
HISTORY & CONCEPTS
HISTORY & CONCEPTS
GLOSSARY
alleles Alternative variant forms of a gene that result from a mutational change in DNA sequence or expression of the gene. Alleles can be recessive, meaning they only have an effect when there are two copies, or dominant, where a single copy is enough to have an effect.
amino acids Water-soluble organic compounds that are the building blocks to make proteins. There are around 20 amino acids. Ten cannot be made by the human body and so must be included in the diet: they are called essential amino acids.
chromosomes Long strings of double-stranded DNA that carry genes and the genetic information. In eukaryotic cells (those with a discrete nucleus) the chromosomes are in the nucleus and composed of DNA, some RNA and proteins. A prokaryotic cell (one without a discrete nucleus) has a single chromosome made of DNA and a very small amount of protein.
codon Genetic information is coded in triplets of DNA that encodes triplets of messenger RNA (mRNA) nucleotides. Each string of three mRNA nucleotides is called a codon and each codon codes for a different amino acid used to make the corresponding protein.
double helix The double-stranded structure of DNA. The two strands of DNA are wrapped around each other like a twisted cord.
DNA Deoxyribonucleic acid is a long molecule that carries the genetic information and transmits inherited traits. DNA is found in the cells of all prokaryotes and eukaryotes.
gametes Specialized cells for sexual reproduction. Male gametes are sperm cells and female gametes are egg cells.
genes Units of heredity, located on a chromosome. Genes consist of DNA, except in some viruses where they are made of RNA. Particular genes control specific cellular processes – for example, genes can control cell division and cell suicide.
genome Complete set of genetic material in an organism or a cell. Genomics is the study of an organism’s genome, focusing on its evolution, function and structure.
locus (pl. loci) Position of a gene on a chromosome. Different alleles of the same gene occupy the same locus.
mutation Change in DNA sequence or gene structure that can result from substitutions of bases of DNA or the rearrangement, deletion or addition of sections of genes or chromosomes.
nucleotides Building blocks used to make DNA or RNA. Strings of nucleotides are called nucleic acids. In DNA there are four nucleotides (referred to by the letters T, C, G and A) and in RNA there are four ribonucleotides (U, C, G and A). Nucleotides are also called bases. DNA bases can be paired: A pairs with T, and C pairs with G.
polymer Long molecule made up of simpler building blocks (monomers). DNA is a polymer chain made up of strings of nucleotides. Proteins are polymer chains made up of strings of amino acids. Proteins are sometimes called polypeptide chains.
replication Process of exactly copying DNA, usually for doubling the DNA before a cell divides to make two new cells. Replication involves an enzyme machine called DNA polymerase that copies each strand of the DNA to make a precise complementary copy of the DNA molecule.
RNA Ribonucleic acid, a molecule that is made in all living cells and is important for synthesizing proteins and for regulating genes. RNA is normally made by copying one strand of the DNA. Messenger RNA (mRNA) is a copy of DNA that contains the information to make a protein. In some viruses, RNA rather than DNA functions as the carrier of genetic information.
species Group of organisms whose members can interbreed and produce fertile offspring. ‘Species’ is the eighth category in the scientific classification system, beneath ‘genus’.
transcription Process for turning DNA genetic information into RNA. This is done by an enzyme machine called RNA polymerase, which builds an RNA polymer using the DNA as a template.
translation Process for making proteins using genetic information in messenger RNA. The ribosome is a large protein machine that moves along the mRNA and reads mRNA codons. The ribosome links amino acids to make a protein chain.
MENDEL’S LAWS OF HEREDITY
the 30-second theory
Gregor Mendel uncovered the laws of heredity while experimenting with garden peas. He bred plant lineages in isolation for many generations so that their offspring had various identical visible properties. Then he crossbred plants with different visible properties, for example plants with purple flowers with plants with white flowers. In the first generation, only plants with purple flowers were obtained. After crossbreeding these plants again he observed that one-quarter of the new plants had white flowers and three-quarters of them had purple flowers. To explain this, Mendel concluded that they resulted from the transmission of pairs of factors, which determined visible traits according to the laws of probability. The character purple flowers, which dominates in the first generation, is said to be dominant (P) compared to white flowers, which is recessive (p). In humans, blue eye colour is recessive and brown eye colour is dominant. Mendel’s factors are now known as alleles, which are variations in the DNA sequence that specifies traits. By extension, one can speak of dominant or recessive alleles. These alleles are alternative sequences of a locus (Latin for ‘place’), which in many cases can be loosely equated to a gene. More than two alternative alleles can exist in a population.
