same cell? Specific cis-elements within an mRNA are known to affect its stability. The most common location for such elements is within the 3′ UTR, although they exist elsewhere. Whole-genome studies have revealed many highly conserved 3′ UTR motifs, but their roles remain mostly unknown. Many are likely to be target sites for miRNA base pairing. Others are binding sites for RBPs, some of which have known functions in stability. Rates of deadenylation can vary widely for different mRNAs, and sequences that affect this rate have been described.
Destabilizing elements (DEs) have been the most widely studied. The criterion for defining a destabilizing sequence element is that its introduction into a more stable mRNA accelerates its degradation. Removal of an element from an mRNA does not necessarily stabilize it, indicating that an individual mRNA can have more than one DE. To complicate their identification further, the presence of a DE does not guarantee a short half-life under all conditions, because other sequence elements in the mRNA can modify its effectiveness.
The most well-studied type of DE is the AU-rich element (ARE), found in the 3′ UTR of up to 8% of mammalian mRNAs. AREs are heterogeneous, and a number of subtypes have been characterized. One type consists of the pentamer sequence AUUUA present once or repeated multiple times in different sequence contexts. Another type does not contain AUUUA and is predominantly U-rich. A large number of ARE-binding proteins with specificity for certain ARE types and/or cell types have been identified. How do AREs work to stimulate rapid degradation? Many ARE-binding proteins have been found to interact with one or more components of the degradation machinery, including the exosome, deadenylases, and decapping enzyme, suggesting that they act by recruiting the degradation machinery. The exosome can bind some AREs directly. The AREs of a number of mRNAs have been shown to accelerate the deadenylation step of decay, although it is not likely that they all work this way. Another way they might act is by facilitating efficient engagement of the mRNA into processing bodies.
Many AU-rich DEs and other kinds of destabilizing elements have been identified in the mRNAs of budding yeast and other model organisms. For example, the previously mentioned Puf proteins of yeast bind to specific UG-rich elements and accelerate the degradation of target mRNAs. In this case, the destabilizing mechanism is accelerated deadenylation by recruitment of the CCR4-NOT deadenylase. A genomics analysis of yeast 3′ UTRs has identified 53 sequence elements that correlate with the half-lives of mRNAs containing them, suggesting the number of different destabilizing elements may be large. FIGURE 20.9 summarizes the known actions of destabilizing elements.

FIGURE 20.9 Mechanisms by which destabilizing elements (DEs) and stabilizing elements (SEs) function. Effects of DEs and SEs on mRNA stability are mediated primarily through the proteins that bind to them. One exception is a DE that acts as an endonuclease target site.
Stabilizing elements (SEs) have been identified in a few unusually stable mRNAs. Three mRNAs studied in mammalian cells have stabilizing pyrimidine-rich sequences in their 3′ UTRs. Proteins that bind to this element in globin mRNA have been shown to interact with PABPs, suggesting they might function to protect the poly(A) tail from degradation. In some cases, an mRNA can be stabilized by inhibition of its DE. For example, certain ARE-binding proteins act to prevent the ARE from destabilizing the mRNA, presumably by blocking the ARE-binding site. An example of regulated mRNA stabilization occurs for the mammalian transferrin mRNA. It is stabilized when its 3′ UTR iron-response element (IRE), consisting of multiple stem-loop structures, is bound by a specific protein, as shown in FIGURE 20.10. The affinity of the IRE-binding protein for the IRE is altered by iron binding, exhibiting low affinity when its iron-binding site is full and high affinity when it is not. When the cellular iron concentration is low, more transferrin is needed to import iron from the bloodstream, and under these conditions the transferrin mRNA is stabilized. The IRE-binding protein stabilizes the mRNA by inhibiting the function of destabilizing sequences in the vicinity. Interestingly, the same IRE-binding protein also binds an IRE in ferritin mRNA and regulates this mRNA in a very different way. Ferritin is an iron-binding protein that sequesters excess cellular iron. The IRE-binding protein binds IRE stem-loops in the 5′ UTR of ferritin when iron is low and blocks the interaction of the cap-binding complex with ferritin mRNA. Thus, translation of ferritin mRNA is prevented when cellular iron levels are low—the conditions under which transferrin mRNA is stabilized and translated.

FIGURE 20.10 Regulation of transferring mRNA stability by iron (Fe) levels. The IRE in the 3′ UTR is the binding site for a protein that stabilizes the mRNA. The IRE-binding protein is sensitive to iron levels in the cell, binding to the IRE only when iron is low.
Many cis-element–binding proteins are subject to modifications that are likely to affect their functions, including phosphorylations, methylations, conformational changes due to effector binding, and isomerizations. Such modifications may be responsible for changes in mRNA degradation rates induced by cellular signals. mRNA decay can be altered in response to a wide variety of environmental and internal stimuli, including cell cycle progression, cell differentiation, hormones, nutrient supply, and viral infection. Microarray studies have shown that almost 50% of changes in mRNA levels stimulated by cellular signals are due to mRNA stabilization or destabilization events, not to transcriptional changes. How these changes are effected remains largely unknown.
20.8 Newly Synthesized RNAs Are Checked for Defects via a Nuclear Surveillance System
All newly synthesized RNAs are subject to multiple processing steps after they are transcribed (see the chapter titled RNA Splicing and Processing). At each step, errors may be made. Whereas DNA errors are repaired by a variety of repair systems (see the chapter titled Repair Systems), detectable errors in RNA are dealt with by destroying the defective RNA. RNA surveillance systems exist in both the nucleus and cytoplasm to handle different kinds of problems. Surveillance involves two kinds of activities: one to identify and tag the aberrant substrate RNA, and another to destroy it.
