CHAPTER II
Information Medicine in Clinical Practice
The application of new paradigm medicine to clinical practice is based on the realization that the ordering-structuring factor in the universe Bohm called “in-formation,” and traditional wisdom named chi or qi (or prana or etheric factor), is the key to maintaining the integrity and safeguarding the health of organisms. Clinical experience confirms the healing power of nature’s in-formation, and the importance of using it in addition to, or in concert with, artificial compensatory methods.
The information contextualized by cells and cellular systems is not limited to the classically recognized biophysiological aspects of the body. The information that directs choices in the organism is suprabiological: in the last count, it is the effect of the attractor that forms and structures processes in the universe. Consequently medicine must not limit itself to the classical biochemical approaches.
It is now recognized that the causes of disease may be more than bacterial and viral: there may be deficiencies and imbalances in the organism that are not evident on the biochemical level. Subliminal psychoemotional causes could also manifest as disease. And there may be personal lifestyle and environmental factors that impact negatively on the organism. These indicate inadequate in-formation by the attractor that structures systems in space and time.
In-formation guides formative processes throughout the organism, as well as between organisms. It acts on the epigenetic system, the system that guides the activation and suppression of particular genes—which in turn regulates the functioning of the whole organism. In acting on the epigenetic system, in the final count it is the in-formation received by the organism that determines the response of the organism to the signals that reach it in its environment. In the embryo it determines the process whereby stem cells differentiate into normal cells (kidney, lung, liver, brain, and still other cells). Cellular differentiation involves a complex series of precisely sequenced steps, at the end of which every cell in the developing organism shares the same genetic code. Following cell differentiation in the embryo, the two codes remain aligned and use the same system to receive and decode messages. This enables the diverse types of cells in the embryo to “understand” each other.
Molecules and other factors of the microenvironment carry information, and cells process that information, decoding and integrating it both in regard to its form and its content. The messages elicit responses, and the responses communicate information to all the cells of the body, whether they are proximal or distant.
Communication among cells takes place when the cells are integrated in a body. Through the exchange of messages, cells constitute an integral cognitive network. This network processes information in all species of organism, including the simplest unicellular species.
As already noted, the living organism is more than the sum of its parts. The integrality of the organism ensures that the various chemical and physical-chemical reactions are not expressions of separate events but result from the fine-tuning of the organism to its intrinsic and extrinsic (internal and external) environment. It allows molecules and cells to adopt behaviors corresponding to their location within the system.
The differentiation of DNA is indispensable for the reprogramming of dysfunctional or downright malignant tumor cells toward benign behavior. To understand why, we need to look at what is happening in the body following the absorption of synthetic molecules—structures that do not exist in nature.
Let us suppose that an unnatural and for the organism unfamiliar substance produces toxic effects. Although it comes into contact with this substance for the first time, the liver, which is the organ of detoxification, is able not only to recognize the form of the message received from the unfamiliar substance but to understand the nature of its content. If the substance is toxic, cells in the liver activate one of the systems of detoxification. These activate in turn biotransformative processes that render the substance harmless. This process can take place because the differentiation of the cells that produces a new organism is a process of “cellular cognition.”
Liver cells, for example, the same as other cells in the organism, have alternatives, and they make their choice among them on the basis of the signals received from the rest of the body. If these signals were not contextualized, liver cells would not process the information, and in the absence of that information they would not make the right choices; they may be killed by the toxins. Decontextualized cells die easily, while those within the organism’s network are relatively robust. It is the whole system that makes cells intelligent and capable of processing information of whole-system significance. This directs molecules and cells to adopt viability-enabling behaviors.
For the organism as an integral cognitive system, it is the context that matters. The chemical and physical-chemical reactions that occur in the body are more than the expression of mechanical events governed by blind cause-effect reactions. Cells communicate with each other cognitively and adopt behaviors commensurate with the needs of the whole system. The genetic and the epigenetic systems use the same codes to process the messages.
REPROGRAMMING CANCER STEM-LIKE CELLS
The whole organism, with all its subsystems and organs, is an integral cognitive network, and in the case of a disease such as cancer, information in that system is blocked: it does not govern the behavior of a group of malignant cells. The dialogue between the organism and that group of cells is arbitrarily uncontextualized—interrupted. The signification codes have changed; they are not the same as those in healthy, normal cells.
Cancer tumor cells organize their own behavior at the expense of the whole organism. Normally differentiated cells cooperate, and their behavior is in harmony with cells in the rest of the organism, whereas cancer cells, not being adequately in-formed, may develop malignancy and adopt counter-functional behaviors. However, this flaw can be corrected. If the organism is given access to the in-formation that maintains or contributes to the health of the organism, it can reprogram its malfunctioning cells.
It is known that during the phase of organogenesis (when the organs are forming) in the embryo, regulatory substances are present that correct alterations caused by carcinogens in the embryo’s stem cells. These substances act on the epigenetic code that regulates gene expression in the embryo. They are stem cell differentiation stage factors (SCDSFs), and they slow or stop the growth of a variety of tumors.
The behavior of tumor cells cannot be reduced to a problem of cells and genes. An explanation based solely on molecular mechanisms resulting from genetic activity and related growth factors does not clarify the nature of cancer. Cancer is a complex system where contact and communication with the flow of information among organs and systems of organs is interrupted, and the in-formation that structures and governs the living organism is inadequately accessed. As a result the health, and even the survival, of the organism is at risk.
The key discovery of information medicine is that substances produced in the dialogue between mother and embryo are effective instruments for inducing the differentiation of mutant and degenerative stem cells in the embryo. These substances are precisely tuned to the embryo’s developmental stage. When extracted and applied to stem cells in other organisms, including the human, they regulate the developmental program of stem cells in those organisms. Clinical evidence shows that cells obtained at precisely selected phases of organogenesis differentiate and reprogram cancer cells in the host organism, inducing malignant cells to mutate into normal healthy cells.
The new discovery has been tested both in vitro (in the laboratory) and in vivo (under natural conditions). The testing has confirmed the discovery’s effectiveness in cases of serious tumoral and degenerative diseases. Here we review some strands of the evidence, prefacing our review by a summary of the processes of embryonic differentiation.
Embryonic Differentiation Processes
Shortly after fertilization, generally in the middle-blastula-gastrula period, the processes of differentiation begin. There are basic features of the process:
- Every cell nucleus contains the complete genome produced in the fertilized egg. In molecular terms, the DNAs of all the differentiated cells are identical.
- Inactive genes in the differentiated cells are not destroyed or mutated; they retain the potential for being expressed.
- Only a small percentage of the genome is expressed in each differentiated cell, and the portion of the synthesized RNA is specific for each cell type.
Briefly, the differentiation, which leads totipotent embryonic stem cells to specialization, consists of the differential regulation of genes, restricting the genome that is expressed. The gene configurations of the cells, which arise after each stage of differentiation, differ from the progenitor cells for many expressed genes.
The differentiation of the cells is controlled by regulators: generally, these are factors that cooperate in a network that promotes and controls the differentiation of each type of cell. All cells communicate with each other through the network.
When a new organism is formed, regulation and control in each cell does not govern merely that particular cell but connects with the control system of all the cells in the organism. Systemic integration begins with embryonic development; it is what makes that development goal oriented. It takes place in all living species. Whenever a new organism is formed, its unique species code is the basis of its development.
During embryo development, cell multiplication and differentiation (or specialization) processes take place. These processes occur through a series of complex stages that take place at mind-boggling speed.
The Beginning of Cell Differentiation in Various Types of Stem Cells
The first stage in the development of the embryo is the segmentation phase. It is a multiplicative phase, in which all the cells are totipotent, meaning that every cell can develop into a complete new organism. This phase is followed by a stage in which the first process of cellular differentiation begins; that is, the development of the blastocyst (a structure made up of an internal mass of cells that develops into an embryo from a hollow cavity surrounded by a layer of cells; in mammals, this becomes the embryonic membrane and placenta).
In this stage, not all embryonic cells are totipotent: from the cells surrounding the blastocyst, those that develop the embryonic membranes begin to differentiate. The totipotent cells of the internal mass of the blastocyst also begin to differentiate into three pluripotent stem daughter cells, which represent three primary germ cell layers; that is, the ectoderm (which develops into the skin, including mammary tissue and the nervous system), the endoderm (which will develop into the tissue of the digestive system, including the digestive glands, liver, spleen, etc.), and the mesoderm (which will develop into bones, muscle, connective tissue, and blood vessels). The cell’s loss of totipotency and gain of specialized functions is the consequence of an asymmetric process of division (the mother cells on the one hand give rise to identical daughter cells and to other cells that began to differentiate into three pluripotent stem cells).
Subsequent divisions create different types of stem cells that, according to their degree of specialization, can be defined as: multipotent; b) oligopotent; c) progressively differentiating cells; and d) completely differentiated cells. It should be pointed out that these cells gradually acquire specialized functions and lose their ability to multiply. Indeed, completely differentiated cells cannot multiply any longer. There are cells in some tissues of the adult organisms that continue to multiply and then differentiate, such as bone marrow cells, cells that develop into blood cells, skin germ cells, and intestinal villus cells. Many cells in some tissues are in fact stem cells preserved in the adult organism.
To understand how differentiation takes place from totipotent stem cells, we have to follow the chain of events through which information is exchanged between the cells. In brief, the heart of the process is the transmission of information during the processes of replication and differentiation, making it possible for the genetic code, which is the same in all the cells of the organism, to carry out required functions in each of the specialized cells.