3-SECOND THRASH
The random encounter of an egg and a sperm cell carrying only one allele of each gene explains why the alleles split off independently. This is known as Mendel’s first law of inheritance.
3-MINUTE THOUGHT
When we consider the separation of alleles that specify different characteristics, things get more complicated. When the relevant genes are located on different chromosomes, or are distant enough on the same chromosome, they split off and lead to more complex proportions than one-quarter/three-quarters. This is Mendel’s second law. Both laws went unnoticed until they were rediscovered at the end of the nineteenth century.
RELATED TOPICS
See also
DNA CARRIES THE GENETIC INFORMATION
3-SECOND BIOGRAPHIES
GREGOR MENDEL
1822–84
Moravian-Silesian monk who discovered the laws of genetic inheritance
HUGO DE VRIES
1848–1935
Dutch botanist who rediscovered Mendel’s laws of heredity in the 1890s
30-SECOND TEXT
Reiner Veitia
Two recessive alleles (pp) result in a recessive trait, whereas two dominant alleles (PP) or a dominant and a recessive allele (Pp or pP) will result in the organism expressing a dominant trait.

DARWIN & THE ORIGIN OF SPECIES
the 30-second theory
Where do we come from? Why do we have limbs and eyes? Such questions were considered to be outside the realm of science until Charles Darwin published his magnum opus On the Origin of Species in 1859. Darwin’s view of life is now called the theory of evolution. Briefly, it states that some of the traits that differ between individuals within a population can be transmitted to the next generation. The individuals who are best adapted to the environment in which they live are most likely to survive, reproduce and pass on their heritable characters to future generations. In this way, populations change over time, adapting to their environment; this can eventually lead to new species. Darwin’s ideas conflict with the intuition that humans are distinct from other animals or with the belief that species remain invariant over time. His book ignited huge philosophical and religious debates, some of which are still ongoing. The discovery of genes, genetics and DNA in the 1920s–60s provided new support for Darwin’s theory. This led to today’s modern theory of evolution, which is central to our understanding of the living world.
3-SECOND THRASH
Darwin’s book is a masterpiece of observation and creative thinking; it profoundly changed how humanity conceives its origins.
3-MINUTE THOUGHT
Like other scientific explanations, the theory of evolution is challenged and refined by new facts. Although the core of the theory presented by Darwin is still valid today, certain parts have been refuted (for example, the diversification of species resembles a meshwork rather than a branching tree, as he suggested) and others (such as the origin of life) remain enigmatic.
RELATED TOPIC
See also
3-SECOND BIOGRAPHIES
ALFRED RUSSEL WALLACE
1823–1913
British naturalist who conceived the theory of evolution at the same time as Darwin
THEODOSIUS DOBZHANSKY
1900–75
Russian-American geneticist, famous for stating that ‘nothing in biology makes sense except in the light of evolution’
JERRY COYNE
1949–
American biologist who actively promotes the theory of evolution in books and blogs
30-SECOND TEXT
Virginie Courtier-Orgogozo
Darwin's theory of evolution by natural selection is one of the most revolutionary ideas in the history of science.

DNA CARRIES THE GENETIC INFORMATION
the 30-second theory
The history of the discovery of deoxyribonucleic acid (DNA) can be traced back to the work of Friedrich Miescher, who isolated a substance he termed ‘nuclein’ from the nuclei of white blood cells in the late 1880s. This substance is composed of proteins and what we now know as DNA. Its former name, coined by Richard Altmann, was the generic ‘nucleic acid’. Later on, Frederick Griffith showed that a substance derived from disease-causing (pathogenic) bacteria could change non-pathogenic bacteria into virulent forms. Griffith’s experiment was continued by Oswald Avery, Colin MacLeod and Maclyn McCarty. They destroyed everything but the DNA of pneumonia-producing bacteria. After this drastic treatment, DNA could still transform non-pathogenic into pathogenic bacteria. Only the destruction of DNA prevented this transformation and this demonstrated that it was the DNA that carried the genetic information. In the meantime, Phoebus Levene had identified the components of DNA: the bases adenine, cytosine, guanine, thymine, a sugar molecule and a phosphate group. All these discoveries paved the way for the unravelling of the chemical structure of DNA by Rosalind Franklin, Maurice Wilkins, James Watson and Francis Crick in the early 1950s.
