For quite a few years, Xist was considered an anomaly, a strange molecular outlier with an extraordinarily unusual impact on gene expression. Even when Tsix was identified, it was possible to think that junk RNAs were restricted to the vital but unique process of X inactivation. It is only in recent years that we have begun to recognise that the human genome expresses thousands of this type of molecule, and that they are surprisingly important in normal cellular function.
We now categorise Xist and Tsix as members of a large class known as the long non-coding RNAs. The term is a somewhat misleading one, because of course what it means is non-coding with respect to proteins. As we shall see, the long non-coding RNAs do code for functional molecules. The functional molecules are the long non-coding RNAs themselves.
Long non-coding RNAs are defined rather arbitrarily as molecules which are greater than 200 bases in length, and which don’t code for proteins, making them different from messenger RNA. 200 bases is the lower size limit, but the biggest long non-coding RNAs can be 100,000 bases in length. There are lots of them, although no agreement yet on the precise number. Estimates range from 10,000 to 32,000 in the human genome.1,2,3,4 But although there are a lot of long non-coding RNAs, they don’t tend to be expressed to as high a level as the classical messenger RNAs which code for proteins. Normally, the expression level of a long non-coding RNA is less than 10 per cent of the level of an average messenger RNA.5
This relatively low abundance of any one long non-coding RNA is one of the reasons why we have tended to disregard this type of molecule until fairly recently. Essentially, when the expression of RNA molecules from cells was analysed, the long non-coding RNAs simply could not be detected very reliably because the technology wasn’t sensitive enough. However, now that we know about their existence, we might think we should be able to analyse the genome of any organism, including humans, and predict their existence from the DNA sequence. We are, after all, pretty good at doing that for protein-coding genes.
But there are a number of aspects that make this difficult. We can identify putative protein-coding genes because of a number of features. They have certain sequences near the beginning and end of the genes that help us to find them. They also encode predicted runs of amino acids, which again give us confidence that a protein-coding gene may be present. Finally, most protein-coding genes are pretty similar if you look at a specific gene in different species. This means that if we identify a classical gene in an animal such as a pufferfish, it’s easy to use that sequence as a basis for analysing the human genome to see if we can predict the presence of a similar gene in ourselves.
However, long non-coding RNAs don’t have such strong sequence indicators as protein-coding genes, and they are also poorly conserved across species. Consequently, knowing the sequence of a long non-coding RNA in another species may not help us to identify a functionally related sequence in the human genome. Less than 6 per cent of a specific class of long non-coding RNAs in zebrafish, a common model system, have clearly equivalent sequences in mice and humans.6 Only about 12 per cent of the same class of long non-coding RNAs that are found in humans and mice can be detected elsewhere in the animal kingdom.7,8 The relatively poor conservation of long non-coding RNAs was confirmed in a recent study comparing expressed long non-coding RNAs from various tissues of different tetrapod species. Tetrapod refers to all land-living vertebrates along with those that have ‘returned to the sea’ such as whales and dolphins. This paper reported that there were 11,000 long non-coding RNAs that were only found in primates. Only 2,500 were conserved across tetrapods, of which a mere 400 were classified as ancient, by which the authors meant that they had originated over 300 million years ago, around the time when amphibians and other tetrapods diverged. The authors suspected that the ancient long non-coding RNAs are the ones that are most actively regulated in all organisms, and are probably mostly involved in early development.9 Most vertebrates look very similar during the earliest stages of embryogenesis, so it may make sense that we and all our distant cousins are using similar pathways to get started.
The generally poor conservation across species has led some authors to speculate that the long non-coding RNAs are not very important. The rationale behind this is that if they were significant they would be more constrained to remain similar during evolution and the development of species; whereas instead, the sequences coding for these ‘junk’ RNAs are evolving much more rapidly than the ones that encode proteins.