The Epigenetic Code
It has become clear that in addition to the genetic code, consisting of genes that codify proteins, there is also a tightly knit network of molecules and a substantial portion of DNA that perform regulatory functions. Together the network and the regulating DNA make up the “epigenetic code.” This code is a precise system of regulation of gene expression active during embryonic development, and thereby determining which genes remain active and which do not; which protein products are synthesized and which are not; which molecular communication mechanisms remain operational and which do not; and in what way codifying genes interact with their products.
This is the code that makes differentiation and specialization possible as the embryo develops. This is the code that enables a totipotent embryonic stem cell to become a liver cell, kidney cell, brain cell, lung cell, and so on. Just as the conductor of an orchestra decides how a piece is to be played, so the epigenetic code decides how the codifying DNA within each cell should be read. In this way, when the differentiation process has been completed, all the differentiated cells have the same DNA as their base, but the part of active codifying genes in each differently specialized cell is specifically different and represents only a fraction of the entire DNA that it is able to codify. With the exception of very few cases, the differences between specialized cells are epigenetic and not genetic. Today the study of epigenetics is changing the face of biology. The twenty-first century will be the century of epigenetics, shifting the spotlight until now directed on the genetic code.
We are now realizing the extent of these changes: in fact, the prospects for the future in the therapeutic field are likely to stem from this branch of research rather than from genetics, where results linked to genetic manipulation are ethically questionable.
Let us at this point examine the various stages of epigenetic regulation to which totipotent embryonic stem cells are subject until they become fully differentiated cells. As already mentioned, in a multicellular organism with specialized cells and tissue, each cell possesses all the genes of that organism. The difference between specialized cells is due to the specific activity of the genes that continue to function even after the specific and selective silencing to which many of them are subject during the differentiation of the embryonic cell. This process of differentiation is very precise and finely tuned. So during this process certain proteins must therefore be synthesized at the right moment and in the right cell: gene expression in this way is strictly controlled and the possibility of error is very low. Unlike DNA replication, which generally in each cell is regulated according to the “all or nothing” principle, gene expression is a highly selective process.
Chromatin Remodeling
One of the first places in which the regulatory process occurs is at chromatin level, which is the material contained in the cell nucleus and composed of dense DNA wrapped around a center made up of proteins called histones, which play an important role in compacting nucleic acid.
Just under two DNA folds, composed of 146 nucleotides, wrap around a center composed of eight histones to form a structure that resembles a pearl in a necklace and is called a “nucleosoma.” With the help of other histone proteins that link the centers of the various nucleosomes with the DNA interposed between them, the string of nucleosomes folds and becomes a chromatin fiber that is further compacted. The degree of chromatin density impacts accessibility to important factors for the transcription of the genes contained therein. The more compact the chromatin, the less accessible the transcription factors. One way to keep a gene from transcribing is through methylation whereby a group of CH3 is attached to some of cells’ bases. In this case the DNA sequence is not modified, but the chromatin is compacted, thereby determining the probability of transcription, which at this point is lower. Conversely, adding acetylic groups (C2H5) to certain amino-acids of histones usually leads to a looser chromatin structure, thereby increasing the likelihood of transcription. Removing the acetylic groups from the histone proteins, like methylating their amino acids, renders the chromatin denser and does not allow the DNA to be transcribed. The remodeling of the chromatin can involve an entire chromosome; for example, one of the X chromosomes determining gender. It is widely known that normal female mammals have two X chromosomes while normal male mammals have an X chromosome and a Y chromosome. Between males and females there is a sizable difference in terms of the “dosage” of genes linked to the X chromosome. In other words, each cell of a female has two copies of genes present in the X chromosome and can potentially produce twice the proteins codified by these genes compared to male cells. This does not occur because during cell differentiation one of the two maternal X chromosomes is methylated and its chromatin is condensed in such a way that the DNA sequences become inaccessible to the transcription molecular machinery.
Transcriptional and Post-Transcriptional Regulation
Transcription is the first step of gene expression, in which a particular segment of DNA is copied into RNA. Both DNA and RNA are nucleic acids, which use base pairs of nucleotides as a complementary language. The transcription machinery is highly complex and is made up of proteins called transcription factors, which are necessary to an enzyme called RNA polymerases to form a complex that determines the starting point of transcription. This complex binds to a DNA sequence called a promoter and is the region where the transcription of the codifying region of the gene is promoted. At the opposite end of this ladder is a region of the DNA called terminator, the point at which transcription is terminated. In this way there are precise start and stop signals built into the transcription of DNA into RNA.
At the transcriptional level there are many regulatory events that significantly change gene expression. Regulating regions have been recently discovered that are immediately grouped at the origin of the promoter. Various regulating proteins can link up to these regions and can activate transcription. Far from the promoter we find intensifying sequences (intensifiers). These sequences bind activating proteins and this greatly stimulates the transcription complex.
In DNA, there are also negative regulating regions, silencing sequences, which have an effect opposite to that of the intensifiers. Silencers arrest transcription and link the proteins called suppressors. In some cases, gene expression is regulated by the movement of a gene to a new position on the chromosome. As a result, the DNA is rearranged. This is important, among other things, for producing highly variable proteins, such as those that make up the repertoire of human antibodies.
Rearrangement is also important in determining certain forms of cancer in which inactive genes can move next to active promoters. Another type of regulation is known as “genetic amplification.” This occurs, for example, in the mature egg cells of amphibians and fishes. After fertilization, a trillion ribosomes are necessary for protein synthesis. Cells that differentiate into egg cells initially contain less than one thousand copies of genes that codify for the ribosomal RNA. It would take about fifty years to synthesize all this RNA. Egg cells have solved this problem by selectively increasing the group of genes for ribosomal RNA, thereby hugely increasing the quantity. In fact this group of genes rises from 0.2% to 68% of total codifying DNA. These million copies that transcribe very quickly synthesize in just a few days the trillion ribosomes needed for the next protein synthesis.
In terms of post-transcriptional regulation processes, there is an extremely important mechanism in evolved organisms, which is linked to a process known as “alternative RNA splicing.” In the DNA, codifying sequences, called exons, are next to noncodifying sequences, known as introns. Transcribed introns appear in the primary RNA transcriptions called pre-mRNA, which undergo a splicing process before arriving in the cytoplasm in large complexes of RNA and proteins called spliceosomes in which the introns of primary transcription of the RNA are eliminated and the exons are grouped together. This messenger RNA (mature mRNA) is translated into polypeptides (proteins). Splicing is often much more complex, as it can make many different cuts that are able to give rise to multiple messenger RNA and consequently to differing protein.
Another post-transcriptional regulatory event is the one linked to RNA editing, which takes place essentially either by inserting new nucleotides into the mRNA or by chemically modifying a nucleotide. In both cases the synthesized protein is modified. Even the survival time of messenger RNA in the cytoplasm undergoes post-transcriptional regulation. The less time mRNA spends in the cytoplasm, the fewer the codified proteins synthesized by mRNA.
Finally, regulation mechanisms that are important in terms of transcription and in terms of post-transcription, but also in terms of translation of the proteins, are those linked to the action of RNA regulators. In particular, the so-called micro RNAs (miRNAs) have great importance, because they recognize the sequence of target messenger RNAs and mate with them, thereby keeping them from being translated into proteins. In this case, we can speak of translational regulation. MiRNAs are involved in cell differentiation, and they have a significant impact on the regulation of gene expression and therefore on the development of an organism. Studies in which a part of the machinery of miRNA processing was destroyed have shown that an organism cannot survive without the regulator role that miRNA plays. Another bizarre trait of these small RNAs is their ability to move to other cells, even different types, and thus cause genetic silencing from a distance.
Translational and Post-Translational Regulation
Having addressed above how specific microRNA uses the translational regulation mechanism to block the translation of specific mRNA (which obviously prevents the corresponding proteins from being synthesized), we are now focusing on translational regulation events. These latter events are often connected to the concentration of specific proteins that need to be synthesized. When their concentration is low, the speed at which messenger RNA already present in the cytoplasm translates them increases. Inversely, when their concentration is high, translation slows down.
Lastly, proteins can be regulated after they have been synthesized. An important post-translational regulation event is tied to the control of survival time of proteins within the cell itself. It is well known how some proteins involved in cellular division, like cyclins, are hydrolyzed at just the right moment so that sequencing can develop correctly over time. In many instances a protein that is to be degraded is linked to a polypeptide chain composed of seventy-six amino acids called ubiquitin (so-called because it is ubiquitous). This complex in turn binds to another large complex made up of a dozen polypeptides called proteasomes, which make up a sort of molecular destruction chamber in which the degrading protein is destroyed. The fact that so many proteins are concentrated within the cell is not determined by the differing speeds of transcription of the genes, but rather by how fast proteasomes destroy the molecules.
In short, cellular differentiation consists of a differential, specific, and selective gene regulation process that essentially restricts the expressed genome. A cell’s gene expression after each stage of differentiation is different from the progenitor cell in regard to hundreds of expressed genes. This is possible, as we mentioned earlier, because a gene, which in the past was thought to be the control center independent and autonomous from protein synthesis, in actual fact is directly and indirectly controlled by a regulation network and by synthesized proteins. In the embryo, the intense and extended interaction between the nucleus and the cytoplasm and the cytoplasm and the microenvironment is a prime example of complexity. An embryo under development and differentiation is an excellent example of what complexity researchers have termed “a complex adaptive system.”
The above system is made up of a network of multiple cells that act in parallel and have differing levels of organization. They are constantly subjected to revision and control. In addition, this system has an implicit prediction written in its genetic code and in the epigenetic regulatory events. Lastly, the system is in continuous transition and changes constantly. In this way, the totipotent stem cells that are produced first give rise to pluripotent stem cells, then to multipotent stem cells, and then oligopotent cells, until a new organism is formed. At this point, it is clear that stem cell differentiation stage factors (SCDSFs) present at the different stages of cell differentiation represent the epigenetic code, the system that can transmit whole-system information—that is, “in-formation” to the organism. This has remarkable healing properties.