3-SECOND THRASH
Experiments in the 1940s formally demonstrated that DNA is the molecule that carries the genetic information of most known organisms.
3-MINUTE THOUGHT
The history of the discovery of DNA and its structure is tainted by injustice. The results of Avery, MacLeod and McCarty were largely unrecognized and unaccepted. In another famous example, Watson and Crick built their famous double-helix model based on pictures of DNA's structure that were obtained by Rosalind Franklin and Maurice Wilkins. Franklin died at 37 and her vital contribution to the story has been downplayed until recently.
RELATED TOPICS
See also
3-SECOND BIOGRAPHIES
JOHANNES FRIEDRICH MIESCHER
1844–95
Swiss physician and biologist who first identified nuclein and nucleic acids
OSWALD AVERY, JR.
1877–1955
Canadian-born American physician who demonstrated that DNA is genetic material
PHOEBUS LEVENE
1863–1940
American biochemist who identified the components of DNA
30-SECOND TEXT
Reiner Veitia
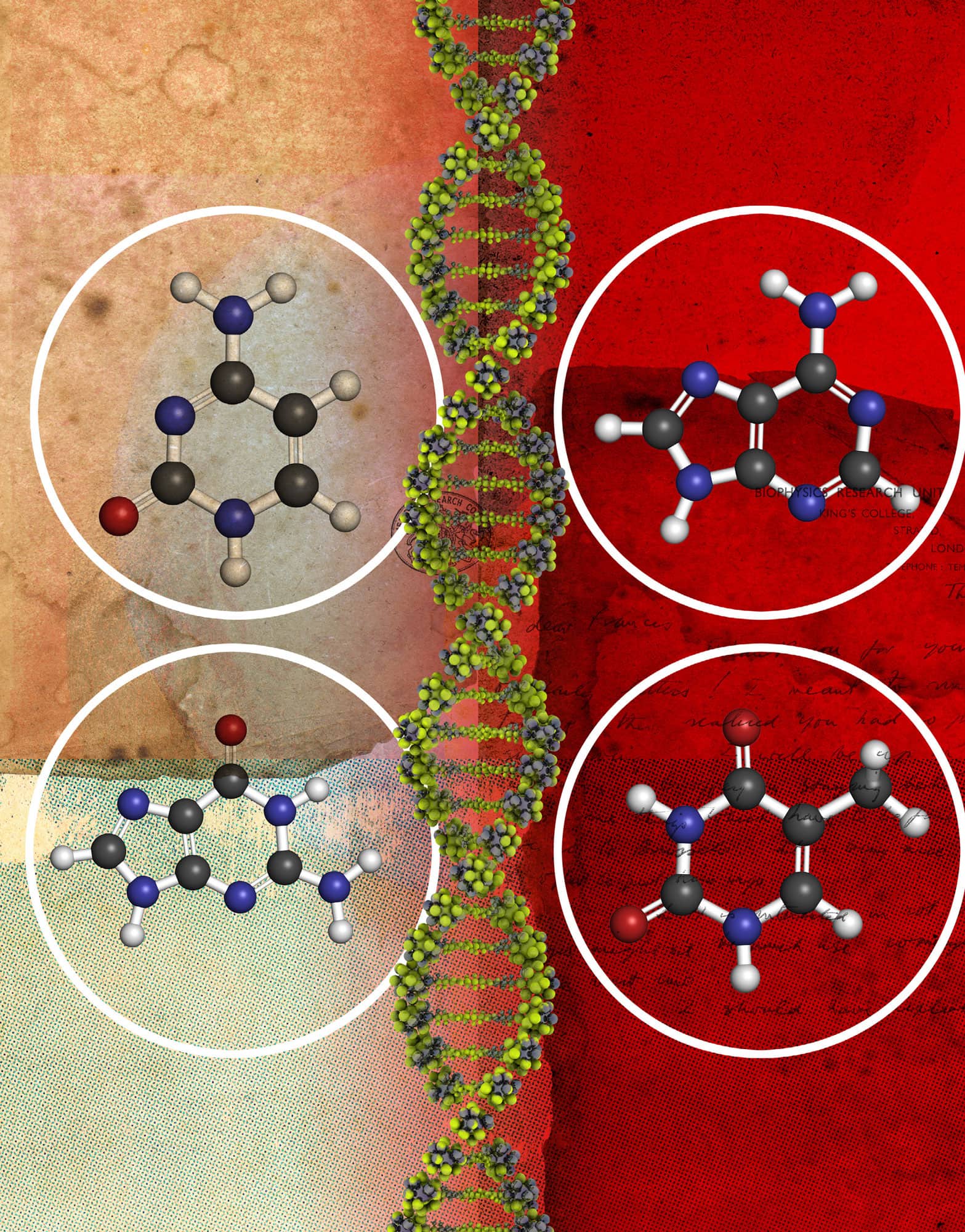
DNA's basic components include the four bases adenine, cytosine, guanine and thymine.
