 9500 nucleotides. Tightly bound to p7 nucleocapsid proteins and late assembly protein p6.
9500 nucleotides. Tightly bound to p7 nucleocapsid proteins and late assembly protein p6.Icosahedral with the following components (see Fig. 38.1):
• Envelope: a lipid bilayer formed from host cell lipids and viral proteins. Nine to ten spikes (gp160) are embedded in the envelope, involved in binding and membrane fusion. These spikes are composed of 2 loosely attached viral surface glycoprotein, gp120, and transmembrane glycoprotein, gp41.
• Matrix: encapsulated by the envelope, made up of viral protein p17.
• Core: Conical and comprises:
• RNA dimer—two identical copies of single-stranded RNA, each containing  9500 nucleotides. Tightly bound to p7 nucleocapsid proteins and late assembly protein p6.
9500 nucleotides. Tightly bound to p7 nucleocapsid proteins and late assembly protein p6.
• Capsid protein p24 encapsulates the ribonucleoprotein core, which contains three enzymes; reverse transcriptase (p51), integrase (p32), and protease (p11).
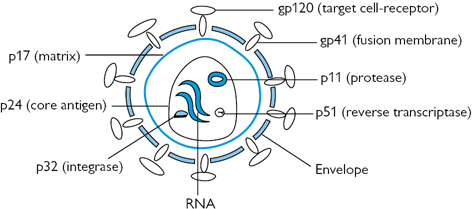
Fig. 38.1 HIV structure: gp and p refer to glycoprotein and protein, respectively, and the numerical values (×103) indicate molecular weight
• Genetic information is stored as RNA.
• Gene maps for HIV1 and HIV2 are similar, except that HIV2 has vpx instead of vpu.
• Both sides of the HIV provirus are flanked by a repeated sequence, known as the long-terminal repeat (LTR).
• HIV genes and their major functions are outlined in Table 38.1.
Table 38.1 HIV genes and their major functions
| Major structural proteins | Gag | Encodes for capsid, matrix, and nucleocapsid |
| Pol | Encodes for viral enzymes | |
| Env | Encodes for envelope glycoproteins | |
| Regulatory proteins | Tat | Regulates HIV transcription |
| Rev | Stimulates HIV proteins production and suppresses expression of HIV regulatory genes | |
| Accessory proteins | vpu | Helps the assembly and budding of new virus particles and enhances the degradation of CD4 proteins. |
| vpr | Accelerates HIV proteins production, nuclear localization of PIC and slows host cell growth cycle. | |
| vif | Promotes infectivity by interfering with cellular defences. | |
| nef |  Regulates cell surface expression of CD4 and stimulates HIV infectivity Regulates cell surface expression of CD4 and stimulates HIV infectivity |
Replication occurs in the following sequence: binding  fusion and entry
fusion and entry  reverse transcription
reverse transcription  integration
integration  proviral transcription and translation
proviral transcription and translation  assembly and budding
assembly and budding 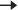 maturation.
maturation.
Glycoprotein120 binds to the extracellular component of the CD4 receptor (expressed in helper T-cells, macrophages, monocytes, microglial, dendritic, and Langerhans’ cells). This results in conformational change facilitating 2° interaction with a second receptor (a chemokine co-receptor) CCR5 or CXCR4.
The distal tips of gp41 are inserted in to the cellular membrane followed by conformational changes folding in half to form coiled loops. This process pulls the viral and cellular membranes together, and fusing them with subsequent uncoating of the viral capsid, and the release of the HIV RNA dimer and proteins into the host cell cytoplasm.
In this process, viral ssRNA is transcribed by the viral reverse transcriptase (RT) into double-stranded DNA (dsDNA). HIV uses cellular tRNALys3 as a primer. The primer hybridizes to a specific site (primer binding site, PBS) of the virus RNA genome to initiate reverse transcription. Transcription takes place in 5' 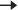 3' direction to form a minus-strand complementary DNA (cDNA). The RNA:DNA hybrid is then degraded by RNase H domain of reverse transcriptase. DNA:tRNA is transferred to the 3'-end of the template to complete synthesis of the first strand DNA. The rest of viral ssRNA is degraded by RNase H, except for a purine-rich sequence (named polypurine tract, ppt), which subsequently serves as a primer for initiation of the second strand ssDNA synthesis. RT then elongates the ppt primer from the 3'-end of the template. tRNA is then degraded by RNase. PBS from the second strand hybridizes with the complementary PBS of the first strand. Extension of both strands produces a complete viral dsDNA.
3' direction to form a minus-strand complementary DNA (cDNA). The RNA:DNA hybrid is then degraded by RNase H domain of reverse transcriptase. DNA:tRNA is transferred to the 3'-end of the template to complete synthesis of the first strand DNA. The rest of viral ssRNA is degraded by RNase H, except for a purine-rich sequence (named polypurine tract, ppt), which subsequently serves as a primer for initiation of the second strand ssDNA synthesis. RT then elongates the ppt primer from the 3'-end of the template. tRNA is then degraded by RNase. PBS from the second strand hybridizes with the complementary PBS of the first strand. Extension of both strands produces a complete viral dsDNA.
Reverse transcription yields a pre-integration complex (PIC), composed of linear dsDNA, cellular host proteins, transcriptase, nucleocapsid, matrix, vpr, and integrase. PIC enters the nucleus through the nuclear pore complex.