Studies on the functioning of the epigenetic code revealed that this code is present in its totality and with all its various functions the moment life appears in the embryo.
This code—that is the “in-formation” of the embryo—can be studied and understood in its global functions, even if divided into the various stages of differentiation. When all of the code, that is, the entire “in-formation” that regulates the genes of all the cells that make up the organism, is before us, we have what has been called the Code of Life.
The possibility of studying the epigenetic code in its entirety exists only in the embryo, and only in the period of organogenesis, when starting from a single totipotent stem cell, through various stages that form all the types of stem cells: pluripotent, multipotent, oligopotent, definitively differentiating cells, and finally complete differentiated cells.
When organogenesis is over, it is no longer possible to study the entire epigenetic code in its various functions. This is because, when organogenesis is completed, the epigenetic code subdivides in the various organs and tissues, and in every organ serves to control and regulate the gene expression of the cells. At that point, it is no longer possible to study all the various functions of the Code of Life.
However, research prior to the completion of organogenesis permits studying the entire Code of Life. This research allowed us to discover how to prolong the life span and to stop the senescence of cells, how to regenerate the tissues and reinforce the cellular and organic vitality of the organism, how to slow the growth of imperfectly differentiated cells, and how to induce altered stem cells to differentiate into normal cells—or undergo programmed cell death.
REVIEW OF THE EXPERIMENTAL EVIDENCE
Here we record various experiments with stem cell differentiation stage factors (SCDSFs). These factors are accessible at different stages of the differentiation of the cells of the embryo of the Zebrafish. The choice of the tiny Zebrafish may appear arbitrary, but it has sound reasons: notwithstanding their size and relative simplicity, Zebrafish have very largely the same proteins as humans, and access to their genetic material is simple and safe. In addition, it is easy to discover the exact time of the fertilization of Zebrafish eggs. This is important for standardizing the experiments.
Results of the Experiment In Vitro
Six different human tumor lines (including a malignant brain tumor, primary liver tumor, colon and breast cancer, kidney tumor, and acute lymphoblastic leukemia) have been treated with factors obtained from Zebrafish embryos in four different developmental stages: a) morula stage, characterized by cell-multiplicatory events only, and therefore constituted of totipotent embryonic stem cells; b) medium-blastula-gastrula stage, where the totipotent embryo stem cells differentiate into pluripotent stem cells; c) the five somite stage; and d) the twenty somite stage, where important differentiation events of the intermediate and final embryo differentiation process take place.
All cell lines have shown a significant slowdown in growth when treated with factors drawn during the above-mentioned cell-differentiation stages, with inhibition percentages ranging from 73 percent of the malignant brain tumor to 26 percent of the melanoma. No effects have been registered with factors extracted in the morula stage, except for a weak tumoral growth.1
The above data confirms the hypothesis that during the stages of differentiation, factors are present in the embryo that address tumoral cells and reorient them toward the normal path. These factors appear in the very first phases of differentiation, and they are absent in stages of mere multiplication.
Several studies were carried out in order to understand which molecular events are involved in the tumor-growth inhibition mechanism. It appears that molecules that have a fundamental role in the cellular cycle regulation process, such as p53 and pRb, are involved through transcriptional and post-translational regulation events. More precisely, a p53 transcriptional regulation (meaning the activation of the tumor-suppressing gene with the synthesis of p53 protein) was obtained, highlighted by a considerable increase of the p53 protein’s concentration in the cells of some tumor lines, such as melanoma and brain tumor. This has been measured through the quantitative as well as the immune-histochemical method, after treatment with the cell-differentiation factors.2
The slowdown of tumor growth on other tumor lines, such as kidney cancer, is due to the post-translational regulation of a protein (pRb), which is named restriction factor of cell cycle—this indicates the regulation of the already translated protein of the retinoblastoma—and leads to a change in the relation between the protein’s phosphorylated and non-phosphorylated shape.3 It is well known that the non-phosphorylated shape stops the cellular cycle, preventing the transcription of the E2F-1 gene. The latter is facilitated when the protein is phosphorylated.
Finally, programmed cell-death events (apoptosis) as well as cell-differentiation events were studied, in order to understand the consequences of the tumor-cell regulation cycle by the differentiation factors. The analysis was carried out on colon adenocarcinoma cells, demonstrating the activation of an apoptotic pathway as well as a cell-differentiation pathway. Within a culture of colon tumor cells, there was a significant increase in apoptosis, as well as in the concentration of cell-differentiation markers.4 Therefore, the molecular mechanisms at the basis of the slow-down of tumor growth due to treatment with SCDSFs can be summed up as follows: cessation of the cell cycle in accordance with the type of tumor, genetic damage repair, and cell re-differentiation—or the apoptosis (death) of the tumor cells (if repair is no longer possible).
Clinical Results
Results from Clinical Trials on Intermediate-Advanced Hepatocellular Carcinoma (HCC)
A randomized controlled trial was conducted from January 1, 2001, to April 31, 2004, on 179 patients affected by HCC in the intermediate-advanced stage, for which no further treatment was possible, such as liver resection or transplantation, chemo-embolization, ablation with radiofrequency, or other type of treatments. A product was administered fine-tuned on the basis of the above studies. The dosage was thirty sublingual drops three times a day. (The sublingual solution was chosen because the active fraction is composed of low molecular weight proteins and nucleic acids.)
Objective tumor response and median overall survival and performance status have been evaluated. Results show that 19.8 percent of the patients experienced a regression and 16 percent of the patients experienced stabilization with an overall survival after forty months of more than 60 percent of the patients who responded, compared with 10 percent of the nonresponding patients.
A wide improvement of performance status has been registered in a great majority of patients (82.6 percent), including in those who are in the progressive phase of the disease.5 A recent study of reprogramming normal and cancerous stem cells confirms the role of SCDSFs in producing a complete response in 13.1 percent patients experiencing primitive, intermediate, or advanced liver cancer.6
Results of Experiments on Neurodegenerative Diseases
It has been shown that the completeness of the information and the redundancy of factors of the epigenetic code are able to significantly prevent the degeneration of the nervous cells; for example, the cells of the hippocampus. This happens because initially the redundancy of all the factors present from the beginning of the differentiation process first expands the number of stem cells and then differentiates them in the specific tissue. In other words, what we have understood by conducting the experiments on the hippocampal cells, the cells that first undergo neurodegeneration in Alzheimer’s disease, is that if we want to prevent the degeneration of these cells, we must exactly simulate the program that life adopts to self-organize and realize itself.
First we must, as already mentioned above, administer the substances that keep the stem genes turned on, allowing the survival of the few stem cells left active in the brain of an Alzheimer patient and therefore keeping their number intact, but then we must administer the substances that are able to differentiate stem cells in specific nervous cells. To obtain this result, we must exactly simulate the process that originates life and administer all the factors that are able to make the whole process of the cells perform.
With various collaborators we are now studying how the different specific molecules that make up the epigenetic code are able to repair the tissues and therefore can be used in regenerative medicine, in particular in pathologies where stem cell transplantation is required. These epigenetic regulators can in fact enhance the positive effects related to stem cell transplantation and in the future replace the same transplant, considering that it has been shown that the beneficial effects due to the stem cell transplantation are not due to the transplanted cells, but to the factors that they produce. And the factors that they produce are the same factors which constitute the epigenetic code.7
Results of Clinical Studies on Psoriasis
Two clinical trials8, 9 were conducted to evaluate the efficacy of a topical formulation of Zebrafish embryo extracts with Boswellia serrata, 18-beta glycyrrhetinic acid, Zanthoxylum alatum, 7-deidrocolesterolo, and vitamin E in cases of psoriasis. Results show a very high clinical objective improvement, with a reduction of keratosis and itch after 20 to 30 days from the beginning of the treatment.
Results of Anti-Aging Studies
Studies by Biava et al. on antiaging processes demonstrate that the very early stage of cell differentiation is able to activate the same genes that Shinya Yamanaka introduced with a retrovirus in a completely differentiated cell, reprogramming it into so-called induced pluripotent stem cells, an accomplishment for which he received the 2012 Nobel Prize in Physiology and Medicine. Biava and collaborators are able to activate the same genes without physical manipulation, on the basis of epigenetic “in-formational” regulation. In this case, the cells maintain their cyclicity, whereas in the case of the physical manipulation of the DNA, as in the experiments of Yamanaka, the cells lose their cyclicity. This is important, because in in-formational regulation the cells maintain their physiological characteristics, whereas those characteristics are lost in DNA manipulation.
The above experiments demonstrate that one can maintain not only the functions of the genes that halt the sectioning of telomeres, thereby increasing the cell’s life span, but also activate other genes that stop the senescence of the cells. In this way, the cells increase not only their life span, but maintain themselves in a youthful stage (see the paper included in part two: The Research: “Stem Cell Differentiation Stage Factors from Zebrafish Embryo: A Novel Strategy to Modulate the Fate of Normal and Pathological Human (Stem) Cells).”
CONCLUSIONS BASED ON THE EXPERIMENTS
The use of stem cell differentiation stage factors in anticancer therapy enabled the development of a new model of cancer.10 In that model, cancer cells are undifferentiated cells, in which mutation or epigenetic alterations are present that block the cells in a multiplication phase between two stages of differentiation.
