On 26 June 2000, it was announced that the initial draft of the sequence of the human genome had been completed. In February 2001, the first papers describing this draft sequence in detail were released. It was the culmination of years of work and technological breakthroughs, and more than a little rivalry. The National Institutes of Health in the USA and the Wellcome Trust in the UK had poured in the majority of the approximately $2.7 billion1 required to fund the research. This was carried out by an international consortium, and the first batch of papers detailing the findings included over 2,500 authors from more than 20 laboratories worldwide. The bulk of the sequencing was carried out by five laboratories, four of them in the US and one in the UK. Simultaneously, a private company called Celera Genomics was attempting to sequence and commercialise the human genome. But by releasing their data on a daily basis as soon as it was generated, the publicly funded consortium was able to ensure that the sequence of the human genome entered the public domain.2
An enormous hoopla accompanied the declaration that the draft human genome had been completed. Perhaps the most flamboyant statement was from US President Bill Clinton, who declared that ‘Today we are learning the language in which God created life’.3 We can only speculate on the inner feelings of some of the scientists who had played such a major role in the project as a politician invoked a deity at the moment of technological triumph. Luckily, researchers tend to be a shy lot, especially when confronted by celebrities and TV cameras, so few expressed any disquiet publicly.
Michael Dexter was the Director of the Wellcome Trust, which had poured enormous sums of money into the Human Genome Project. He was not much less fulsome, albeit somewhat less theistic, when he defined the completion of the draft sequence as ‘The outstanding achievement not only of our lifetime, but in terms of human history’.4
You might not be alone in thinking that perhaps other discoveries have given the Human Genome Project a run for its money in terms of impact. Fire, the wheel, the number zero and the written alphabet spring to mind, and you probably have others on your own list. It could also be claimed that the human genome sequence has not yet delivered on some of the claims that were made about how quickly it would impact on human disease. For instance, David Sainsbury, the then UK Science Minister, stated that ‘We now have the possibility of achieving all we ever hoped for from medicine’.5
Most scientists knew, however, that these claims should be taken with whole shovelfuls of salt, because we have been taught this by the history of genetics. Consider a couple of relatively well-known genetic diseases. Duchenne muscular dystrophy is a desperately sad disorder in which affected boys gradually lose muscle mass, degenerate physically, lose mobility and typically die in adolescence. Cystic fibrosis is a genetic condition in which the lungs can’t clear mucus, and the sufferers are prone to severe life-threatening infections. Although some cystic fibrosis patients now make it to the age of about 40, this is only with intensive physical therapy to clear their lungs every day, plus industrial levels of antibiotics.
The gene that is mutated in Duchenne muscular dystrophy was identified in 1987 and the one that is mutated in cystic fibrosis was identified in 1989. Despite the fact that mutations in these genes were shown to cause disease over a decade before the completion of the human genome sequence, there are still no effective treatments for these diseases after 20-plus years of trying. Clearly, there’s going to be a long gap between knowing the sequence of the human genome, and developing life-saving treatments for common diseases. This is especially the case when diseases are caused by more than one gene, or by the interplay of one or more genes with the environment, which is the case for most illnesses.
But we shouldn’t be too harsh on the politicians we have quoted. Scientists themselves drove quite a lot of the hype. If you are requesting the better part of $3 billion of funding from your paymasters, you need to make a rather ambitious pitch. Knowing the human genome sequence is not really an end in itself, but that doesn’t make it unimportant as a scientific endeavour. It was essentially an infrastructure project, providing a dataset without which vast quantities of other questions could never be answered.
There is, of course, not just one human genome sequence. The sequence varies between individuals. In 2001, it cost just under $5,300 to sequence a million base pairs of DNA. By April 2013, this cost had dropped to six cents. This means that if you had wanted to have your own genome sequenced in 2001, it would have cost you just over $95 million. Today, you could generate the same sequence for just under $6,000,6 and at least one company is claiming that the era of the $1,000 genome is here.7 Because the cost of sequencing has decreased so dramatically, it’s now much easier for scientists to study the extent of variation between individual humans, which has led to a number of benefits. Researchers are now able to identify rare mutations that cause severe diseases but only occur in a small number of patients, often in genetically isolated populations such as the Amish communities in the United States.8 It’s possible to sequence tumour cells from patients to identify mutations that are driving the progression of a cancer. In some cases, this results in patients receiving specific therapies that are tailored for their cancer.9 Studies of human evolution and human migration have been greatly enhanced by analysing DNA sequences.10
Honey, I lost the genes
But all this was for the future. In 2001, amidst all the hoopla, scientists were poring over the data from the human genome sequence and pondering a simple question: where on earth were all the genes? Where were all the sequences to code for the proteins that carry out the functions of cells and individuals? No other species is as complex as humans. No other species builds cities, creates art, grows crops or plays ping-pong. We may argue philosophically about whether any of this makes us ‘better’ than other species. But the very fact that we can have this argument is indicative of our undoubtedly greater complexity than any other species on earth.