Although this is a fair point, it’s perhaps an over-simplification. Long non-coding RNA molecules may be long in terms of the number of bases they contain, but that doesn’t necessarily mean they are elongated stringy molecules in the cell. This is because long RNA molecules can fold onto themselves, forming three-dimensional structures. The bases in RNA pair up, following similar rules to the way in which the two strands of DNA are bonded together. RNA is a single-stranded molecule, so its bases pair up over relatively short distances, bending the molecule into complex stable shapes. These 3D structures may be very important in the function of the long non-coding RNA, and it’s possible that the 3D structure is conserved across species, even if the base sequence is not.10 This is shown in Figure 8.1. Unfortunately, predicting similar structures is difficult to do using sequence data, limiting the usefulness of this technique in helping us to find functionally conserved long non-coding RNAs.
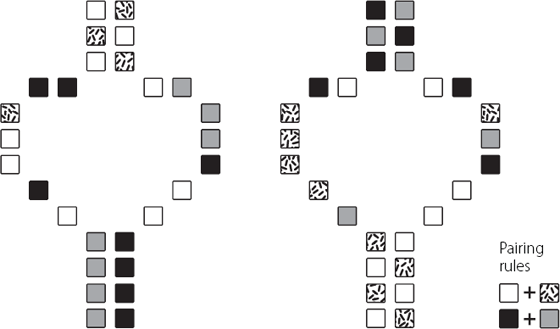
Figure 8.1 Representation of how two single-stranded long non-coding RNA molecules with different base sequences can form the same shape as each other. The shapes are determined by pairing of the A and U or C and G bases, which are represented by the differently shaded/patterned boxes. The representation is an over-simplification. In reality, the long non-coding RNAs may have multiple regions that can form complex structures. They will also be three-dimensional, rather than the flat shape shown here.
Logs or chips?
Because of the complications that arise if we try to identify long non-coding RNAs from the human genome sequence, most researchers lean towards the more pragmatic approach of identifying long non-coding RNAs by detecting the molecules themselves in cells. But there is a considerable degree of conflict in the scientific community about how to interpret the results. Hardcore junk aficionados might claim that if a sequence is expressed as a long non-coding RNA molecule then that molecule is being expressed for a reason. Other scientists are much more sceptical, positing that the expression of the long non-coding RNAs is essentially what we call a bystander event. This means that the long non-coding RNAs are expressed, but just as a by-product of switching on a ‘proper’ gene.
To understand what’s meant by a bystander event, let’s imagine we are cutting up tree branches with a chainsaw. The major aim of our activity is to create logs that we can use to build a cabin or to provide fuel for a stove. We aren’t trying to create woodchips or sawdust, but this happens anyway as a result of the chainsaw function. It’s not worth our while trying to avoid creating the woodchips. They don’t really interfere with our main aim, and if we do find a way to avoid generating them, it might be at the expense of efficient production of the logs. Just occasionally, we may even find that we have a use for the woodchip by-product, using it to mulch a flowerpot, or provide bedding for our pet snake.
In a similar model, the junk sceptics postulate that expression of long non-coding RNA simply reflects a loosening of repression when genes in a particular region are expressed. In this model, the production of long non-coding RNAs is simply an inevitable consequence of an important process, but essentially harmless and insignificant. The believers counter that that fails to address certain aspects of long non-coding RNA expression. For example, different types of long non-coding RNAs are expressed if we examine samples from different brain regions.11 Enthusiasts for long noncoding RNAs claim this supports their model for the importance of these molecules, because why else would different brain regions switch on different long non-coding RNAs? The sceptics claim that the different long non-coding RNAs are detected simply because various brain regions switch on different classical protein-coding genes. In our chainsaw analogy, this is equivalent to getting different woodchips depending on whether we are sawing up oak branches or pine.
It’s early days but current data suggest that the extremists on both sides should probably relax a little because the reality is likely to lie somewhere between their two positions. The only way we can really test the hypothesis that long non-coding RNAs have functions in the cell is to test each one, in the correct cell type. Although perfectly sensible as an approach, this isn’t as straightforward as it sounds. Partly this is down to sheer numbers. If we detect hundreds or thousands of different long non-coding RNAs in a cell or tissue, we have to make a decision about which one we want to test. But to do that, we already need to have developed a hypothesis about what that specific long non-coding RNA might do in the cell. Without that hypothesis, we won’t know what effects we should be looking for if we interfere with the expression or function of that molecule.