Therefore cancer cells can be defined as “altered stem cells,” in which mutations of DNA or epigenetic alterations are present. These cells, according to their degree of malignancy, are blocked at a particular phase of their development. In support of this model we should note that in tumors with an elevated degree of malignancy, such as acute lymphoblastic and myeloid leukemia, multipotent altered stem cells are present, whereas in tumors with lower malignancy, such as chronic lymphocytic leukemia, incompletely differentiated cells are present, which can then progress toward further differentiation.
This model is supported by the finding that cancer cells and stem cells have common characteristics. The common elements are the presence of oncofetal antigens, maintained during phylogenesis,11 and the specific receptor on the cellular membrane on which the differentiation factors appear to act. As already mentioned, such factors activate metabolic pathways of cellular differentiation, and these pathways lead the cells to differentiate or to die (enter apoptosis). The latter is the usual occurrence in the embryo, where there are many apoptotic pathways.
The problem of cancer stem-like cells is twofold: not only do they present the genetic mutations that (so far as is known) are at the origin of malignancy, but they also manifest important modifications of the epigenetic code. The gene configuration and the metabolism of cancer cells are similar to those in stem cells: both have active proto-oncogenes, both produce embryonic growth factors, and in addition tumor cells present oncofetal antigens expressed by normal stem cells. Both work with an anaerobic metabolism. The difference between them is that tumoral cells, in contrast with normal stem cells, are not able to complete their development and to differentiate, having lost the necessary in-formation in the alteration of their epigenetic code. The correction of this condition calls for the use of the differentiation factors that transform cancer cells into normal cells.
DNA is responsible, via RNA, for the translation of proteins, so that the transcription factors, the microRNAs and the translational and post-translational factors play a fundamental role in the regulation of the genetic code, and thus in the regulation of the life cycle of cells. The epigenetic code can differentiate and regulate normal stem cells as well as cancer stem-like cells, deactivating genes that govern cancer stem-like cells and activating the pathways of differentiation that lead to normal cells.
The results reported here have been confirmed by research at the Children’s Hospital of Chicago. In particular, research confirmed that malignant melanoma reverts to a normal phenotype when it is in the environment of a Zebrafish embryo. At the same time, a growing body of studies highlighted that tumor malignancy is linked to the presence of tumoral stem cells,12 and these appear resistant to conventional therapy, such as chemo-and radiotherapy. In the last few years, scientific studies in this area have been so numerous that it is nearly impossible to take all of them into account. Here we mention only the research that demonstrates the presence of tumoral stem cells in breast cancer,13 lung cancer,14, 15, 16, 17 prostate cancer,18, 19 ovary cancer,20, 21, 22, 23 liver cancer,24, 25, 26, 27, 28, 29 stomach cancer,30, 31, 32, 33, 34 colon cancer,35, 36, 37, 38 pancreas cancer,39, 40, 41 head and neck cancer,42, 43, 44, 45 glioblastoma.46, 47, 48 It is known that malignancy in many hematological tumoral diseases is due to the presence of stem cells.
The results of antiaging treatments and the prevention of neurodegenerative disorders point to the same processes: the differentiation factors are epigenetic regulators with different specific functions. They are like an orchestra that can interpret different symphonies. The use of epigenetic regulation factors can be widely applied in the prevention and treatment of degenerative diseases, not only of the nervous system but also of the cardiovascular-osteoarticular system, diabetes, and others. They are also antiaging factors, improving the overall state of health of the elderly.
Noteworthy is that for the in-formation carried by the regulating factors to be effective, it must be transferred not in individual bits, but in precise instruction packages. In fact, every cell-differentiation process occurs only if multiple genes (rather than a single gene) are simultaneously addressed and regulated. Life is organized on the basis of in-formation programs that provide integral and precise instruction packages: these packages act as integral wholes. If they are fragmented, they lose their effectiveness. Experiments with cellular differentiation factors confirm that the organism is a unitary mind-body system.
Information is crucial to the processes of this integral body-mind system; its regulation is in the form of factors of information. This information is the “in-formation” that “forms” evolutionary processes in the universe. It is the manifest effect of a universal attractor that codes complex and coherent systems for integrality and health. For a human organism maintaining and, if needed, restoring health calls for effective contact with this attractor, ensuring the effective reception of the in-formation that orients living systems toward health and normalcy. Stem cell differentiation stage factors (SCDSFs) are factors of information that ensure such contact for systems suffering from diseases due to imperfect forms of cellular differentiation.
ADDENDUM: DECLARATION OF A COMMITTEE OF ONCOLOGISTS
Stem Cell Differentiation Factors as Integrative Treatments in Oncology: Their Role in Increasing the Efficacy of Chemotherapy and in Reducing its Adverse Effects
Edited by Michele Carruba, Chairman of the Scientific Committee, Director of the Department of Pharmacology of the University of Milan
Parallel to the sequencing of the genetic code, another area of research, that of epigenetics, emerged. There are therefore two types of code: the genetic and the epigenetic code that is above the genetic code and regulates its functioning. In the field of epigenetic approaches, and in particular in the field of cancer, the research on the reprogramming of tumor stem cells with embryo differentiation factors has been consolidating for several years. In fact, it has been shown that the differentiation factors of stem cells extracted from a Zebrafish egg, which has over 90 percent of common proteins with human ones, are able to normalize the cell cycle of cancerous cells.
These are the same mechanisms that in nature are active during the phases of organogenesis, when all the stem cell differentiation processes that lead to the formation of tissues and organs take place. In these phases, where there is a high risk of developing errors in replication, differentiation factors play an important role in correcting those cells that are making mistakes. Reconfirmation of this mechanism occurred when cancer cells were implanted into an embryo during the organogenesis phase: differentiated and apoptotic processes were observed on the implanted tumor cells. On the contrary, when tumor cells were implanted into an embryo after the organogenesis phase, they continued to proliferate. One can therefore talk of epigenetic reprogramming of diseased cells through the integration of those peptides that are able to restore the cell within its normal physiology.
These researches, begun in the late 1980s by Biava et al.49 were then developed by the Children’s Hospital in Chicago, Northwestern University, La Sapienza University of Rome, and other institutions. It emerges that differentiation factors are able, in association with standard chemotherapy treatments, to slow and often block the cell cycle of tumor cells, either by activating in a transcriptional way the p53 oncorepressor gene, or by regulating in a post-translational way the protein of retinoblastoma (pRb), which also has a block cell cycle activity. At the same time, a series of regulating gene cascades are also activated by the same differentiation factors as they attempt to repair cellular damage at the origin of malignancy: if the alterations are not too serious, they are effectively repaired; if mutations are too serious and are not repaired, the genes of programmed cell death (apoptosis) are activated and the cancerous cells die.
In fact, after treatment with stem cell differentiation factors, tumor cells leave the cellular multiplication cycle. A number of important researches on the synergy between chemotherapeutic agents and differentiation factors have already been carried out in 2011. For example, research conducted at Current Pharmaceutical Biotechnology at La Sapienza University in Rome obtained in vitro the slowdown of growth of CaCo2 colon cancer cells: a 35 percent slowdown was obtained by treatment with Fluorouracil alone, while a 98 percent slowdown was obtained with the simultaneous administration of 5-Fluorouracil and differentiation factors. Richard Ablin (PSA discoverer) reported in an article published in 2014 a greatly beneficial effect on the synergy between differentiation factors and surgical ablation or other traditional prostate cancer treatments. Stem cell differentiation factors also demonstrated improved performance and quality of life in 82 percent of patients treated; it also reduced adverse side effects of chemotherapy.
The figures on pages 42–49 are the main images related to the different studies carried out on embryo differentiation factors. The state of the art of research includes both in vitro work on molecular dynamics involving stem differentiation factors and in vivo work on mice and on humans. It remains necessary to implement in vivo research, especially the clinical kind. Specifically, studies have been conducted on the following aspects:
- Downloading and/or blocking of the cell cycle
- Activating in a transcriptional way the oncosupressor gene p53
- Regulating in a post-translational way the retinoblastoma protein (pRb)
- Slowing the growth of tumor cell lines
- Animal studies
- Protein analysis of Zebrafish egg extract
- Clinical study of 200 patients to evaluate possible side effects
- Randomized clinical study in 179 patients with intermediate or advanced hepatocarcinoma
Activation of Oncosuppressor p53
It has been shown that in the embryonic microenvironment there are factors that regulate the expression of the p53 oncorepressor, activating it, and post-translationally pRb. In fact, with the differentiation of stem cell differentiating factors of different tumor lines, a block of cell cycle was observed in G1-S phase.