Due to its poor proofreading ability, reverse transcriptase has a high error rate when transcribing RNA into DNA. This allows mutations to accumulate, which in turn, have a role in the development of drug resistance and escape from immune surveillance.
Integrase binds to the end of each LTR of the HIV double-stranded cDNA. The final outcome, after a series of reactions, is a ‘cut-and-paste’ clipping of the host DNA and joining the proviral genome to the clipped ends.
Proviral genome remained latent until CD4 cell is activated. Certain cellular transcription factors (NF-kB is the most important) must be present for active virus production, which are then up-regulated when T-cells get activated. Early in the transcription process, short completely spliced mRNAs that codes for Tat and Rev are produced. As transcription rate increases, larger incompletely spliced mRNAs are produced. These code for Env, vif, vpr, and vpu. Late in the process, full length transcripts are produced, which act as virion genomic RNA. Spliced mRNAs are exported from the nucleus and translated in the endoplasmic reticulum into structural proteins Gag and Env, and others into the regulatory proteins Tat and Rev. Gag proteins bind to copies of the virus RNA genome to package them into new virus particles.
Assembly occurs at the plasma membrane. Gag (and Gag-Pro-Pol) polyprotein mediates the essential events in virion assembly, including binding the plasma membrane, making the protein–protein interactions necessary to create spherical particles, concentrating the viral Env protein, and packaging the genomic RNA. Conformational change(s) within Gag couples membrane binding, virion assembly, and RNA packaging. Although Gag itself can bind membranes and assemble into spherical particles, the budding event that releases the virion from the plasma membrane is mediated by host machinery
Viral maturation begins concomitant with (or immediately following) budding and is driven by viral protease cleavage of the Gag and Gag-Pro-Pol polyproteins at 10 different sites, ultimately producing the fully processed MA, CA, NC, p6, PR, RT, and IN proteins. Over the course of maturation, these processed proteins rearrange to create the mature infectious virion.
Mature virions are released ready to infect new cells and begin the replication cycle once again. This process is very active, 108 –1010 viral particles produced each day.
• The primary cellular receptor for HIV entry is CD4. Expression of CD4 on a target cell is necessary, but not sufficient for HIV entry.
• CD4 antigen is the principal receptor, mainly expressed on the surface of helper T lymphocytes. It is also expressed, but to a lesser degree in CD4 dendritic cells including Langerhans’ cells and CD4 monocytes, macrophages, and microglial cells.
• Co-receptors: several described, act as co-factors that allow HIV entry when co-expressed with CD4 on a cell surface. CCR5 and CXCR4 are the most important, and they act in concert with CD4 to facilitate HIV and cell membrane fusion. CCR5, which is expressed on macrophages and on some T cells. M-tropic HIV isolates appear to use some T cells. CXCR4 is expressed on T cells, but not usually on macrophages. Other chemokine co-receptors, e.g. CCR3, expressed on eosinophils and microglia, is used by some strains of HIV for infection of the microglia resulting CNS pathology.
• Age:  age is associated with
age is associated with  progression.
progression.
• Co-infection: may affect immune system resulting in  progression (e.g. tuberculosis and hepatitis C). CMV associated with
progression (e.g. tuberculosis and hepatitis C). CMV associated with  progression in haemophiliacs.
progression in haemophiliacs.
• Gender: ♀ appear to have higher VLs at any CD4 level and may progress more rapidly.
• Psychosocial factors: depression, impaired intellectual functioning, drug use, social deprivation may be associated with  progression.
progression.
• 4–15% of Caucasians have a deletion in CCR5 gene resulting in a mutant co-receptor (CCR5∆32). The effect is a truncated co-receptor not expressed at the cell surface. People of Northern European descent have the higher frequency of these deletions. Homozygous individuals (1–2% of the Caucasian population) are almost resistant to HIV infection and heterozygotes are slow progressors. Other alleles of CCR5 genes were described in Africans and Asians, and have different effects on co-receptor expression.
• CCR2 (a minor co-receptor) deletion (CCR-V641) is widespread in all ethnic groups and results in slower progression to AIDS
• certain HLA types associated with  or
or  progression
progression
• possession or lack of certain genes—e.g. low copy number of CCL3L1 associated with  susceptibility.
susceptibility.
• Nutrition: poor premorbid state associated with  progression.
progression.
• Pharmacological variability: individual drug metabolism and elimination modify response to therapy.
Changes in the phenotype and the genotype of the virus enable it to ‘escape’ control by the immune system. In late HIV infection switching from CCR5 to CXCR4 tropism leads to infection of both active and resting immune cells resulting in  disease progression. Mutations may alter viral ‘fitness’ influencing pathogenicity. Gene mutation involving nef is associated with
disease progression. Mutations may alter viral ‘fitness’ influencing pathogenicity. Gene mutation involving nef is associated with  progression and some drug-resistant mutations (e.g. M184V, which induces lamivudine resistance) may
progression and some drug-resistant mutations (e.g. M184V, which induces lamivudine resistance) may  viral fitness.
viral fitness.
Drug susceptibility depends largely on HIV genotypic and phenotypic characteristics with genotype mutations rendering some drugs ineffective. Other factors, such as efflux pumps, may also be involved.