The destroyer is the nuclear exosome. The nuclear exosome core is almost identical to the cytoplasmic exosome, though it interacts with different protein cofactors. It removes nucleotides from targeted RNAs by 3′ to 5′ exonuclease activity. The nuclear exosome has multiple functions involving RNA processing of some noncoding RNA transcripts (snRNA, snoRNA, and rRNA) and complete degradation of aberrant transcripts. The exosome is recruited to its processing substrates by protein complexes that recognize specific RNA sequences or RNA–RNP structures. For example, Nrd1–Nab3 is a sequence-specific protein dimer that recruits the exosome to normal sn/snoRNA processing substrates. This protein pair binds to GUA[A-G] and UCUU elements, respectively. The Nrd1–Nab3 cofactor is also involved in transcription termination of these nonpolyadenylated Pol II–transcribed RNAs, suggesting that the processing exosome may be recruited directly to the site of their synthesis.
Aberrantly processed, modified, or misfolded RNAs require other protein cofactors for identification and exosome recruitment. The major nuclear complex performing this function in yeast is called TRAMP (an acronym for the component proteins), and it exists in at least two forms, differing in the type of poly(A) polymerase present. The TRAMP complex acts in several ways to effect degradation:
It interacts directly with the exosome, stimulating its exonuclease activity.
It includes a helicase, which is probably required to unwind secondary structure and/or move RNA-binding proteins from structured RNP substrates during degradation.
It adds a short 3′ oligo(A) tail to target substrates. The oligo(A) tail is thought to make the targeted RNP a better substrate for the degradation machinery in the same way that the oligo(A) tail functions in bacteria.
FIGURE 20.11 summarizes the roles of TRAMP and the exosome. It has become clear that RNA degradation in bacteria and archaea and nuclear RNA degradation in eukaryotes are evolutionarily related processes. Their similarity suggests that the ancestral role of polyadenylation was to facilitate RNA degradation, and that poly(A) was later adapted in eukaryotes for the oddly reverse function of stabilizing mRNAs in the cytoplasm.

FIGURE 20.11 The role of TRAMP and the exosome in degrading aberrant nuclear RNAs. Defective RNPs are tagged by protein cofactors, which then recruit the nuclear exosome. The cofactor in yeast cells is the complex TRAMP. The poly(A) polymerase (PAP, or Trf4) in TRAMP adds a short poly(A) tail to the 3′ end of the targeted RNA.
What are the substrates for TRAMP–exosome degradation? The TRAMP complex is remarkable in that it recognizes a wide variety of aberrant RNAs synthesized by all three transcribing polymerases. It is not known how this is accomplished given that the targeted RNAs share no recognizably common features. Some researchers favor a kinetic competition model, hypothesizing that RNAs that do not get processed and assembled into final RNP form in a timely manner will become substrates for exosome degradation. This mechanism avoids the need to posit specific recognition of innumerable possible defects.
What kinds of abnormalities condemn pre-mRNAs to nuclear destruction? Two kinds of substrates have been identified. One type is unspliced or aberrantly spliced pre-mRNAs. Components of the spliceosome retain such transcripts either until they are degraded by the exosome or until proper splicing is completed, if possible. It is thought that the kinetic competition model probably applies here, too. A pre-mRNA that is not efficiently spliced and packaged is at increased risk of being accessed by the exosome degradation machinery. The basis for recognition of aberrantly spliced pre-mRNAs is not known. The second type of pre-mRNA substrate is one that has been improperly terminated, lacking a poly(A) tail. Whereas polyadenylation is protective in true mRNAs, it may actually be destabilizing for cryptic unstable transcripts (CUTs). These non-protein-coding RNAs (also discussed in the Regulatory RNA chapter) are transcribed by RNA Pol II and do not encode recognizable genes; however, they frequently overlap with (and sometimes regulate) protein-coding genes. These transcripts are polyadenylated by a component of the TRAMP complex (Trf4). They are distinguished from other transcripts of unknown function by their extreme instability, normally being degraded by the TRAMP–exosome complex immediately after synthesis, possibly targeted by the Trf4-dependent polyadenylation. In fact, the existence of these transcripts was first convincingly demonstrated in yeast strains with impaired nuclear RNA degradation. More than three-quarters of RNA Pol II transcripts may be composed of noncoding RNAs and be subject to rapid degradation by the exosome! Some CUTs appear to arise from spurious transcription initiation, and the short-lived RNA products themselves typically do not appear to have a function (i.e., these RNAs do not typically act in trans). However, some examples indicate that the transcription process itself may play a role in regulating nearby or overlapping coding genes (one example is described in the Regulatory RNA chapter).
20.9 Quality Control of mRNA Translation Is Performed by Cytoplasmic Surveillance Systems
Some kinds of mRNA defects can be assessed only during translation. Surveillance systems have evolved to detect three types of mRNA defects that threaten translational fidelity and to target the defective mRNAs for rapid degradation. FIGURE 20.12 shows the substrates for each of these three systems. All three systems involve abnormal translation termination events, so it is useful to review what happens during normal termination (see the Translation chapter for a more detailed description). When a translating ribosome reaches the termination (stop) codon, a pair of release factors (eRF1 and eRF2 in eukaryotes) enters the ribosomal A site, which is normally filled by incoming tRNAs during elongation. The release factor complex mediates the release of the completed polypeptide, followed by the mRNA, remaining tRNA, and ribosomal subunits.

FIGURE 20.12 Substrates for cytoplasmic surveillance systems. Nonsense-mediated decay (NMD) degrades mRNAs with a premature termination codon (PTC) position ahead of its normal termination codon (TC). Nonstop decay (NSD) degrades mRNAs lacking an in-frame termination codon. No-go decay (NGD) degrades mRNAs having ribosome stalled in the coding region.