Through cytofluorometry and immunohistochemistry, a significant increase in the concentration of p53 protein has been demonstrated in specific lines of cellular tumor lines such as multiform glioblastoma, melanoma, and hepatocarcinoma treated with stem differentiation factors. This increase is a consequence of the transcriptional regulation of the p53 oncosupressor gene. The two images on page 42 show the concentration of the p53 protein before and after the treatment.50
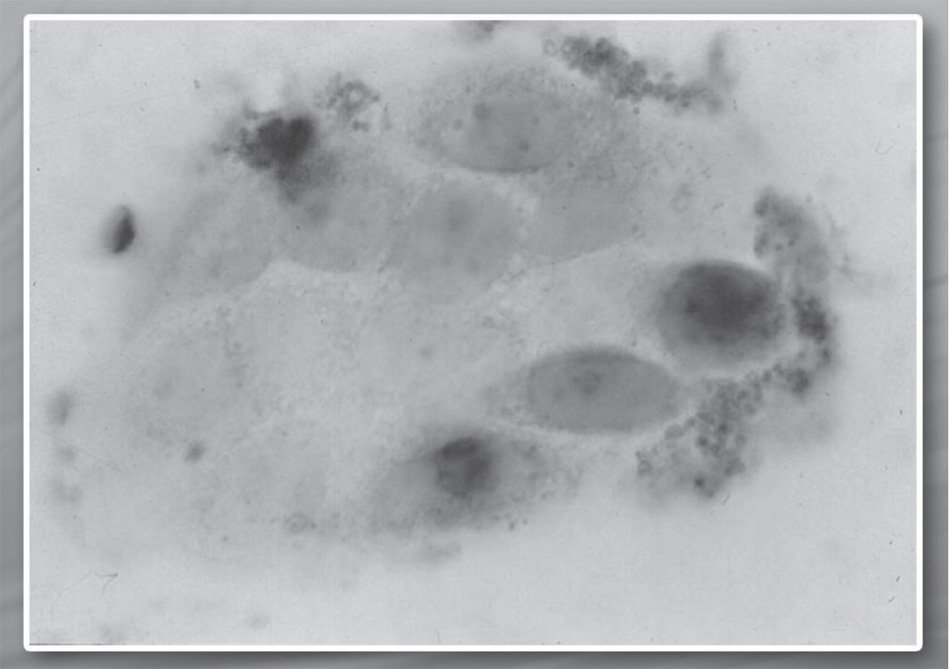
Fig. 2.1. Concentration of p53 protein (dark colored) in glioblastoma cells before the treatment with SCDSFs
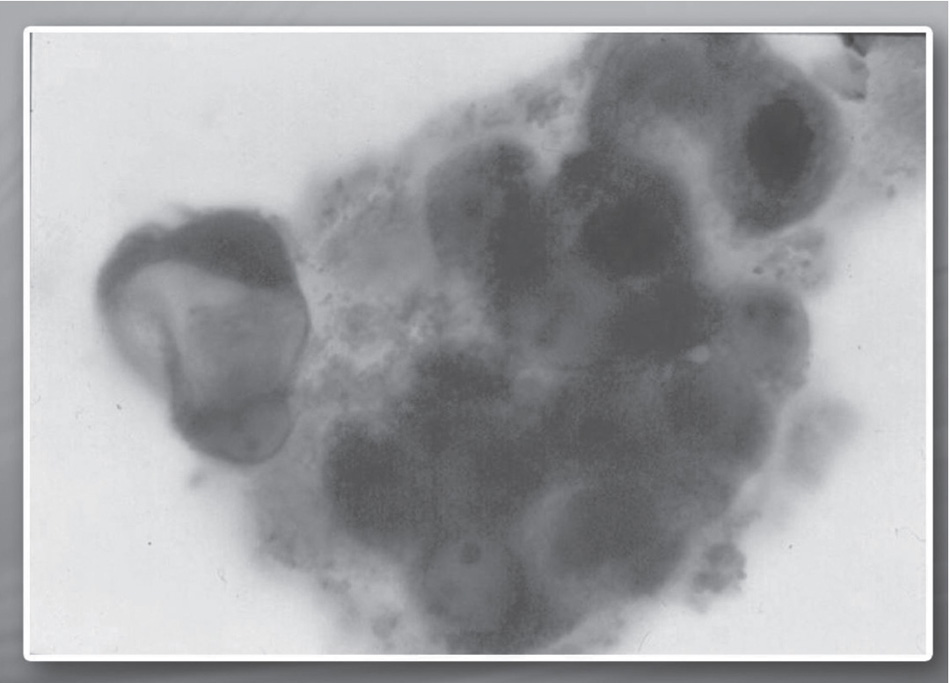
Fig. 2.2. Concentration of p53 protein (dark colored) in glioblastoma cells after the treatment with SCDSFs
Post-Translational Regulation of the Retinoblastoma Protein
By treating cancer cell lines such as those of kidney adenocarcinoma, a post-translational regulation of retinoblastoma protein (pRb) was observed, known for its cell cycle block function. This adjustment involves modification of the relationship between the phosphorylated form and the nonphosphorylated form of the pRb protein in favor of the nonphosphorylated form. This form blocks the cell cycle, stopping the transcription of the E2F-1 gene.51
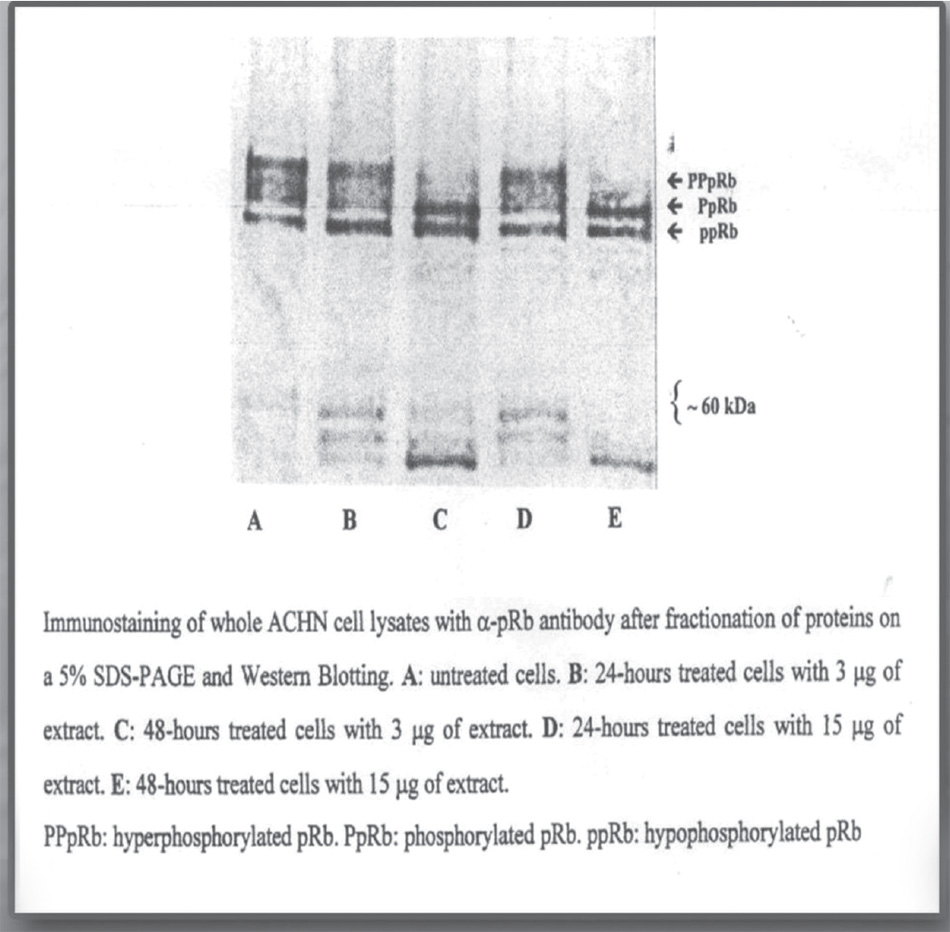
Fig. 2.3. Post-translational modification of the retinoblastoma protein (pRb) induced by in vitro administration of Zebrafish embryonic extracts on human kidney adenocarcinoma cell line
Slow-down of the Growth of Tumor Cell Lines
These blocking mechanisms of the cell cycle have been observed in several tumor cell lines. Specifically, tumor cell lines were investigated of: glioblastoma, melanoma, breast cancer, lymphoblastic leukemia, and kidney adenocarcinoma. The following figures illustrate the cell proliferation curves of different human tumor lines after in vitro treatment with Zebrafish embryonic extracts.52