Another complication is that many of the long non-coding RNAs are found in the same region as classical protein-coding genes. Sometimes they may be in exactly the same position, but encoded on the opposite strand, just as we saw for Xist and Tsix in Chapter 7. Others may be found within the stretches of junk that lie between two amino acid-coding regions in a single gene, which we first encountered in Friedreich’s ataxia in Chapter 2 (see page 18). There are lots of ways in which the long non-coding RNAs may be co-located in the same region as protein-coding genes and this creates substantial experimental difficulties if trying to investigate function.
Usually the functions of genes are tested by mutating them. There are all sorts of mutations that can be introduced but the most commonly used will either switch the gene off or will lead to it being expressed at a higher level than normal. But because so many of the long non-coding RNAs overlap with protein-coding genes, it’s hard to mutate one without mutating the other at the same time. We then face the problem of knowing whether the effects we see are due to the change in the long non-coding RNA or in the protein-coding gene.
A frivolous analogous example may help to visualise this problem. A PhD student was investigating how frogs hear. He had developed an experimental system where he surgically removed certain parts of a frog and then monitored if it could hear a loud noise, in this case a gunshot. One day he rushed in to his supervisor’s office, yelling that he had worked out how frogs hear. ‘They hear with their legs!’ he told his bemused supervisor. When she asked how he could be so sure he said, ‘It’s simple. Normally if I fire the gun, the frog hears it and jumps in fright. But when I remove the frog’s legs it doesn’t jump anymore when I fire the gun, so it must hear through its legs.’*
Theoretically, of course, it’s also possible that some of the unexpected effects sometimes encountered when we mutate protein-coding genes have been due to unrecognised changes in co-located long non-coding RNAs which we hadn’t even realised were present at the time the experiment was carried out.
Because of this potential collateral damage to protein-coding genes, many researchers are focusing their efforts on a subset of long non-coding RNAs which don’t overlap these regions. There’s plenty of choice, as there are at least 3,500 long noncoding RNAs in this category. There is a tendency in the literature to refer to these more distant long non-coding RNAs as a special class, and they have been given a separate name.**12 But it’s worth remembering that if we do this, we are classifying these molecules by what they are not, i.e. they aren’t co-located with protein-coding genes. This could mean that we lump together large numbers of long non-coding RNAs in one class when really they may turn out to be functionally quite distinct from each other.
The rush to create categories and nomenclature has been, and continues to be, a real problem in the whole field of genome analysis because it tends to lock us in to definitions before we really have enough biological understanding to create relevant categories. Imagine if you had never seen a movie, and then you were treated to a week of films. Let’s imagine you see Top Hat; Singin’ in the Rain; The Good, the Bad and the Ugly; High Noon; The Sound of Music; The Magnificent Seven; Cabaret; True Grit; Unforgiven and West Side Story. If asked to categorise movies, you would say they come in two flavours: musicals and westerns. That’s fine, but what happens in the following week if you are shown Bridget Jones’s Diary and Gravity? Or Paint Your Wagon, Seven Brides for Seven Brothers and Calamity Jane, all of which are song-and-dance films involving cowboys? You’ll be stuck trying to shoehorn movies into genre definitions you developed before you understood the cinematic landscape. For a similar reason, we’ll try to avoid too many definitions of individual classes of long non-coding RNAs and just focus on what we really know experimentally.