THE DOUBLE HELIX
the 30-second theory
The function of DNA depends on its structure. DNA is composed of building blocks called nucleotides, which consist of a deoxyribose sugar, a phosphate group and one of four bases: adenine (A), thymine (T), guanine (G) and cytosine (C). Nucleotides link together in long chains known as polymers, and each is specifically paired with a nucleotide on the opposite strand: A always bonds with T, and C bonds with G. In the early 1950s there was a race to work out how these base pairs fit together into a three-dimensional structure. Rosalind Franklin, working with Maurice Wilkins at King’s College London, beamed X-rays through crystals of the DNA molecule to gain insights into its structure. This X-ray diffraction technique produced an image that suggested DNA molecules form a helical shape. James Watson and Francis Crick, who were working at the Cavendish Laboratory in Cambridge, saw this image and realized that it provided a critical clue to the structure of DNA. They developed a chemical model for the DNA molecule, and in 1953 were the first to propose that its structure is that of a double helix. Further research into the structure revealed the mechanism for base pairing, and explained how genetic information can be stored and copied in living cells.
3-SECOND THRASH
The discovery of the molecular structure of DNA was a landmark moment in genetic and molecular biology research.
3-MINUTE THOUGHT
Francis Crick and James Watson first described the structure of a double-stranded DNA molecule as a ‘double helix’ in the journal nature in 1953. Two linear strands run in opposite directions and are connected into a twisted helical structure. The sequence of the bases in each strand makes a digital code that carries the instructions for life.
RELATED TOPICS
See also
DNA CARRIES THE GENETIC INFORMATION
3-SECOND BIOGRAPHIES
FRANCIS CRICK
1916–2004
British biophysicist who co-discovered DNA’s structure with James Watson
ROSALIND FRANKLIN
1920–58
English chemist who generated the crucial X-ray diffraction images of the DNA molecule