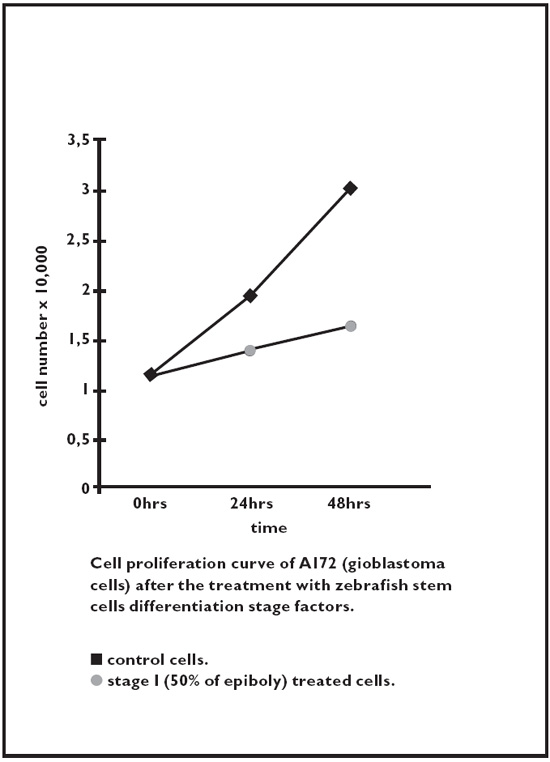
Fig. 2.4. Glioblastoma
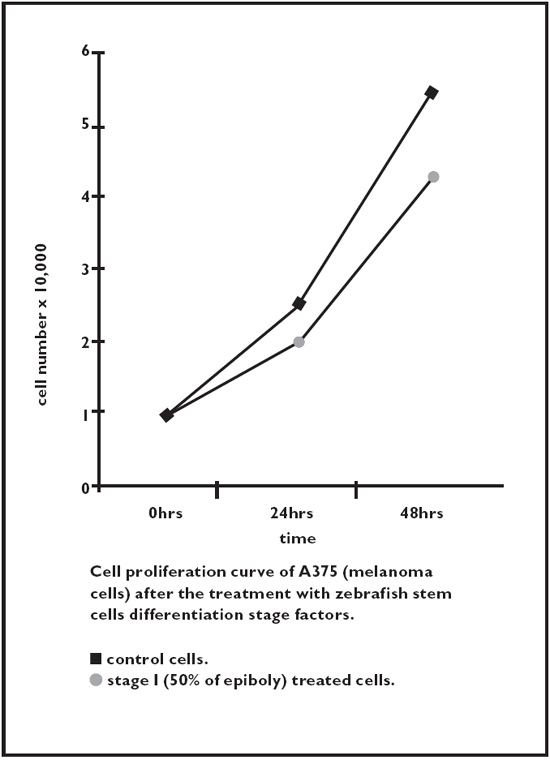
Fig. 2.5. Melanoma
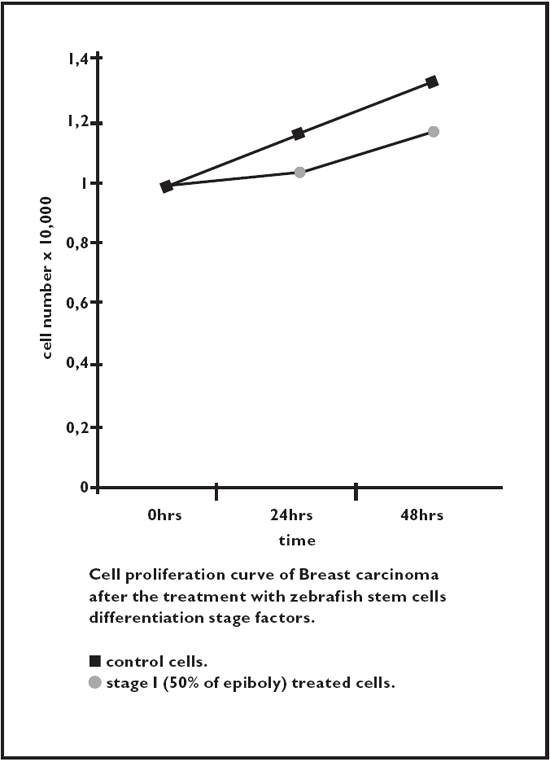
Fig. 2.6. Breast cancer
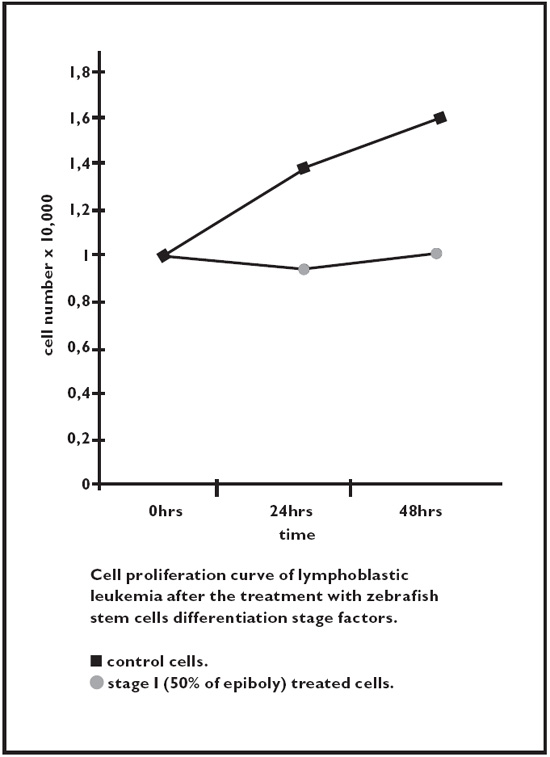
Fig. 2.7. Acute lymphoblastic leukemia
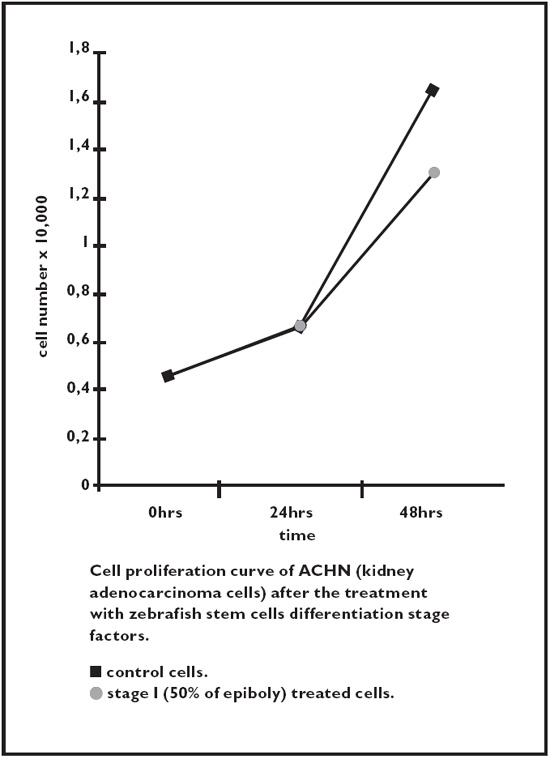
Fig. 2.8. Kidney adenocarcinoma
Animal Studies
The effects of stem differentiation factors on inhibition of tumor growth were in vivo tested on females of singular C57BL / 6 mice from the weight of 18 to 20 grams to which a subcutaneous Lewis primary carcinoma injection was performed. Therefore, both the size of the primary tumor and the survival time of the mice have been evaluated. In terms of development of the primary tumor, an extremely significant difference (P<0.001) was observed between treated and control mice (fig. 2.9) and thus also with regard to the survival ratio, always in favor of the treated mouse.53
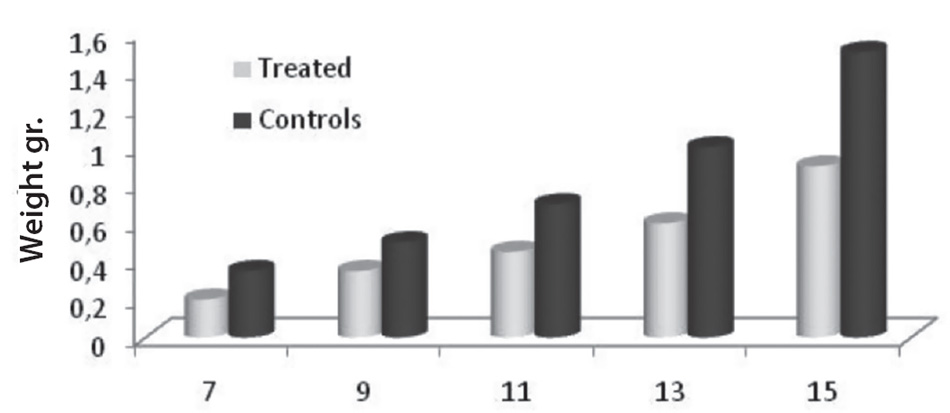
Days after tumor’s implantation
Fig. 2.9. The effects of stem differentiation factors on inhibition of tumor growth
Protein Analysis of Zebrafish Egg Extract
A protein analysis of embryonic extract from Zebrafish eggs was performed. A suspension of the glycerol-alcohol solution was analyzed with monodimensional gel electrophoresis (SDS-PAGE). As shown in figure 2.10, in all five phases extracted, they can be distinguished by their molecular weight by three major groups: over 45 kDa, about 25 to 35 kDa and below 20 kDa. In any case, the relative amount of protein is different in the five-stage samples.