The importance of a good start in life
Appropriate control of gene expression is required throughout life, but it’s critically important in very early development, because even the slightest shift in events during the first few cell divisions can have dramatic effects. This is particularly true in the zygote, the single cell formed from the fusion of an egg and a sperm. The zygote, and the first few cells generated by division from this progenitor, are known as totipotent. They are able to create all the cells of the embryo and placenta. Researchers would love to work with these cells, but they are tiny in number. Instead, most research is carried out in embryonic stem cells, also known as ES cells. These were originally derived from embryos, many years ago, but we don’t need to access embryos any more to get them, as they can be grown in cell culture. ES cells are from a slightly later stage in development and aren’t quite as unconstrained as the zygote. They are known as pluripotent, as they have the potential to form any cell type in the body, but not placental cells.
In the correct, carefully controlled culture conditions, ES cells divide to generate yet more pluripotent stem cells. But relatively minor changes to the culture conditions lead to a loss of pluripotency. The ES cells begin to differentiate into more specialised cell types. One of the most dramatic changes is when ES cells differentiate into heart cells, which beat spontaneously and in synchrony in a Petri dish. But essentially the ES cells can move down many different development routes, depending on the ways that they are treated.
Researchers manipulated ES cells in culture by knocking down the expression of nearly 150 of the long non-coding RNAs that are located far from any known protein-coding genes. They knocked down the expression of just one long non-coding RNA in each experiment. They found that in dozens of cases, knockdown of just one long non-coding RNA was enough to change the ES cells from being pluripotent to starting to differentiate into other cells. The authors also analysed which genes were expressed before and after they knocked down the long non-coding RNAs. They found that over 90 per cent of the long non-coding RNAs controlled expression of protein-coding genes either directly or indirectly. In many cases, the expression of hundreds of protein-coding genes was affected. These were nearly always genes that were far away on the genome, not the ones that were closest to the long non-coding RNAs that they had knocked down.
The scientists also performed the reciprocal experiment. They treated ES cells with a chemical that is known to cause them to differentiate and then analysed the expression of the specific long non-coding RNA class in which they were interested. They found that expression of about 75 per cent of the long non-coding RNAs dropped as the cells moved from being pluripotent to being committed to a development pathway. The two sets of data are consistent with the idea that the levels of expression of certain long non-coding RNAs act as gatekeepers to maintain ES cells in a pluripotent state.13 This created confidence that these nonprotein-coding RNAs do have a function in the cell, at least during early development.
Some long non-coding RNAs may also affect later developmental stages. We met the HOX genes in Chapter 4. These are the genes that are important for correct patterning of body parts. They’re the ones where mutations in fruit flies can lead to bizarre effects such as legs on the head. HOX genes are found in clusters in the genome, and these regions are extraordinarily rich in long non-coding RNAs. This is in contrast to their lack of ancient viral repeats. Scientists were keen to investigate if the long noncoding RNAs influenced the activity of the HOX genes in the same place in the genome. To test this, researchers used a technique to decrease the expression of a specific long non-coding RNA from the HOX gene region in chick embryos. When they did this, limb development went wrong. The bones towards the ends of the limbs were abnormally short.14 Similarly, knocking out expression of another long non-coding RNA from this genome region in mice resulted in animals with malformations of the bones of the spine and wrists.15 Both sets of data are consistent with the long non-coding RNAs being important regulators of HOX gene expression, and consequently of limb development.
Cancer can in some ways be thought of as the flip side of development. One of the problems in cancer is that mature cells may change and revert to having some of the characteristics of less specialised cells, with a higher capacity to divide uncontrollably. Given that long non-coding RNAs are important in pluripotency and in development, it’s perhaps not surprising that some have now been implicated in cancer.
One large study analysed the expression of long non-coding RNAs in over 1,300 individual tumours from four different cancer types (prostate, ovarian, a type of brain tumour called glioblastoma and a specific form of lung cancer). There were about 100 long noncoding RNAs where high levels of expression were most commonly found in patients who died quickly from the disease. Nine of these long non-coding RNAs showed this association no matter the class of cancer that was assessed, which suggests they may be useful as more general markers for predicting survival chances in a patient.16
For three of the cancer types (prostate cancer was the exception), the same study reported that they could detect long non-coding RNAs that differentiated one sub-class of tumour from another. Although we refer to ovarian cancer, for example, there are different types of ovarian cancer depending on the cell types involved, and this affects the natural history of the tumour in a patient. This in turn can have implications for the disease prognosis and the treatment that a patient should receive. Analysing the expression of specific long non-coding RNAs in a tumour sample may help clinicians in the future to select the most appropriate therapies for an individual patient.