JAMES WATSON
1928–
American biologist and co-discoverer of DNA’s structure
30-SECOND TEXT
Matthew Weitzman
Watson and Crick won a Nobel Prize in 1962 for discovering DNA's double helix structure.

CRACKING THE GENETIC CODE
the 30-second theory
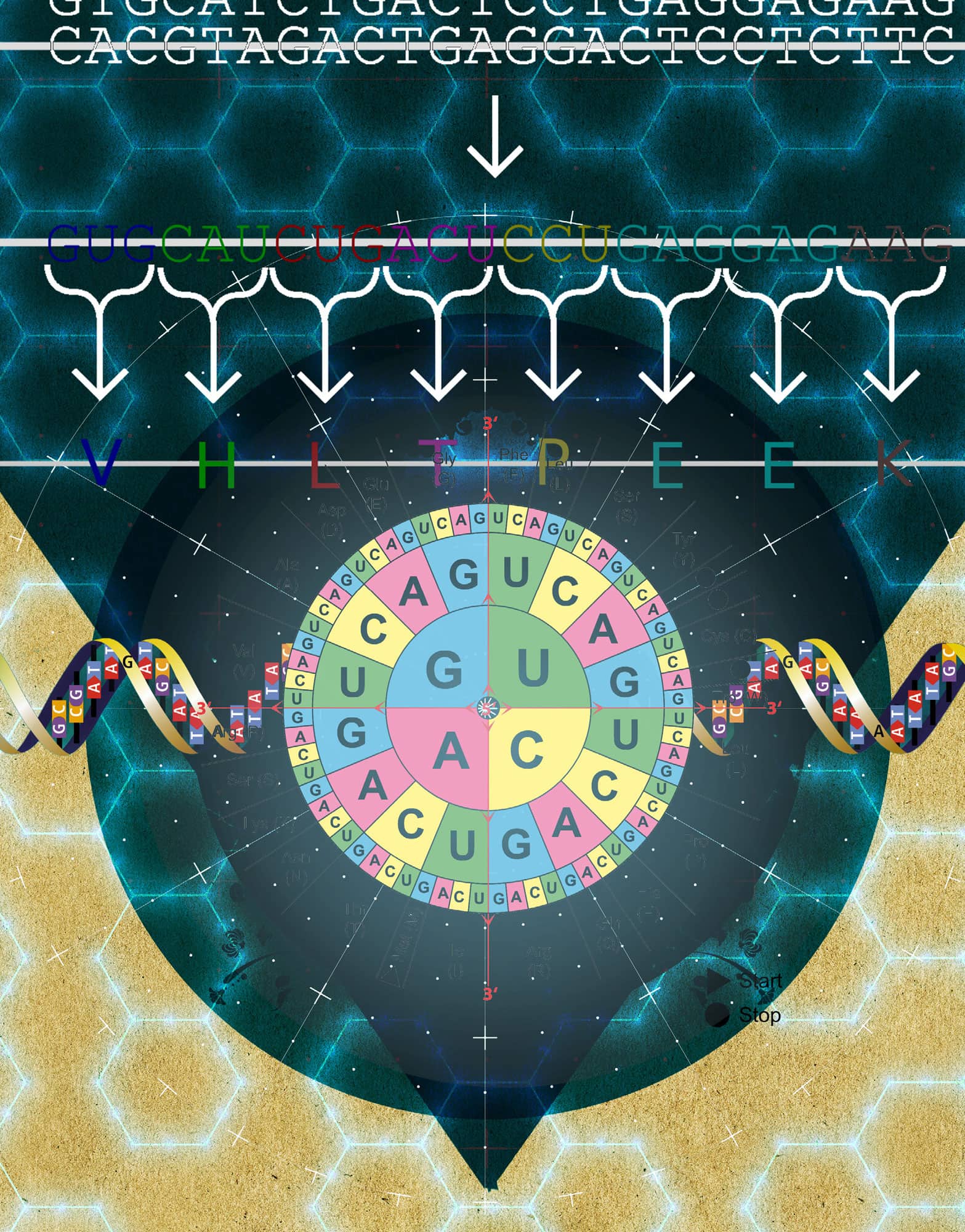
Working out the encryption rules to crack a code is the romantic job of spies and secret agents. But researchers also had to be pretty cunning to work out how information encoded in the DNA sequence is turned into the string of amino acids in proteins. The genetic code is the rulebook for translating DNA information into protein information. The code is extremely similar among all living organisms. As there are four ‘letters’ called nucleotides in DNA (A, G, C and T) and these need to code for 20 amino acids, it quickly became clear that the code had to involve at least three letters. That gave 64 possible combinations, but which one corresponded to which amino acid? In the 1960s researchers performed pioneering experiments to prove that a triplet code (called a codon) was responsible for each amino acid. The code-cracking break came when they used a cell-free system and put in RNA molecules with a long string of just one letter. Using a synthetic chain of nucleotides called ‘poly-uracil’ they deduced that UUU was the code for the amino acid phenylalanine. Then it was just a matter of working out the other combinations. Today, with the full table of 64 combinations, any student can predict a protein sequence from DNA code.
3-SECOND THRASH
DNA information in genes is organized into triplet codons, where three letters of the DNA encode for one amino acid in the resulting protein.
3-MINUTE THOUGHT
Because there are 64 possible combinations of three letters, there is an inherent redundancy. For example, there are four codons that correspond to the amino acid Alanine (GCU, GCG, GCA or GCG). This means that the third letter can change without affecting the code. This is called a silent mutation. Researchers use the word ‘degeneracy’ to refer to this redundancy in the genetic code.