Nonsense-mediated decay (NMD) targets mRNAs containing a premature termination codon (PTC). Its name comes from nonsense mutation, which is only one way that mRNAs with a PTC can be generated. Genes without nonsense mutations can give rise to aberrant transcripts containing a PTC by (1) RNA polymerase error or (2) incomplete, incorrect, or alternative splicing. It has been estimated that almost half of alternatively spliced pre-mRNAs generate at least one form with PTC. About 30% of known disease-causing alleles probably encode an mRNA with a PTC. An mRNA with a PTC will produce C-terminal truncated polypeptides, which are considered to be particularly toxic to a cell due to their tendency to trap multiple binding partners in nonfunctional complexes. The NMD pathway has been found in all eukaryotes.
Targeting of PTC-containing mRNAs requires translation and a conserved set of protein factors. They include three Upf proteins (Upf1, Upf2, and Upf3) and four additional proteins (Smg1, Smg5, Smg 6, and Smg7). Upf1 is the first NMD protein to act, binding to the terminating ribosome—specifically to its release factor complex. UPF attachment tags the mRNA for rapid decay. The specific roles of the NMD factors have not yet been defined, although phosphorylation of ribosome-bound Upf1 by Smg1 is critical. Their combined actions condemn the mRNA to the general decay machinery and stimulate rapid deadenylation. The target mRNAs are degraded by both 5′ to 3′ and 3′ to 5′ pathways.
How are PTCs distinguished from the normal termination codon further downstream? The mechanism has been studied extensively both in yeast and in mammalian cells, where it is somewhat different; these mechanisms are illustrated in FIGURE 20.13. The major signal that identifies a PTC in mammalian cells is the presence of a splice junction, marked by an exon junction complex (EJC) downstream of the premature termination codon. The majority of genes in higher eukaryotes do not have an intron interrupting the 3′ UTR, so authentic termination codons are not generally followed by a splice junction. During the pioneer round of translation for a normal mRNA, all EJCs occur within the coding region and are displaced by the transiting ribosome. During the pioneer round of translation for an NMD substrate, Upf2 and Upf3 proteins bind to the residual downstream EJC(s), targeting it for degradation.

FIGURE 20.13 Two mechanisms by which a termination codon is recognized as premature. (a) In mammals, the presence of an EJC downstream of a termination codon targets the mRNA for NMD. (b) In probably all eukaryotes, an abnormally long 3′ UTR is recognized by the distance between the termination codon and the poly(A)–PABP complex. In either case, the Upf1 protein binds to the terminating ribosome to trigger decay.
Most S. cerevisiae genes are not interrupted by introns at all, so the mechanism for PTC detection must be different. In this case an abnormally long 3′ UTR is the warning sign. This was demonstrated by the finding that extension of the 3′ UTR of a normal mRNA could convert it into a substrate for NMD. A current model proposes that proper translation termination at a stop codon requires a signal from a nearby PABP. Although 3′ UTRs are highly variable in nucleotide length, the physical distance between the termination codon and the poly(A) tail is not strictly a function of length because secondary structures and interactions between bound RBPs can compress the distance. The requirement for PABP was demonstrated in multiple organisms by tethering a PABP close to the PTC, as illustrated in FIGURE 20.14. The mRNA was no longer targeted by NMD. PTC recognition also occurs independently of splicing in Drosophila, Caenorhabditis elegans, plants, and in some mammalian mRNAs, suggesting that the length and structure of the 3′ UTR may be critical for the normal process of translation termination in all eukaryotic organisms.
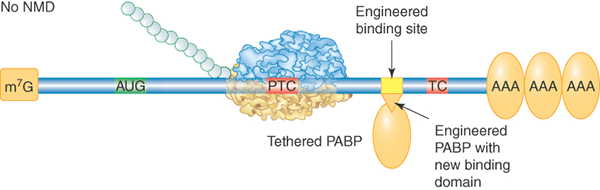
FIGURE 20.14 Effect of tethering a PABP near a premature termination codon. A PABP gene was altered to express a phage RNA-binding domain. Its binding site was engineered into a test NMD-substrate gene. The tethered PABP prevented the usual rapid degradation of this mRNA by NMD. This method has many applications in molecular biology.
Some normal mRNAs are targeted by NMD. These were identified by experiments in which Upf1 levels were reduced, resulting in a subset of transcripts that increased in abundance. The list of normal NMD substrates includes mRNAs with especially long 3′ UTRs, mRNAs encoding selenoproteins (which use the termination codon UGA as a selenocysteine codon), and an unknown number of alternatively spliced mRNAs. Not all targeted mRNAs are predicted to be NMD substrates based on our current understanding. NMD may turn out to be an important rapid decay pathway for a variety of short-lived mRNAs.
Bacteria are also able to rapidly degrade mRNAs with premature termination codons. In the E. coli version of NMD, the endonuclease RNase E cuts the mRNA in the region 3′ to the PTC, which is in an abnormally unprotected state due to premature release of ribosomes. This mechanism probably does not require any additional means to distinguish a PTC from the correct termination codon and would also work for polycistronic mRNAs.