The number of studies that report associations between long non-coding RNA expression and cancer are growing all the time. Intriguing data are also emerging from genetic studies of cancers. Some cancers are caused by a single really strong mutation which is passed on within a family. Probably the best-known example is the mutated BRCA1 gene which puts women at very high risk of aggressive breast cancer. It was knowing that she had a mutation in this gene that led the actress Angelina Jolie to elect for a double mastectomy in 2013. Such very strong single gene mutations are pretty rare in cancer. But studies have shown that quite a number of cancers do have a genetic component. The problem has been that when scientists mapped where the genetic variations were that were associated with cancer risk, they were frequently in regions of the genome where there were no protein-coding genes. Of just over 300 genetic variations linked to cancer, only 3.3 per cent changed amino acids in a protein, and over 40 per cent were located in regions between classical protein-coding genes. In these situations the variations may be affecting not protein-coding genes but long non-coding RNAs. Recent studies have confirmed this is the case for some of these variations in at least two cancer types (papillary thyroid cancer and prostate cancer).17
Encouragingly, we are also beginning to gather functional data that shows in some cases that these relationships are more than just associations, that the long non-coding RNAs are themselves causing alterations in the behaviour of the cancer cells.
There is a long non-coding RNA whose expression is increased in prostate cancer. This over-expression causes decreased expression of key proteins that normally hold cells back from proliferating too fast.18,19 Over-expression of this long non-coding RNA is therefore essentially like releasing the handbrake on a car parked facing down a hill. The long non-coding RNA that causes skeletal deformations when it is knocked out in developing mice is over-expressed in a variety of cancers including liver,20 colorectal,21 pancreatic22 and breast23 and its over-expression is associated with poor prognosis for the patients. Studies using cancer cells in culture in the lab suggest that the over-expression of this long non-coding RNA may make the cells more likely to migrate and invade other parts of the body.
Some of the strongest data confirming that long non-coding RNAs are actively involved in cancer, rather than just carried along for the ride, come from prostate cancer. When prostate cancer begins to develop, its growth depends on the male hormone, testosterone. Testosterone binds to a receptor and this leads to activation of various genes that promote cell proliferation. Testosterone binding to its receptor is like you putting your foot down on the accelerator pedal of your car. Prostate cancer is initially treated using drugs that stop the hormone binding to its receptor. This is like having something between your foot and the accelerator pedal, so that you can’t press down on it to make the car go faster.
But over time, the cancer cell frequently finds a way around this. The hormone receptor finds ways of activating genes irrespective of whether there is testosterone around or not. It’s as if someone has put a bag of sugar on top of the accelerator. The pedal is always pressed down and speeding up the car, even if you have your feet on the dashboard. Two long non-coding RNAs that are highly over-expressed in aggressive prostate cancer have been shown to play a critical role in this process. They assist the receptor, driving gene expression even when there is no hormone around, and accelerating cell proliferation. They play the role of the bag of sugar in the car simile. If expression of these specific long non-coding RNAs is knocked down in cancer models, the tumours show a really dramatic decrease in growth, supporting the critical role of these molecules.24
Another long non-coding RNA has also been implicated in prostate cancer. The higher the levels of this long non-coding RNA, the more aggressive the cancer, the shorter the recurrence time after treatment and the greater the risk of death. Knockdown of this long non-coding RNA has a similar protective effect in cancer models to that described above, but in this case the effects do not seem to be due to interactions with the testosterone receptor.25 This indicates that long non-coding RNAs may affect cancer progression in different ways, even in one tumour type.