RELATED TOPICS
See also
DNA CARRIES THE GENETIC INFORMATION
3-SECOND BIOGRAPHIES
GEORGE GAMOW
1904–68
Ukrainian-American physicist who hypothesized that the genetic code might be made up of three letters (nucleotides) in DNA
MARSHALL W. NIRENBERG
1927–2010
American biochemist who cracked the first codon to begin solving the puzzle of the genetic code
30-SECOND TEXT
Jonathan Weitzman
The three-letter DNA code contains the information for making proteins.
THE CENTRAL DOGMA
the 30-second theory
The ‘central dogma’ of molecular biology describes the transfer of information from DNA to RNA to protein. It was first articulated by Francis Crick to explain information transfer from one polymer molecule with a defined ‘alphabet’ to another. In this way the ordered sequence information is faithfully maintained. Replication is the process of copying DNA, in which information from the parent DNA strand is transferred to the daughter strand. Transcription is the process by which information from DNA is transferred into messenger RNA (mRNA). Translation is the process whereby the sequence of mRNA is used as a template to synthesize proteins. These newly synthesized polypeptide chains are then processed, folded and modified to form functional proteins. The central dogma states that there is a directional flow of sequence information, and that information cannot be transferred from protein to protein or from protein back to nucleic acid. Discoveries from viruses have shown that RNA can be replicated into RNA, and that RNA can be reverse-transcribed back to DNA, but there is no evidence for reversible transfer of information from proteins to DNA.
3-SECOND THRASH
The central dogma of molecular biology describes the flow of genetic information: DNA has the information to make RNA, which has the information to make protein.
3-MINUTE THOUGHT
The central dogma describes the faithful transfer of genetic information from nucleic acid to protein. DNA information can be copied into messenger RNA through the process of transcription. Proteins are synthesized using the information in mRNA as a template in the process termed translation. Today we know that there is a good deal of information in DNA and RNA that is not used to make proteins but instead regulates genome function.
RELATED TOPICS
See also
DNA CARRIES THE GENETIC INFORMATION
3-SECOND BIOGRAPHIES
FRANCIS CRICK
1916–2004
British biophysicist and co-discoverer, with James Watson, of the structure of DNA, who coined the term ‘central dogma’ to summarize the flow of genetic information from DNA to RNA to protein
HOWARD TEMIN
1934–94
American virologist who discovered reverse transcriptase, the enzyme that turns viral RNA into proviral DNA
30-SECOND TEXT
Matthew Weitzman
Genetic information is transcribed from DNA to RNA, and then from RNA to protein.

THE HUMAN GENOME PROJECT
the 30-second theory
The Human Genome Project is probably the biggest collaborative project ever undertaken by biologists. It is biology’s equivalent of the Apollo programme that took humans to the moon. The genome is all the DNA that contains all the genes in a cell. Laboratories from around the world joined forces to map and understand all the genes in humans. Following much debate in the 1980s, the US National Institute of Health (NIH) launched the Human Genome Project in 1990, expecting it to last at least 15 years. The project began by making a map of the 23 human chromosomes. This was followed by ordered sequencing of human DNA in research centres around the world. In 1996, the leaders of the project drafted the ‘Bermuda Principles’ to encourage the sharing of all genetic information. The rapid increase in the efficiency and speed of DNA sequencing technology accelerated the project. Sequencing the human genome became a race in 1998, when the private company Celera Genomics set out to sequence the genome at the same time. The first draft of the human genome sequence was published by both the public and the private projects in 2001. The complete sequence published in 2003 showed there are around 20,000 human genes encoded in a genome that contains three billion letters.
3-SECOND THRASH
The Human Genome Project sequenced all the DNA letters in humans and made the sequence freely available for all to study.
3-MINUTE THOUGHT
The generation of the first human genome took 13 years, involved thousands of researchers around the world and cost billions of dollars. DNA sequencing technology continues to increase in speed and accuracy and decrease in cost and time required. Today it takes just hours to sequence a human genome and costs less than $1,000.
RELATED TOPICS
See also
3-SECOND BIOGRAPHIES
JAMES WATSON
1928–
American molecular biologist and co-discoverer of the DNA helix, who was the first person to have his genome sequenced
J. CRAIG VENTER
1946–
American biotechnologist who founded Celera Genomics, the company that raced the Human Genome Project to the finish line
FRANCIS COLLINS
1950–
American geneticist who led the Human Genome Project at the NIH
30-SECOND TEXT
Jonathan Weitzman
The Human Genome Project is one of the largest biology projects of all time.