What is the molecular explanation for our complexity and sophistication as organisms? There was a reasonable degree of consensus that the explanation would lie in our genes. Humans were expected to possess a greater number of protein-coding genes than simpler organisms such as worms, flies or rabbits.
By the time the draft human genome sequence was released, scientists had completed the sequencing of a number of other organisms. They had focused on ones with smaller and simpler genomes than humans, and by 2001 had sequenced hundreds of viruses, tens of bacteria, two simple animal species, one fungus and one plant. Researchers had used data from these species to estimate how many genes would be found in the human genome, along with data from a variety of other experimental approaches. Estimates ranged from 30,000 to 120,000, revealing a considerable degree of uncertainty. A figure of about 100,000 was frequently bandied about in the popular press, even though this had not been intended as a definitive estimate. A value in the region of 40,000 was probably considered reasonable by most researchers.
But when the draft human sequence was released in February 2001, researchers couldn’t find 40,000 protein-coding genes, let alone 100,000. The scientists from Celera Genomics identified 26,000 protein-coding genes, and tentatively identified an additional 12,000. The scientists from the public consortium identified 22,000 and predicted there would be a total of 31,000 in total. In the years since the publication of the draft sequence, the number has consistently decreased and it is now generally accepted that the human genome contains about 20,000 protein-coding genes.11
It might seem odd that scientists didn’t immediately agree on the numbers of genes as soon as the draft sequence was released. But that’s because identifying genes relies on analysing sequence data and isn’t as easy as it sounds. It’s not as if genes are colourcoded, or use a different set of genetic letters from the other parts of the genome. To identify a protein-coding gene, you have to analyse specific features such as sequences that can code for a stretch of amino acids.
As we saw in Chapter 2, protein-coding genes aren’t formed from one continuous sequence of DNA. They are constructed in a modular fashion, with protein-coding regions interrupted by stretches of junk. In general, human genes are much longer than the genes in fruit flies or the microscopic worm called C. elegans, which are very common model systems in genetic studies. But human proteins are usually about the same size as the equivalent proteins in the fly or the worm. It’s the junk interruptions in the human genes that are very big, not the bits that code for protein. In humans, these intervening sequences are often ten times as long as in simpler organisms, and some can be tens of thousands of base pairs in length.
This creates a big signal-to-noise problem when analysing genes in human sequences. Even within one gene there’s just a small region that codes for protein, embedded in a huge stretch of junk.
So, back to the original problem. Why are humans such complicated organisms, if our protein-coding genes are similar to those from flies and worms? Some of the explanation lies in the splicing that we saw in Chapter 2. Human cells are able to generate a greater variety of protein variants from one gene than simpler organisms. Over 60 per cent of human genes generate multiple splicing variants. Look again at Figure 2.5 (page 18). A human cell could produce the proteins DEPARTING, DEPART, DEAR, DART, EAT and PARTING. It might produce these proteins in different ratios in different tissues. For example, DEPARTING, DEAR and EAT could all be produced at high levels in the brain, but the kidney might only express DEPARTING and DART. And the kidney cells might produce 20 times as much of DART as of DEPARTING. In lower organisms, cells may only be able to produce DEPARTING and PARTING, and they may produce them at relatively fixed ratios in different cells. This splicing flexibility allows human cells to produce a much greater diversity of protein molecules than lower organisms.
The scientists analysing the human genome had speculated that there might be protein-coding genes that are specific to humans, which could account for our increased complexity. But this doesn’t seem to be the case. There are nearly 1,300 gene families in the human genome. Almost all of these gene families occur through all branches of the kingdom of life, from the simplest organisms upwards. There is a subset of about 100 families that are specific to animals with backbones but even these were generated very early in vertebrate evolution. These vertebrate-specific gene families tend to be involved in complex processes such as the parts of the immune system that remember an infection; sophisticated brain connections; blood clotting; signalling between cells.