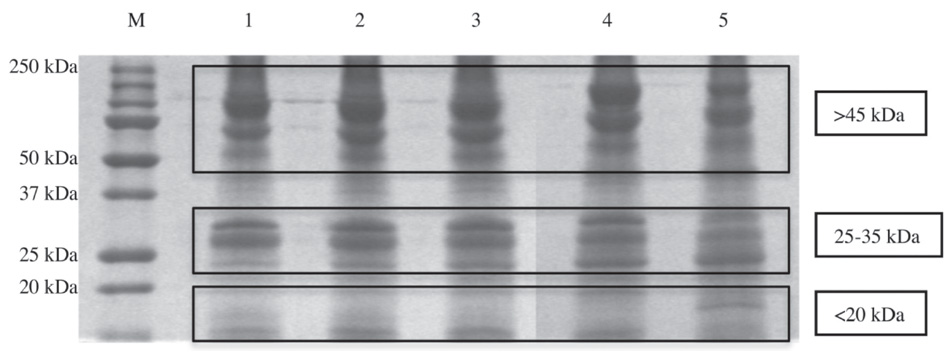
Fig. 2.10. Protein analysis of embryonic extract from Zebrafish eggs: suspension of the glycerol-alcohol solution analyzed with monodimensional gel electrophoresis (SDS-PAGE)
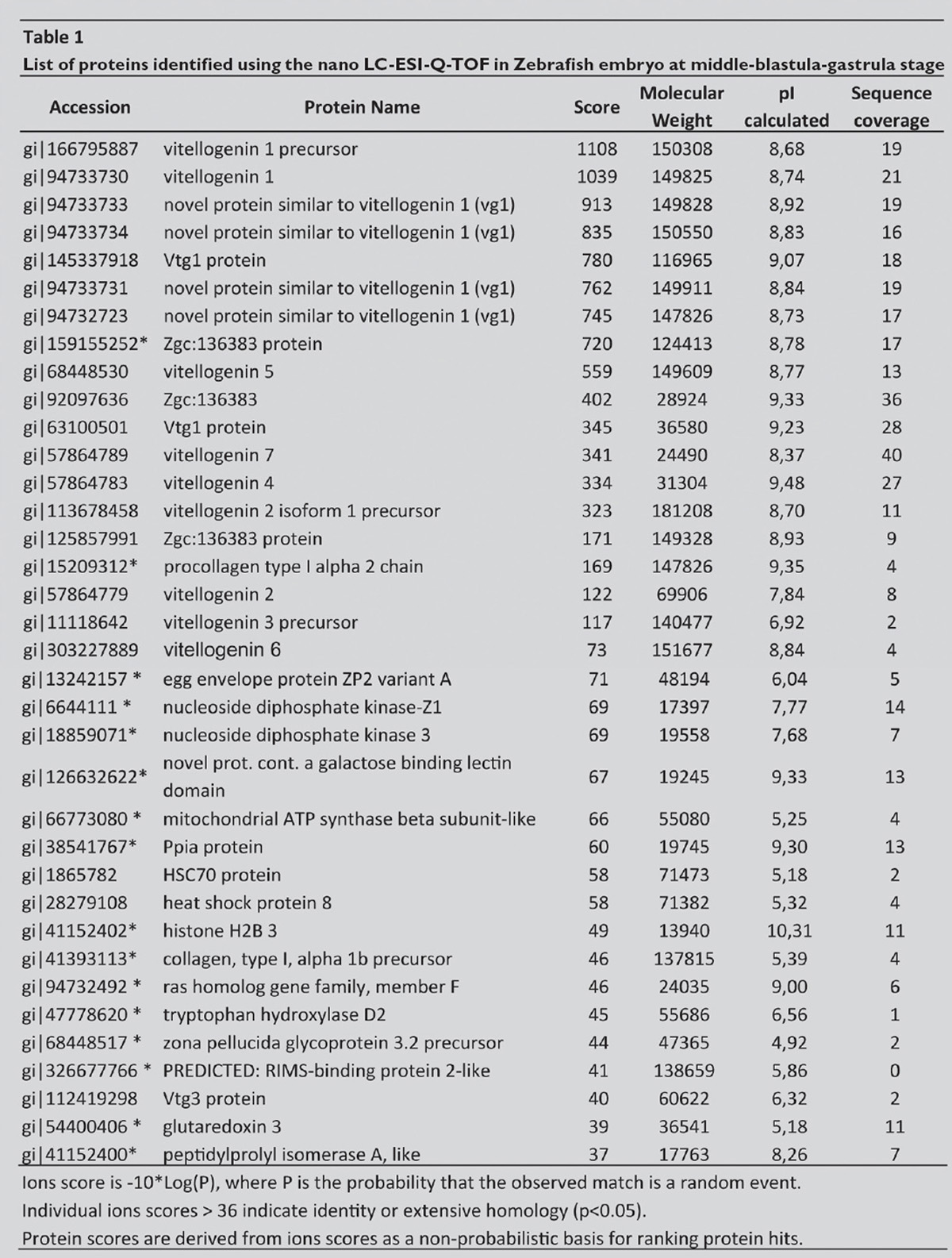
Below is the list of proteins that have been identified by Biava et al. with mass spectrometry analysis (see page 97). The asterisk (*) lists the proteins that have never before been described in the Zebrafish embryo.
Clinical Studies
Two clinical studies have been carried out, first to check the safety and then the effectiveness of integration of standard chemotherapy treatments with stem differentiation factors. A first study was conducted on 200 patients, including 60 with advanced breast cancer, to evaluate possible side effects: for this reason no control group was foreseen. The protocol included administering patients 1 ml three times a day as sublingual drops containing 3 microgram/ml staminal differentiation factors. After three years of treatment, no adverse events were reported in any of the 200 patients treated. In addition, 80 percent of these patients showed an improvement in the performance status, evaluated with the E.C.O.G scale: generally, the status shifted from a 4 to 3 state to one of 2 or 1. In the 60 breast cancer patients there were also four cases of partial regression and 70 percent of survival after three years.
The second study of embryonic differentiation factors with anticancer properties produced preliminary clinical results in the therapy for advanced tumors. A randomized clinical trial was performed on 179 patients with intermediate or advanced hepatocarcinoma. The results showed a statistically significant difference between the group treated with the synergy of differentiation factors and standard treatments and the control group (P = 0.03), a difference in favor of the group treated with integration. There was 19.8 percent of regression (2.5 percent of which was total regression) and 16 percent disease stabilization.54
Conclusions of the Committee of Oncologists
The committee met to evaluate the scientific soundness of the reported investigations, and it confirmed the importance of this course of study and encouraged at the university level the development of further research both on an in vivo model and at a clinical level. There is also the possibility of developing food supplements that inspire these researches to provide physicians with a valuable support to integrate many of the differentiation factors described in the text. Such solutions are to be understood only as integration into standard therapies, and it is hoped that they can be validated in advance by appropriate clinical trials.
References
1. Biava, P.M.; Bonsignorio, D.; Hoxa, M. Cell proliferation curves of different human tumor lines after in vitro treatment with aafish embryonic extracts. J. Tumor Marker Oncol., 2001, 16(3), 195–202.
2. Biava, P.M.; Carluccio, A. Activation of anti-oncogene p53 produced by embryonic extracts in vitro tumor cells. J. Tumor Marker Oncol., 1977, 12(4), 9–15.
3. Biava, P.M.; Bonsignorio, D.; Hoxa, M.; Impagliazzo, M.; Facco, R.; Ielapi, T.; Frati, L.; Bizzarri, M. Post-translational modification of the retinoblastoma protein (pRb) induced by in vitro administration of Zebrafish embryonic extracts on human kidney adenocarcinoma cell line. J. Tumor Marker Oncol., 2002, 17(2), 59–64.
4. Cucina, A.; Biava, P.M.; D’Anselmi, F.; Coluccia, P.; Conti, F.; di Clemente, R.; Miccheli, A.; Frati, L.; Gulino, A.; Bizzarri, M. Zebrafish embryo proteins induce apoptosis in human colon cancer cells (Caco2). Apoptosis, 2006, 11(9), 1617–1628.
5. Livraghi, T.; Meloni, F.; Frosi, A.; Lazzaroni, S.; Bizzarri, T. M.; Frati, L.; Biava, P.M. Treatment with stem cell differentiation stage factors of intermediate-advanced hepatocellular carcinoma: an open randomized clinical trial. Oncol. Res., 2005, 15(7–8), 399–408.
6. Livraghi, T. Ceriani, R. Palmisano, A. Pedicini, V. Pich, M. G. Tommasini, M.A. Torzilli, G. Complete response in 5 out of 38 patients with Advanced Hepatocellular Carcinoma treated with stem cell differentiation Stage Factors: Case Report from a single Center. Curr. Pharm. Biotech., 2011, 12(2).
7. Biava, P.M.; Canaider, S.; Facchin, F.; Bianconi, E.; Ljungberg, L.; Rotilio, D.; Burigana, F.; and Ventura, C. Stem Cell Differentiation Stage Factors from Zebrafish embryo: a novel strategy to modulate the fate of normal and pathological (stem) cells. Curr. Pharm. Biotechnol. 2015, 16(9), 782–92.
8. Harak, H.; Frosi, A.; Biava, P.M. Studio clinico sull’efficacia e tollerabilita’ di una crema per uso topico nel trattamento della psoriasi. La Med. Biol., 2012, 3, 27–31.
9. Calzavara-Pinton, P.; Rossi, M. A topical remedy in association with phototherapy. Efficacy evaluation in patients suffering from moderate psoriasis. Hi.tech dermo, 2012, 1, 41–47.
10. Biava, P.M.; Bonsignorio, D. Cancer and cell differentiation: a model to explain malignancy. J. Tumor Marker Oncol., 2002, 17(2), 47–54.
11. Biava, P.M; Monguzzi, A; Bonsignorio, D; Frosi, A; Sell, S; Klavins J.V. Xenopus laevis Embryos share antigens with Zebrafish Embryos and with human malignant neoplasms. J. Tumor Marker Oncol., 2001, 16(3), 203–206.
12. Lawson, J.C.; Blatch, G.L.; Edkins, A.L. Cancer stem cells in breast cancer and metastasis. Breast Cancer Res. Treat., 2009, 118(2), 241–254.
13. Luo, J.; Yin, X.; Ma, T.; Lu, J. Stem cells in normal mammary gland and breast cancer. Am. J. Med. Sci., 2010, 339(4), 366–370.
14. Spiro, S. G.; Tanner, N. T.; Silvestri, G. A.; Janes, S. M.; Lim, E.; Vansteenkiste, J.F.; Pirker, R. Lung cancer: progress in diagnosis staging and therapy. Respirology, 2010, 15(1), 44–50.
15. Gorelik, E.; Lokshin, A.; Levina, V. Lung cancer stem cells as a target for therapy. Anticancer Agents Med. Chem., 2010, 10(2), 164–171.
16. Sullivan, J.P.; Minna, J.D.; Shay, J.W. Evidence for self-renewing lung cancer stem cells and their implications in tumor initiation, progression and targeted therapy. Cancer Metastasis Rev., 2010, 29(1), 61–72.
17. Westhoff, B.; Colaluca, I.N.; D’Ario, G.; Donzelli, M.; Tosoni, D.; Volorio, S.; Pelosi, G.; Spaggiari, L.; Mazzarol, G.; Viale, G.; Pece, S.; Di Fiore, P.P. Alterations of the Notch pathway in lung cancer. Proc. Natl. Acad. Sci. USA, 2009, 106(52), 22293–22298.
18. Lang, S.H.; Anderson, E.; Fordham, R.; Collins, A.T. Modeling the prostate stem cell niche: an evaluation of stem cell survival and expansion in vitro. Stem. Cells Dev., 2010, 19(4), 537–546.
