Several powerful laboratory techniques and associated computer software programs are available for conducting genetic parentage analyses (i.e., for ascertaining who parented whom in nature) (Avise 2004b). Especially for species with pregnancy or brooding, the logic of such analyses is straight-forward. Typically, each embryo within an egg case or a nest or inside a gestating parent is genotyped at each of several highly polymorphic Mendelian loci, as is the custodial adult. Then, by comparing the diploid genotype of each offspring with that of its known or suspected biological parent, at each locus the allele that came from the nonpregnant parent usually becomes evident by subtraction (or default). An example of this approach is diagrammed in the accompanying figure (modified from Avise 2002). In this case, a pregnant female and the 19 embryos she carried were genotyped at a locus that proved to display six different alleles (A–F) within the brood. Because the known dam was heterozygous for the A and D alleles, as expected about 50% of the progeny proved to carry maternal allele A, and the other 50% carried maternal allele D. Barring de novo mutation, this also means that all other alleles (B, C, E, and F) in the brood must be of paternal origin. Thus, at least two different males must have mated with the female and genetically contributed to this litter (assuming that each sire was heterozygous for a distinct pair of alleles not shared by the mother).
By extending this approach to multiple polymorphic loci, the multilocus genotype of the sire(s) can often be deduced, and such genetic data thereby permit estimates of the numbers of fathers and their proportionate contributions to each brood. For male-pregnant species such as pipefishes, the same logic applies except that in this case the biological sire is known, and the primary intent of the genetic assays is to deduce the maternity of each brood. By accumulating such genetic information for many broods or litters, researchers can estimate the genetic mating system of a population (chapter 7).
Similar kinds of logic apply to genetic paternity exclusions in nest-tending or externally brooding species when particular offspring sometimes display alleles incompatible with those of their male custodian. Cuckoldry (stolen fertilizations by other males), nest piracy, and egg theft are among the behavioral possibilities that may account for such cases of male foster parentage, and these can often be distinguished in particular instances by considering details of the natural-history setting (Avise et al. 2002). Finally, for nonpregnant species in which neither parent is available for genetic examination (as is generally true for organisms that lack parental care), the statistical exclusionary power and the capacity to draw definitive conclusions about genetic parentage are diminished considerably.
In practice, the methodology of parentage analysis is not quite as simple as implied earlier. For example, technical laboratory difficulties sometimes arise in identifying polymorphic loci with sufficient resolving power for the task at hand. The most popular molecular markers in recent years have been provided by microsatellite loci, which sometimes display a dozen or more codominant alleles per locus in natural populations of many species. Each microsatellite locus consists of multiple tandem-repeat units (each of which is 2–5 base pairs long), in which the different alleles are distinguished by how many such units they display. Statistical challenges also exist with respect to extracting reliable parentage information from the molecular data. Many statistical methods have been developed in recent years, each tailored to particular biological settings (reviewed in Jones and Arden 2003; Jones et al. 2010). Notwithstanding such laboratory and statistical complications, the fact remains that such difficulties are greatly alleviated for the many brooding species in which one of the genetic parents of each offspring is known or suspected at the outset from independent biological information (the pregnancy itself).
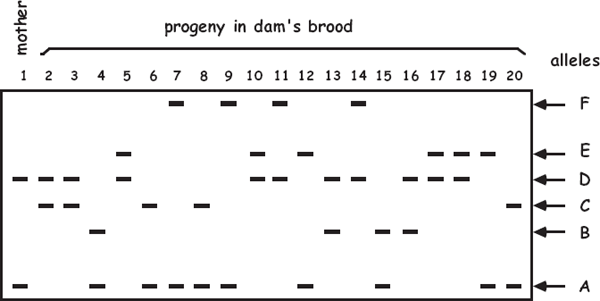
Diagram of a microsatellite gel showing, at one locus, a mother’s genotype (leftmost column) and genotypes of 19 offspring in her brood. See the text for further explanation.
Molecular-parentage analyses have been conducted on numerous litters from many vertebrate and invertebrate species that display either standard pregnancy or some other form of brooding by a “pregnant” parent. The following table summarizes genetic findings from representative surveys.
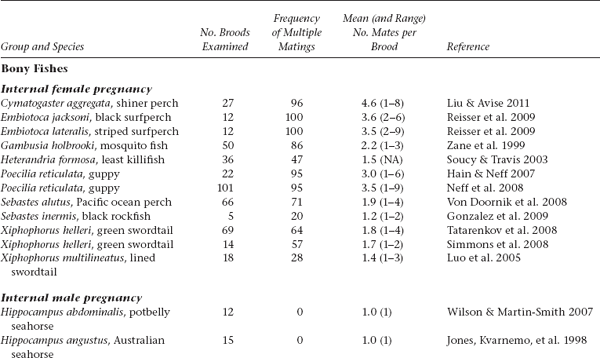
APPENDIX TABLE 1 Examples of reported numbers of successful mates and frequencies of multiple mating by the pregnant gender in various animal groups.
All data summarized in this table came from studies that used highly polymorphic molecular markers (typically microsatellite loci) to deduce genetic parentage within the broods carried by pregnant females (or in some cases carried or tended by pregnant males). For a more exhaustive compilation for fishes, see Coleman and Jones 2011.