Nonstop decay (NSD) targets mRNAs that lack an in-frame termination codon (middle panel in Figure 20.12). Failure to terminate results in a ribosome translating into the poly(A) tail and probably stalling at the 3′ end. NSD substrates are generated mainly by premature transcription termination and polyadenylation in the nucleus. Such prematurely polyadenylated transcripts are surprisingly common. Analysis of random cDNA populations derived from yeast and human mRNAs suggests that 5% to 10% of polyadenylation events may occur at upstream “cryptic” sites that resemble an authentic polyadenylation signal. Targeting nonstop substrates involves a set of factors called the SKI proteins. The ribosome is released from the mRNA by the action of Ski7. Ski7 has a GTPase domain similar to eEF3 and probably binds to the ribosome in the A site to stimulate release. The subsequent recruitment of the other SKI proteins and the exosome results in 3′ to 5′ decay of the mRNA. Decay of nonstop substrates can also occur in the absence of Ski7 and proceeds by decapping and 5′ to 3′ digestion. Susceptibility to decapping could be due to the pioneer ribosome displacing PABPs as it traverses the poly(A) tail. Rapid decay of nonstop substrates results in not only prevention of toxic polypeptides but also liberation of trapped ribosomes. Interestingly, E. coli uses a specialized noncoding RNA (tmRNA) that acts like both a tRNA and an mRNA to rescue ribosomes stalled on a nonstop mRNA. tmRNA directs the addition of a short peptide that targets the defective protein product for degradation, provides a stop codon to allow recycling of the ribosome, and targets degradation of the defective mRNA by RNAse R.
No-go decay (NGD) targets mRNAs with ribosomes stalled in the coding region codon (bottom panel of Figure 20.12). Transient or prolonged stalling can be caused by natural features of some mRNAs, including strong secondary structures and rarely used codons (whose cognate tRNAs are in low abundance). This newly discovered surveillance pathway has been studied only in yeast and is the least understood of the three. Targeting of the mRNA involves recruitment of two proteins, Dom34 and Hbs1, which are homologous to eRF1 and eRF3, respectively. mRNA degradation is initiated by an endonucleolytic cut, and the 5′ and 3′ fragments are digested by the exosome and Xrn1. Dom34 might be the endonuclease, as one of its domains is nuclease-like. Why would a normal mRNA have hard-to-translate sequences that might condemn it to rapid degradation? Such sequences can be thought of as another kind of destabilizing element. Evolutionary retention of impediments to efficient translation suggests that they serve an important function in controlling the half-life of these mRNAs.
20.10 Translationally Silenced mRNAs Are Sequestered in a Variety of RNA Granules
The occurrence in germ cells and neurons of macroscopic, cytoplasmic particles containing mRNA has been known for many years. RNA granules were considered to be mRNA storage structures unique to these specialized cell types. Recent studies have vastly expanded the known occurrence and probable roles of these and related granules. One similarity among all of the known RNA granules is that they harbor untranslated mRNAs and about 50 to 100 different proteins, depending on granule type. The protein components differ among granule types, though all granules contain sets of proteins that mediate aggregation through self-interaction motifs. RNA granules form by aggregation of mRNPs and protein and are heterogeneous in size. The cytoskeleton and motor proteins also can play roles in assembly and disassembly of granules (as well as their transport).
Germ cell granules (also called maternal mRNA granules) are found in oocytes from a variety of organisms. These granules comprise collections of mRNAs that are held in a state of translational repression until they are activated during subsequent development. Repression is achieved by extensive deadenylation, and activation is achieved by polyadenylation. These granules also may carry mRNAs being transported to specific regions of this large cell (see the next section in this chapter, titled Some Eukaryotic mRNAs Are Localized to Specific Regions of a Cell). Neuronal granules are similar to maternal mRNA granules in that they function in the translational repression and transport of specific mRNAs. These granules are essential for normal neuronal function.
New studies suggest that at least some mRNA degradation occurs within discrete particles throughout the cytoplasm of most or all cell types. These particles, called processing bodies (PBs), are the only granule type that contains proteins involved in mRNA decay, including the decapping machinery and Xrn1 exonuclease. mRNAs silenced via RNAi and miRNA pathways are present in PBs. PABPs are not found in PBs, suggesting that deadenylation precedes mRNA localization into these structures. Processing bodies are dynamic, increasing and decreasing in size and number, and even disappearing, under different cellular and experimental conditions that affect translation and decay. For example, release of mRNAs from polysomes by a drug that inhibits translation initiation results in a large increase in PB number and size, as does slowing degradation by partial inactivation of decay components. Not all resident mRNAs are doomed for destruction, though; some can be released for translation, but which ones and why they are freed is not yet clear. It is not known whether all mRNA degradation normally occurs in these bodies, or even what function(s) they serve. One idea is that concentrating powerful destructive enzymes in isolated locations renders mRNA degradation more safe and efficient. Another is that they serve as temporary storage sites when the capacity of the decay and/or translation machinery is exceeded.
Another mRNA-containing particle related to PBs is called a stress granule (SG). Whereas PBs are constitutive, SGs only accumulate in response to stress-induced inhibition of translation initiation (a response common to probably all eukaryotic organisms). PBs and SGs share some, but not all, protein components. For example, SGs lack components of the RNA decay machinery, which PBs have, but include many translational initiation components that PBs lack. Both types of particle can coexist in one cell, and the size and numbers of both increase under stress conditions. mRNAs may be exchanged between the two types of particles. In the presence of polysome-stabilizing drugs, which trap mRNAs in a static state of translation, both PBs and SGs become smaller or disappear, suggesting that the granule mRNAs are normally in a dynamic equilibrium with the population of mRNAs being translated. SGs share many components with neuronal granules. Of particular interest is the fact that a number of shared RNA-binding proteins, known to be essential to SG formation, have been implicated in neuronal defects.