It’s a little as if our protein-coding genome has been built from a giant LEGO kit. Most LEGO kits, especially the large starter boxes, contain a selection of bricks that are variations on a small number of themes. Rectangles and squares, some sloping pieces, perhaps a few arches. Various colours, proportions and thicknesses, but all basically similar. And from these you can build pretty much all basic structures, from a two-brick step to an entire housing development. It’s only when you need to build something extremely specialist, like the Death Star, that it’s necessary to have very unusual pieces that don’t fit the basic LEGO templates.
Throughout evolution, genomes have developed by building out from a standard set of LEGO templates, and only very rarely have they created something completely new. So we can’t explain human complexity by claiming we have lots of unusual human-specific protein-coding genes. We simply don’t.
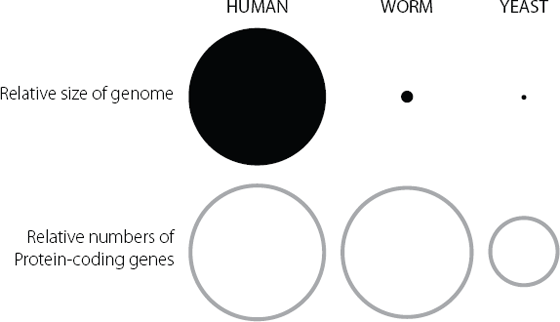
But where this all becomes odd is when we compare the size of the human genome with that of other organisms. Looking at Figure 3.1, we can see that the human genome is much bigger than that of C. elegans and much, much bigger than that of yeast. But in terms of numbers of protein-coding genes, there isn’t anything like as great a difference.
Figure 3.1 In the upper panel, the areas of the circle represent the relative sizes of the genomes in humans, a microscopic worm and single-celled yeast. The human genome is much bigger than those from the simpler organisms. The lower panel represents the relative numbers of protein-coding genes in each of the three species. The disparity here between humans and the other two organisms is much less than in the top panel. The large relative size of the human genome clearly can’t be explained solely in terms of numbers of protein-coding genes.
These data demonstrated convincingly that the human genome contains an extraordinary amount of DNA that doesn’t code for proteins. Ninety-eight per cent of our genetic material doesn’t act as the template for those all-important molecules believed to carry out the key functions of a cell or an organism. Why do we have so much junk?
Poisonous fish and genetic insulation
One possibility is that the question is irrelevant or inappropriate. Maybe the junk has no function or biological significance. It can be a mistake to assume that because something is present, it has a reason to be there. The human appendix serves no useful purpose; it’s just an evolutionary hangover from our ancestral lineages. Some scientists speculated back in 2001 that this might also be true of most of the junk DNA in the human genome.
Part of the rationale for this suggestion lay in an interesting animal, the pufferfish (also known as the blowfish). Pufferfish are remarkable creatures. Because they are slow, clumsy swimmers they are unable to evade predators. If faced with a threat, they rapidly take in huge amounts of water and swell up into a globe, which in some species is covered in spikes. If that isn’t enough to deter a hungry predator, they also contain a toxin which is over a thousand times more powerful than cyanide. This has given the pufferfish a weird notoriety. In Japan it is considered a delicacy (called fugu), but one with a highly chequered history, since inexpert preparation can carry lethal consequences for the diner.
Genetics researchers were very fond of pufferfish, or at least its DNA. The genome of a particular pufferfish called Fugu rubripes is the most compact of any vertebrate. It is only about 13 per cent of the length of the human sequence, but it contains pretty much all the usual vertebrate genes.12 The reason the pufferfish genome is so small is because it doesn’t contain very much junk DNA. In the days when it cost a lot of money to sequence DNA, pufferfish was a very useful species to use when comparing genomes from different organisms. And because its genome contains so little junk, it was relatively easy to identify individual genes, because there weren’t the signal-to-noise issues that were such a problem when annotating the human genome. Scientists were able to spot genes in Fugu rubripes very easily, and then use the sequence data to help them search for similar genes in noisier genomes such as our own.
Because pufferfish have very little junk DNA but are functional and successful organisms, it was suggested that the non-coding regions of the human genome might be ‘simply parasitic, selfish DNA elements that use the genome as a convenient host’.13 But this isn’t necessarily a logical projection. Just because something has no apparent function in a specific organism, it doesn’t mean it is irrelevant in all species. Because evolution is usually building from a relatively limited repertoire of components (remember the LEGO set), there is a tendency for features to be co-opted for new functions. So, junk DNA could easily have roles in other organisms, especially ones that are more complex.