19. Joung, J.Y.; Cho, K.S.; Kim, J.E.; Seo, H.K.; Chung, J.; Park, W. S.; Choi, M.K.; Lee, K.H. Prostate stem cell antigen mRNA in peripheral blood as a potential predictor of biochemical recurrence of metastatic prostate cancer. J. Surg. Oncol., 2010, 101(2), 145–148.
20. Liu, T.; Cheng, W.; Lai, D.; Huang, Y.; Guo, L. Characterization of primary ovarian cancer cells in different culture systems. Oncol. Rep., 2010, 23(5), 1277–1284.
21. Fong, M.Y.; Kakar, S.S. The role of cancer stem cells and the side population in epithelial ovarian cancer. Histol. Histopathol., 2010, 25(1), 113–120.
22. Murphy, S.K. Targeting ovarian cancer-initiating cells. Anticancer Agents Med. Chem., 2010, 10(2), 157–163.
23. Peng, S.; Maihle, N.J.; Huang, Y. Pluripotency factors Lin 28 and Oct 4 identify a sub-population of stem cell-like cells in ovarian cancer. Oncogene, 2010, 29(14), 2153–2159.
24. Tomuleasa, C.; Soritau, O.; Rus-Ciuca, D.; Pop, T.; Todea, D.; Mosteanu, O.; Pintea, B.; Foris, V.; Susman, S.; Kacso, G.; Irimie, A. Isolation and characterization of hepatic cells with stem-like properties from hepatocellular carcinoma. J. Gastrointestin. Liver Dis., 2010, 19(1), 61–67.
25. Zou, G.M. Liver cancer stem cells as an important target in liver cancer therapies. Anticancer Agents Med. Chem., 2010, 10(2), 172–175.
26. Lee, T.K.; Castilho, A.; Ma, S.; Ng, I.O. Liver cancer stem cells: implications for new therapeutic target. Liver Int., 2009, 29(7), 955–965.
27. Marquardt, J.U.; Thorgeirsson, S.S. Stem Cells in hepatocarcinogenesis: evidence from genomic data. Semin. Liver Dis., 2010, 30(1), 26–34.
28. Kung, J.W.; Currie, I.S.; Forbes, S.J.; Ross, J.A. Liver development, regeneration, and carcinogenesis. J. Biomed. Biotechnol., 2010, 2010, 984248.
29. Gai, H.; Nguyen, D.M.; Moon, Y.J.; Aguila, J.R.; Fink, L.M.; Ward, D.C.; Ma, Y. Generation of murine hepatic lineage cells from induced pluripotent stem cells. Differentiation, 2010, 79(3), 171–181.
30. Correia, M.; Machado, J.C.; Ristimaki, A. Basic aspects of gastric cancer. Helicobacter, 2009, 14(1), 36–40.
31. Takaishi, S.; Okumura,T.; Tu, S.; Wang, S.S.; Shibata, W.; Vigneshwaran, R.; Gordon, S.A.; Shimada, Y.; Wang, T.C. Identification of gastric cancer stem cells using the surface marker CD44. Stem Cells, 2009, 27(5), 1006–1020.
32. Nishii, T.; Yashiro, M.; Shinto, O.; Sawada, T.; Ohira, M.; Hirakawa, K. Cancer stem cell-like SP cells have a high adhesion ability to the peritoneum in gastric carcinoma. Cancer Sci., 2009, 100(8), 1397–1402.
33. Chen, Z.; Xu, W.R.; Qian, H.; Zhu, W.; Bu, X.F.; Wang, S.; Yan, Y.M.; Mao, F.; Gu, H.B.; Cao, H.L.; Xu, X.J. Oct4, a novel marker for human gastric cancer. J. Surg. Oncol., 2009, 99(7), 414–419.
34. Kang, D.H.; Han, M.E.; Song, M.H.; Lee, Y.S.; Kim, E.H.; Kim, H.J.; Kim, G.H.; Kim, D.H.; Yoon, S.; Baek, S.Y.; Kim, B.S.; Kim, J.B.; Oh, S.O. The role of hedgehog signaling during gastric regeneration. J. Gastroenterol., 2009, 44(5), 372–379.
35. Yeung, T.M.; Ghandhi, S.C.; Wilding, J.L.; Muschel, R.; Bodmer, W. F. Cancer stem cells from colorectal cancer derived cell lines. Proc. Natl. Acad. Sci. USA, 2010, 107(8), 3722–3727.
36. Yeung, T.M.; Ghandhi, S.C.; Wilding, J.L.; Muschel, R.; Bodmer, W. F. Cancer stem cells from colorectal cancer derived cell lines. Proc. Natl. Acad. Sci. USA, 2010, 107(8), 3722–3727.
37. Gulino, A.; Ferretti, E.; De Smaele, E. Hedgehog signaling in colon cancer and stem cells. EMBO Mol. Med., 2009, 1(6–7), 300–302.
38. Thenappan, A.; Li, Y.; Shetty, K.; Johnson, L.; Reddy, E.P.; Mishra, L. New therapeutic targeting colon cancer stem cells. Curr. Colorectal Cancer Rep., 2009, 5(4), 209.
39. Rasheed, Z.A.; Yang, J.; Wang, Q.; Kowalski, J.; Freed, I.; Murter, C.; Hong. S.M.; Koorstra, J. B.; Rajeshkumar, N.V.; He, X.; Goggins, M.; Iacobuzio-Donahue, C.; Berman, D.M.; Laheru, D.; Jimeno, A.; Hidalgo, M.; Maitra, A.; Matsui, W. Prognostic significance of tumorigenic cells with mesenchymal features in pancreatic adenocarcinoma. J. Natl. Cancer Inst., 2010, 102(5), 340–351.
40. Puri, S.; Hebrok, M. Cellular plasticity within pancreas-lessons learned from development. Dev. Cell, 2010, 18(3), 342–356.
41. Quante, M.; Wang,T. C. Stem cells in gastroenterology and hepatology. Nat. Rev. Gastroenterol. Hepatol., 2009, 6(12), 724–737.
42. Ailles, L.; Prince, M. Cancer stem cells in head and neck squamous cell carcinoma. Methods Mol. Biol., 2009, 568, 175–193.
43. Zhang, P.; Zhang, Y.; Mao, L.; Zhang, Z.; Chen, W. Side population in oral squamous cell carcinoma possesses tumor stem cell phenotype. Cancer Lett., 2009, 277(2), 227–234.
44. Brunner M.; Thurnher, D.; Heiduschka, G.; Grasl, M.Ch.; Brostjan, C.; Erovic, B.M. Elevated levels of circulating endothelial progenitor cells in head and neck cancer patients. J. Surg. Oncol., 2008, 98(7), 545–550.
45. Zhang, Q.; Shi, S.; Yen, Y.; Brown, J.; Ta, J. Q.; Le, A. D. A subpopulation of CD133(+) cancer stem-like cells characterized in human oral squamous cell carcinoma confer resistance to chemotherapy. Cancer Lett., 2010, 289(2), 151–160.
46. Sato, A.; Sakurada, K.; Kumabe, T.; Sasajima, T.; Beppu, T.; Asano, K.; Ohkuma, H.; Ogawa, A.; Mizoi, K.; Tominaga, T.; Kitanaka, C.; Kayama, T.; Tohoku Brain Tumor Study Group. Association of stem cell marker CD133 expression with dissemination of glioblastoma. Neurosurg. Rev., 2010, 33(2), 175–183.
47. Di Tomaso, T.; Mazzoleni, S.; Wang, E.; Sovena, G.; Clavenna, D.; Franzin, A.; Mortini, P.; Ferrone, S.; Doglioni, C.; Marincola, F. M.; Galli, R.; Parmiani, G.; Maccalli, C. Immunobiological characterization of cancer stem cells isolated from glioblastoma patients. Clin. Cancer Res., 2010, 16(3), 800–813.
48. Ji, J.; Black, K.L.; Yu, J.S. Glioma stem cell research for the development of immunotherapy. Neurosurg. Clin. N. Am., 2010, 21(1), 159–166.
49. Biava, P. M.; Fiorito, A.; Negro, C.; Mariani, M. Effects of treatment with embryonic and uterine tissue homogenates on Lewis lung carcinoma development. Cancer Letters, 1988, 41(3); 265–70.
50. Biava, P.M.; Carluccio, A. Activation of anti-oncogene p53 produced by embryonic extracts in vitro tumor cells. J. Tumor Marker Oncol., 1977, 12(4), 9–15.
51. Biava, P.M.; Bonsignorio, D.; Hoxa, M.; Facco, R.; Ielapi, T.; Frati, L.; Bizzarri, M. “Post-translational modification of the retinoblastoma protein (pRb) induced by in vitro administration of Zebrafish embryonic extracts on human kidney adenocarcinoma cell line.: J. Tumor Marker Oncology, 2002, 17(2); 59–64.)
52. Biava, P.M.; Bonsignorio, D.; Hoxa, M. Cell proliferation curves of different human tumor lines after in vitro treatment with Zebrafish embryonic extracts. J. Tumor Marker Oncol., 2001, 16(3), 195–202.
53. Biava, P.M.; Bonsignorio, D.; Hoxha, M.; Impagliazzo, M.; Frosi, A.; Larese, F.; Negro, C. Mother-embryo cross-talk: the anti-cancer substances produced by mother and embryo during cell differentiation. A review of experimental data. J. Tumor Marker Oncol., 2002, 17, 55–58.
54. Livraghi, T.; Meloni, F.; Frosi, A.; Lazzaroni, S.; Bizzarri, M; Frati, L.; Biava, P.M. Treatment with stem cell differentiation stage factors of intermediate-advanced hepatocellular carcinoma: an open randomized clinical trial. Oncol. Res., 2005, 15, 399–408.