20.11 Some Eukaryotic mRNAs Are Localized to Specific Regions of a Cell
The cytoplasm is a crowded place occupied by a high concentration of proteins. It is not clear how freely polysomes can diffuse, and most mRNAs are probably translated in random locations that are determined by their point of entry into the cytoplasm and the distance that they may have moved away from it. Some mRNAs are translated only at specific sites, though—their translation is repressed until they reach their destinations. The regulated localization has been described for more than 100 specific mRNAs, a number that certainly represents a small fraction of the total. mRNA localization serves a number of important functions in eukaryotic organisms of all types. Three key functions are illustrated in FIGURE 20.15 and described below:
Localization of specific mRNAs in the oocytes of many animals serves to set up future patterns in the embryo (such as axis polarity) and to assign developmental fates to cells residing in different regions. These localized maternal mRNAs encode transcription factors or other proteins that regulate gene expression. In Drosophila oocytes, bicoid and nanos mRNAs are localized to the anterior and posterior poles, respectively, and their translation following fertilization results in gradients of their protein products. The gradients are used by cells in early development for the specification of their anterior–posterior position in the embryo. Bicoid encodes a transcription factor, and nanos encodes a translational repressor. Some localized mRNAs encode determinants of cell fate. For example, oskar mRNA localizes in the posterior of the oocyte and initiates the process leading to development of primordial germ cells in the embryo. It is estimated that during Drosophila development 70% of mRNAs are expressed in specific spatial domains.
mRNA localization also plays a role in asymmetric cell divisions; that is, mitotic divisions that result in daughter cells that differ from one another. One way this is accomplished is by asymmetric segregation of cell-fate determinants, which may be proteins and/or the mRNAs that encode them. In Drosophila embryos, prospero mRNA and its product (a transcription factor) are localized to a region of the peripheral cortex of the embryo. Later in development, oriented cell division of neuroblasts ensures that only the outermost daughter cell receives prospero, committing it to a ganglion mother-cell fate. Asymmetric cell division is also used by budding yeast to generate a daughter cell of a different mating type than the mother cell, an event described later in this section.
mRNA localization in adult, differentiated cell types is a mechanism for the compartmentalization of the cell into specialized regions. Localization may be used to ensure that components of multiprotein complexes are synthesized in proximity to one another and that proteins targeted to organelles or specialized areas of cells are synthesized conveniently nearby. mRNA localization is particularly important for highly polarized cells such as neurons. Although most mRNAs are translated in the neuron cell body, many mRNAs are localized to its dendritic and axonal extensions. Among those is β-actin mRNA, whose product participates in dendrite and axon growth. β-actin mRNA localizes to sites of active movement in a wide variety of motile cell types. Interestingly, localization of mRNA at neuronal postsynaptic sites seems to be essential for modifications accompanying learning. In glial cells, the myelin basic protein (MBP) mRNA, which encodes a component of the myelin sheath, is localized to a specific myelin-synthesizing compartment. Plants localize mRNAs to the cortical region of cells and to regions of polar cell growth.

FIGURE 20.15 Three main functions of mRNA localization.
In some cases, mRNA localization involves transport from one cell to another. Maternal mRNPs in Drosophila are synthesized and assembled in surrounding nurse cells and are transferred to the developing oocyte through cytoplasmic canals. Plants can export RNAs through plasmodesmata and transport them for long distances via the phloem vascular system. mRNAs are sometimes transported en masse in mRNP granules. The compositions of these granules are not yet well defined.
Three mechanisms for the localization of mRNA have been well documented:
The mRNA is uniformly distributed but degraded at all sites except the site of translation.
The mRNA is freely diffusible but becomes trapped at the site of translation.
The mRNA is actively transported to a site where it is translated.
Active transport is the predominant mechanism for localization. Transport is achieved by translocation of motor proteins along cytoskeletal tracks. All three molecular motor types are exploited: dyneins and kinesins, which travel along microtubules in opposite directions, and myosins, which travel along actin fibers. This mode of localization requires at least four components: (1) cis-elements on the target mRNA, (2) trans-factors that directly or indirectly attach the mRNA to the correct motor protein, (3) trans-factors that repress translation, and (4) an anchoring system at the desired location.
Only a few cis-elements, sometimes called zipcodes, have been characterized. They are diverse, include examples of both sequence and structural RNA elements, and can occur anywhere in the mRNA, though most are in the 3′ UTR. Zipcodes have been difficult to identify, presumably because many consist of complex secondary and tertiary structures. A large number of trans-factors have been associated with localized mRNA transport and translational repression, some of which are highly conserved in different organisms. For example, staufen, a double-stranded RBP, is involved in localizing mRNAs in the oocytes of Drosophila and Xenopus, as well as the nervous systems of Drosophila, mammals, and probably worms and zebrafish. This multitalented factor has multiple domains that can couple complexes to both actin- and microtubule-dependent transport pathways. Almost nothing is known about the fourth required component—anchoring mechanisms. Two examples of localization mechanisms are discussed in the following paragraphs.
The localization of β-actin mRNA has been studied in cultured fibroblasts and neurons. The zipcode is a 54-nucleotide element in the 3′ UTR. Cotranscriptional binding of the zipcode element by the protein ZBP1 is required for localization, suggesting that this mRNA is committed to localization before it is even processed and exported from the nucleus. Interestingly, β-actin mRNA localization is dependent on intact actin fibers in fibroblasts and intact microtubules in neurons.