Long RNAs and the brain
It isn’t just cancer specialists who are interested in the functions of these molecules. More long non-coding RNAs are expressed in the brain than any other tissue (with the possible exception of the testes).26 Some have been conserved from birds to humans, with expression patterns that occur in the same regions and at the same developmental stages. These may have conserved functions, perhaps in normal brain development. However, many of the long non-coding RNAs expressed in the brain are specific to humans or primates, and this has led researchers to wonder if they could be responsible, at least in part, for the hugely complex cognitive and behavioural functions found in higher primates.27
A long non-coding RNA has been identified that influences how the cells in the brain form connections with each other.28 Another long non-coding RNA, which has evolved since we diverged from the other great apes, may be involved in regulating a gene that is required for the unique developmental processes that generate the human cortex.29
The examples above all suggest that long non-coding RNAs play beneficial roles in the brain. But they may also be implicated in pathology as well as in health. Alzheimer’s disease is the devastating dementia which is usually associated with ageing. Because the human population is generally living longer, Alzheimer’s disease is becoming increasingly common. The World Health Organization estimates that over 35 million people worldwide are suffering from dementia, and that this number will double by 2030.30 There is no cure, and even the drugs that are available, which slow down the clinical progression, don’t stop it altogether, let alone reverse it. The emotional and economic costs of this condition are enormous, but progress in treating it is horribly slow. This is partly because our understanding of what exactly is going wrong in the brain cells of sufferers is still poor.
We are fairly confident that we know that at least one important step in the process is the production of insoluble plaques in the brain, which can be detected at autopsy. These are made of mis-folded proteins, one of the most important of which is called beta-amyloid. This is generated when an enzyme called BACE1 slices up a larger protein. A long non-coding RNA is produced from the same place in the genes as BACE1, but from the opposite DNA strand, rather like the relationship between Xist and Tsix.
The long non-coding RNA and the standard BACE1 messenger RNA bind to each other. This makes the BACE1 messenger RNA more stable so it stays in the cell for longer. Because it stays around for longer, the cell can generate more copies of the BACE1 protein. This leads to increased production of the beta-amyloid that is essential for the formation of the plaques.31
It’s been reported that the levels of this long non-coding RNA are increased in the brains of patients with Alzheimer’s disease, but it’s difficult to interpret these data. This could just be a consequence of increased expression in that region generally. Remember the earlier analogy – the more you chop up logs, the more sawdust you create. But researchers managed to find a way of specifically decreasing the expression of just the long non-coding RNA in a mouse model which frequently develops Alzheimer’s pathology. The knockdown of the long non-coding RNA resulted in decreased BACE1 protein and fewer beta-amyloid plaques. This supports the idea that the long non-coding RNA may play a causative role in this devastating disease.32
It’s not just the central nervous system that can be influenced by long non-coding RNAs. Neuropathic pain is a condition in which the sufferer feels pain, even when there is no physical stimulus. It’s caused by abnormal electrical activity in the nerves that conduct signals from the periphery of the body into the central nervous system (brain and spinal cord). It can be a very distressing condition for sufferers, and normal painkillers such as aspirin or paracetamol don’t really help. It’s often not clear why the nerves are behaving abnormally. Recent work has suggested that in some cases it may be due to increased levels of a long non-coding RNA changing the expression levels of one of the electrical channels. It does this by binding to the messenger RNA molecule that codes for the channel, altering its stability and hence changing the amount of protein produced.33
The types of conditions in which it’s been claimed long non-coding RNAs play a role is growing all the time.34 But the controversy remains over how functional and important these long non-coding RNAs really are. Can they really be as important as proteins? Perhaps on an individual basis the answer is usually ‘no’ unless we are dealing with a molecule as unequivocally vital as Xist. But thinking about the impact in terms of single long non-coding RNAs may be missing the point.
A recent commentary suggested that ‘A distinct possibility is that many of the long transcripts are, at best, nudgers and tweakers of genome management, rather than switches per se.’35 But the greatest complexity and options for flexibility come not from on/off or black/white but from subtle changes in sound levels or from multiple shades of grey. Biologically, we may owe an awful lot to our nudges and tweaks.