It is also worth bearing in mind that there is a functional cost for a cell in containing so much junk DNA. Humans all start life as one cell, formed when an egg fuses with a sperm. That single starting cell divides to form two cells. The two cells divide to form four, and the process continues. An adult human is composed of about 50–70 trillion cells. That’s a lot of cells to visualise, so try it this way. If each cell was a dollar bill, and we stacked 50 trillion dollar bills on top of each other, they would stretch from the Earth to the moon and halfway home again.
It takes about 46 cycles of cell division, at a minimum, to create that many cells. And every time a cell divides, it first has to copy all its DNA. If less than 2 per cent of the DNA is important, why would evolution maintain the other 98 per cent if it is simply functionless junk? As we have already acknowledged, the greatest evidence in favour of evolution of species lies in all those things we are stuck with because of our forebears (such as the appendix). But using huge amounts of resources to reproduce 49 ‘useless’ base pairs for every one that performs a function seems like taking redundancy a bit far.
One of the first theories for why the human genome contains so much DNA arose even before the draft human genome sequence had been completed, when researchers already recognised that there was a significant part of our genome that didn’t code for protein. It’s the insulation theory.
Imagine you own a watch. Not just any old watch, but a phenomenally expensive watch such as a vintage Patek Philippe of the type that sells for a couple of million dollars. Now imagine there is a large and very angry baboon in the vicinity, carrying a really heavy stick. You have to put your watch in a room and you are given a choice. You can’t stop the baboon going into any of the rooms, but you can decide on the room where you want to leave the watch. The choices are:
A. A small room with nothing else in it but a table, on which you have to leave the watch.
B. A large room containing 50 rolls of loft insulation, each roll being 5m in length and 20cm deep, and you can hide the watch deep in any one of the 50 rolls.
It’s not that difficult to work out which to choose to maximise the chances of the watch escaping damage, is it? And the insulation theory of junk DNA was built on the same premise. The genes that code for proteins are incredibly important. They have been subjected to high levels of evolutionary pressure, so that in any given organism, the individual protein sequence is usually as good as it’s likely to get. A mutation in DNA – a change in a base pair – that changes the protein sequence is unlikely to make a protein more effective. It’s more likely that a mutation will interfere with a protein’s function or activity in a way that has negative consequences.
The problem is that our genome is constantly bombarded by potentially damaging stimuli in our environment. We sometimes think of this as a modern phenomenon, especially when we consider radiation from disasters such as those at the Chernobyl or Fukushima nuclear plants. But in reality this has been an issue throughout human existence. From ultraviolet radiation in sunlight to carcinogens in food, or emission of radon gas from granite rocks, we have always been assailed by potential threats to our genomic integrity. Sometimes these don’t matter that much. If ultraviolet radiation causes a mutation in a skin cell, and the mutation results in the death of that cell, it’s not a big deal. We have lots of skin cells; they die and are replaced all the time, and the loss of one extra is not a problem.
But if the mutation causes a cell to survive better than its neighbours, that’s a step towards the development of potential cancer, and the consequences of that can be a very big deal indeed. For example, over 75,000 new cases of melanoma are diagnosed every year in the United States, and there are nearly 10,000 deaths per year from the condition.14 Excessive exposure to ultraviolet radiation is a major risk factor. In evolutionary terms, mutations would be even worse if they occurred in eggs or sperm, as they may be passed on to offspring.
If we think of our genome as constantly under assault, the insulation theory of junk DNA has definite attractions. If only one in 50 of our base pairs is important for protein sequence because the other 49 base pairs are simply junk, then there’s only a one in 50 chance that a damaging stimulus that hits a DNA molecule will actually strike an important region.
It’s also consistent with why the human genome contains so much junk DNA compared with the relatively tiny amounts present in less complex species such as the worm and yeast, as we saw in Figure 3.1. Worms and yeast have short life cycles, and can produce large numbers of offspring. The cost–benefit equation for them is different from that of a species such as humans, who take a long time to reproduce and only have small numbers of offspring. For worms and yeast there probably isn’t much point putting a large amount of effort into protecting the protein-coding genes so extensively. Even if a few of their offspring carry mutations that make them less fit for their environment, the majority will probably be OK. But if you get very few shots at passing your genetic material on to the next generation, protecting those important protein-coding genes makes good evolutionary sense.
Nature, as we have seen, is nothing if not adaptive, and so even though the insulation theory makes good sense, it raises another couple of questions. Is insulation the only role of junk DNA?; and where did all this insulating material come from in the first place?