Genetic analysis of ASH1 mRNA localization in yeast has provided the most complete picture of a localization mechanism to date and is illustrated in FIGURE 20.16. During budding, the ASH1 mRNA is localized to the developing bud tip, resulting in Ash1 synthesis only in the newly formed daughter cell. Ash1 is a transcriptional repressor that disallows expression of the HO endonuclease, a protein required for mating-type switching (see the chapter titled Homologous and Site-Specific Recombination). The result is that mating-type switching occurs only in the mother cell. The ASH1 mRNA has four stem-loop localization elements in its coding region to which the protein She2 binds, probably in the nucleus. The protein She3 serves as an adaptor, binding both to She2 and to the myosin motor protein Myo4 (also called She1). A Puf protein, Puf6, binds to the mRNA, repressing its translation. The motor transports the ASH1 mRNP along the polarized actin fibers that lead from the mother cell to the developing bud. Additional proteins are required for proper localization and expression of the ASH1 mRNA. More than 20 yeast mRNAs use the same localization pathway.


FIGURE 20.16 Localization of ASH1 mRNA. Newly exported ASH1 mRNA is attached to the myosin motor Myo4 via a complex with the She2 and She3 proteins. The motor transports the mRNA along actin filaments to the developing bud.
Localization mechanisms that do not involve active transport have been clearly demonstrated for only a few localized mRNAs in oocytes and early embryos. The mechanism of local entrapment of diffusible mRNAs requires the participation of previously localized anchors, which have not been identified. In Drosophila oocytes, diffusing nanos mRNA is trapped at the posterior germ plasm, a specialized region of the cytoplasm underlying the cortex. In Xenopus oocytes, mRNAs localized to the vegetal pole are first trapped in a somewhat mysterious, membrane-laden structure called the mitochondrial cloud (MC), which later migrates to the vegetal pole, carrying mRNAs with it. The mechanism of localized mRNA stabilization has been described for an mRNA that also localizes to the posterior pole of the Drosophila embryo. Early in development, the hsp83 mRNA is uniformly distributed through the embryonic cytoplasm, but later it is degraded everywhere except at the pole. A protein called smaug is involved in destabilizing the majority of the hsp83 mRNAs, most likely by recruiting the CCR4-NOT complex. How the pole-localized mRNAs escape is not known.
Summary
Cellular RNAs are relatively unstable molecules due to the presence of cellular ribonucleases. Ribonucleases differ in mode of attack and are specialized for different RNA substrates. These RNA-degrading enzymes have many roles in a cell, including the decay of messenger RNA. The fact that mRNAs are short-lived allows rapid adjustment of the spectrum of proteins synthesized by a cell by regulating gene transcription rates. Messenger RNAs of different sequences exhibit very different susceptibilities to nuclease action, with half-lives varying by 100-fold or more.
mRNA associates with a changing population of proteins during its nuclear maturation and cytoplasmic life. A very large number of RBPs exist, most of which remain uncharacterized. Many proteins with nuclear roles are shed before or during mRNA export to the cytoplasm. Others accompany the mature mRNA and have cytoplasmic roles. mRNAs are associated with distinct, but overlapping, sets of RBPs with roles in translation, stability, and localization. The group of mRNAs that share a particular type of RBP has been called an RNA regulon.
Degradation of bacterial mRNAs is initiated by removal of a pyrophosphate from the 5′ terminus. This step triggers a cycle of endonucleolytic cleavages, followed by 3′ to 5′ exonucleolytic digestion of released fragments. The 3′ stem-loop on many mRNAs protects them from 3′ attack. The 3′ to 5′ exonuclease activity is facilitated by polyadenylation of 3′ ends, forming a platform for the enzyme. The main proteins involved in mRNA degradation function as a complex called the degradosome.
Degradation of most eukaryotic mRNAs in yeast, and probably in mammals, requires deadenylation as the first step. Extensive shortening of the poly(A) tail allows one of two degradation pathways to proceed. The 5′ to 3′ decay pathway involves decapping and 5′ to 3′ exonuclease digestion. The 3′ to 5′ decay pathway is catalyzed by the exosome, a large exonuclease complex. Translation and decay by the 5′ to 3′ pathway are competing processes because the translation initiation complex and the decapping enzyme both bind to the cap. Particles called processing bodies (PBs) contain mRNAs and proteins involved in both decay and translational repression and are thought to be the sites of mRNA degradation.
Four other pathways for mRNA degradation have been described that target specific mRNAs. Each uses the same degradation machinery as the deadenylation-dependent pathways but is initiated differently. They are initiated by: (1) deadenylation-independent decapping, (2) addition of a 3′ poly(U) tail, (3) sequence- or structure-specific endonucleolytic cleavage, and (4) base pairing of microRNAs.
Differences in the characteristic half-lives of mRNAs are due to specific cis-elements within an mRNA. Destabilizing elements and stabilizing elements have been described. They are most commonly located in the 3′ UTR and act by serving as binding sites for proteins or microRNAs. AU-rich elements (AREs) destabilize a large number of mRNAs in mammalian cells. Proteins that bind to destabilizing elements probably act primarily by recruiting some component(s) of the degradation machinery. mRNA stability can be regulated in response to cellular signals by modification of binding proteins.
Quality-control surveillance systems operate in both the nucleus and cytoplasm that target defective RNAs for degradation. In the nucleus, the exosome has a role in both processing of certain normal RNAs and destruction of abnormal ones. Defective RNAs are identified by a variety of exosome cofactors that then recruit the exosome. The major cofactor in yeast cells is the TRAMP complex, which has homologs in other eukaryotic organisms. RNA Pol II transcripts that are substrates for nuclear degradation include those that are not spliced correctly or lack normal poly(A) tails. The majority of RNA Pol II transcripts may be cryptic unstable transcripts (CUTs).
A variety of mRNAs are targeted by cytoplasmic surveillance systems. All three systems involve abnormal translation-termination events. Nonsense-mediated decay (NMD) targets mRNAs with premature termination codons. A conserved set of factors (the UPF and SMG proteins) are involved in identifying and committing an NMD substrate to the general decay machinery. A premature termination codon is recognized during the pioneer round of translation by a downstream exon junction complex (EJC) or by an unusually distant 3′ mRNA terminus. NMD also is involved in degrading certain normal unstable mRNAs. Nonstop decay (NSD) targets mRNAs lacking an in-frame termination codon and requires a conserved set of SKI proteins to force release of the trapped ribosome and recruit degradation machinery. No-go decay (NGD) targets mRNAs with stalled ribosomes in their coding regions and causes ribosome release and degradation.
Some mRNAs are localized to specific regions of cells and are not translated until their cellular destinations are reached. Localization requires cis-elements on the target mRNA and trans-factors to mediate the localization. Localization serves three main functions. First, in oocytes it serves to set up future patterns in the embryo and to assign developmental fates to cells residing in different regions. Second, in cells that divide asymmetrically it is a mechanism to segregate protein factors to only one of the daughter cells. Third, in some cells, especially polarized cell types, it is a mechanism to establish subcellular compartments. Three mechanisms for localization are known: (1) degradation of the mRNA at all sites other than the target site; (2) selective anchoring of diffusing mRNA at the target site; and (3) directed transport of the mRNA on cytoskeletal tracks. The third mechanism is the most common method and exploits actin- and microtubule-based molecular motors.
References
General
Houseley J., and Tollervey, D. (2009). The many pathways of RNA degradation. Cell 136, 763–776.
20.2 Messenger RNAs Are Unstable Molecules
Research
Dölken, L., Ruzsics, Z., Rädle, B., Friedel, C. C., Zimmer, R., Mages, J., Hoffmann, R., Dickinson, P., Forster, T., Ghaza, P., and Koszinowski, U. H. (2008). High-resolution gene expression profiling for simultaneous kinetic parameter analysis of RNA synthesis and decay. RNA 14, 1959–1972.
Foat, B. C., Houshmandi, S. S., Olivas, W. M., and Bussemaker, H. J. (2005). Profiling condition-specific, genome-wide regulation of mRNA stability in yeast. Proc. Natl. Acad. Sci. USA 102, 17675–17680.
20.3 Eukaryotic mRNAs Exist in the Form of mRNPs from Their Birth to Their Death
Reviews
Keene, J. D. (2007). RNA regulons: coordination of post-transcriptional events. Nat. Rev. Genet. 8, 533–543.
Moore, M. J. (2005). From birth to death: the complex lives of eukaryotic mRNAs. Science 309, 1514–1518.
Research
Hogan, D. J., Riordan, D. P., Gerber, A. P., Herschlag, D., and Brown, P. O. (2008). Diverse RNA-binding proteins interact with functionally related sets of RNAs, suggesting an extensive regulatory system. PLoS Biol. 6(10), e255.
20.4 Prokaryotic mRNA Degradation Involves Multiple Enzymes
Reviews
Belasco, J. G. (2010). All things must pass: contrasts and commonalities in eukaryotic and bacterial mRNA decay. Nat. Rev. Mol. Cell Biol. 11, 467–478.
Carpousis, A. J. (2007). The RNA degradosome of Escherichia coli: an mRNA-degrading machine assembled on RNase E. Annu. Rev. Microbiol. 61, 71–87.
Condon, C. (2007). Maturation and degradation of RNA in bacteria. Curr. Opin. Microbiol. 10, 271–278.
Deana, A., and Belasco, J. G. (2005). Lost in translation: the influence of ribosomes on bacterial mRNA decay. Genes Dev. 19, 2526–2533.
Research
Bernstein, J. A., Khodursky, A. B., Lin P. H., Lin-Chao, S., and Cohen, S. N. (2002). Global analysis of mRNA decay and abundance in Escherichia coli at single-gene resolution using two-color fluorescent DNA microarrays. Proc. Natl. Acad. Sci. USA 99, 9697–9702.
Celesnik, H., Deana, A., and Belasco, J. G. (2007). Initiation of RNA decay in Escherichia coli by 5′ pyrophosphate removal. Mol. Cell 27, 79–90.
Mohanty, B. K., and Kushner, S. R. (2006). The majority of Escherichia coli mRNAs undergo post-transcriptional modification in exponentially growing cells. Nucleic Acids Res. 34(19), 5695–5704.
20.5 Most Eukaryotic mRNA Is Degraded via Two Deadenylation-Dependent Pathways
Reviews
Franks, T. M., and Lykke-Andersen, J. (2008). The control of mRNA decapping and P-body formation. Mol. Cell 32, 605–615.
Parker, R., and Sheth, U. (2007). P Bodies and the control of mRNA translation and degradation. Mol. Cell 25, 635–646.
Parker, R., and Song, H. (2004). The enzymes and control of eukaryotic mRNA turnover. Nat. Struct. Mol. Biol. 11, 121–127.
Research
Sheth, U., and Parker, R. (2003). Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 300, 805–808.
Zheng, D., Ezzeddine, N., Chen, C. Y., Zhu, W., He, X., and Shyu, A. B. (2008). Deadenylation is prerequisite for P-body formation and mRNA decay in mammalian cells. J. Cell Biol. 182, 89–101.
20.6 Other Degradation Pathways Target Specific mRNAs
Reviews
Filipowicz, W., Bhattacharyya, S. N., and Sonenberg, N. (2008). Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat.Rev. Genet. 9, 102–114.
Garneau, N. L., Wilusz, J., and Wilusz, C. J. (2007). The highways and byways of mRNA decay. Nat. Rev. Mol. Cell Biol. 8, 113–126.
Research
Choe, J., Cho, H., Lee, H. C., and Kim, Y. K. (2010). MicroRNA/Argonaute-2 regulates nonsense-mediated messenger RNA decay. EMBO Rep. 11, 380.
Guo, H., Ingolia, N. T., Weissman, J. S., and Bartel, D. P. (2010). Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466, 835.
Mullen, T. E., and Marzluff, W. F. (2008). Degradation of histone mRNA requires oligouridylation followed by decapping and simultaneous degradation of the mRNA both 5′ to 3′ and 3′ to 5′. Genes Dev. 22, 50–65.
20.7 mRNA-Specific Half-Lives Are Controlled by Sequences or Structures Within the mRNA
Reviews
Chen, C. Y. A., and Shyu, A. B. (1995). AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem. Sci. 20, 465–470.
Von Roretz, C., and Gallouzi, I. E. (2008). Decoding ARE-mediated decay: is microRNA part of the equation? J. Cell Biol. 181, 189–194.
20.8 Newly Synthesized RNAs Are Checked for Defects via a Nuclear Surveillance System
Reviews
Houseley, J., LaCava, J., and Tollervey, D. (2006). RNA-quality control by the exosome. Nat. Rev. Mol. Cell Biol. 7, 529–539.
Houseley, J., and Tollervey, D. (2008). The nuclear RNA surveillance machinery: the link between ncRNAs and genome structure in budding yeast? Biochem. Biophys. Acta 1779, 239–246.
Villa, T., Rougemaille, M., and Libri, D. (2008). Nuclear quality control of RNA polymerase II ribonucleoproteins in yeast: tilting the balance to shape the transcriptome. Biochem. Biophys. Acta 1779, 524–531.
Research
Arigo, J. T., Eyler, D. E., Carroll, K. L., and Corden, J. L. (2006). Termination of cryptic unstable transcripts is directed by yeast RNA-binding proteins Nrd1 and Nab3. Mol. Cell 24, 735–746.
Davis, C. A., and Ares, M. (2006). Accumulation of unstable promoter-associated transcripts upon loss of the nuclear exosome subunit Rrp6p in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 103, 3262–3267.
Kadaba, S., Wang, X., and Anserson, J. T. (2006). Nuclear RNA surveillance in Saccharomyces cerevisiae: Trf4p-dependent polyadenylation of nascent hypomethylated tRNA and an aberrant form of 5S RNA. RNA 12, 508–521.
20.9 Quality Control of mRNA Translation Is Performed by Cytoplasmic Surveillance Systems
Reviews
Isken, O., and Maquat, L. E. (2007). Quality control of eukaryotic mRNA: safeguarding cells from abnormal mRNA function. Genes Dev. 21, 1833–1856.
McGlincy, N. J., and Smith, C. W. J. (2008). Alternative splicing resulting in nonsense-mediated mRNA decay: what is the meaning of nonsense? Trends Biochem. Sci. 33, 385–393.
Shyu, A. B., Wilkinson, M. F., and van Hoof, A. (2008). Messenger RNA regulation: to translate or to degrade. EMBO J. 27, 471–481.
Stalder, L., and Mühlemann, O. (2008). The meaning of nonsense. Trends Cell Biol. 18(7), 315–321.
Research
Wilson, M. A., Meaux, S., and van Hoof, A. (2008). Diverse aberrancies target yeast mRNAs to cytoplasmic mRNA surveillance pathways. Biochem. Biophys. Acta 1779, 550–557.
20.10 Translationally Silenced mRNAs Are Sequestered in a Variety of RNA Granules
Reviews
Anderson, P., and Kedersha, N. (2009). RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat. Rev. Mol. Cell Biol. 10, 430–436.
Buchan, J. R. (2014). mRNP granules. RNA Biol. 11, 1019–1030.
Erickson, S. L., and Lykke-Anderson, J. (2011). Cytoplasmic mRNA granules at a glance. J. Cell Sci. 124, 293–297.
Thomas, M. G., Loschi, M., Desbats, M. A., and Boccaccio, G. L. (2011). RNA granules: the good, the bad and the ugly. Cell. Signal. 23, 324–334.
20.11 Some Eukaryotic mRNA Are Localized to Specific Regions of a Cell
Reviews
Bullock, S. L. (2007). Translocation of mRNAs by molecular motors: think complex? Semin. Cell Devel. Biol. 18, 194–201.
Buxbaum, A. R., Halmovich, G., and Singer, R. H. (2015). In the right place at the right time: visualizing and understanding mRNA localization. Nat. Rev. Mol. Cell Biol. 16, 95–109.
Du, T. G., Schmid, M., and Jansen, R. P. (2007). Why cells move messages: the biological functions of mRNA localization. Semin. Cell Dev. Biol. 18, 171–177.
Giorgi, C., and Moore, M. J. (2007). The nuclear nurture and cytoplasmic nature of localized mRNPs. Semin. Cell Devel. Biol. 18, 186–193.
Holt, C. E., and Bullock, S. L. (2009). Sub-cellular mRNA localization in animal cells and why it matters. Science 326, 1212–1216.
Martin, K. C., and Ephrussi, A. (2009). mRNA localization: gene expression in the spatial dimension. Cell 136, 719–730.
Research
Blower, M. D., Feric, E., Weis, K., and Heald, R. (2007). Genome-wide analysis demonstrates conserved localization of messenger RNAs to mitotic microtubules. J. Cell Biol. 179, 1365–1373.
Lecuyer, E., Yoshida, H., Parthasarathy, N., Alm, C., Babak, T., Cerovina, T., Hughes, T. R., Tomancak, P., and Krause, H. M. (2007). Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell 131, 174–187.