
In order to effectively make use of DNA testing for genealogy it is essential to understand how DNA is inherited. This chapter will set out the inheritance patterns of the four different types of DNA covered by commercial DNA tests: mitochondrial DNA, Y-chromosome DNA, X-chromosome DNA and autosomal DNA. You do not have to be an expert in genetics to understand these concepts; it is all about grasping the basics and using that information as an aid to your DNA results.
Mitochondrial DNA (mtDNA for short) consists of small circular pieces of DNA present within our cells and is the only type of DNA that does not originate in the nucleus of the cell. It exists in the body in vastly greater quantities and is far more durable than nuclear DNA. We all have hundreds or even thousands of copies of mitochondria per cell as opposed to just forty-six chromosomes per cell as is the case with nuclear DNA.
MtDNA is passed down virtually unchanged for hundreds of years and can reach back around 200,000 years in time, which is a similar reach to Y-DNA and a much longer reach than atDNA. It mutates very slowly, so even exact mtDNA matches can be beyond a genealogically relevant timeframe, meaning it is often not possible to identify a common maternal ancestor. The plentiful nature and longevity of mtDNA is why it has been the preferred testing method for ancient DNA cases e.g. the discovery of Richard III.
The unique inheritance pattern of mtDNA makes it useful for researching the direct maternal line i.e. the mother’s, mother’s, mother’s line. MtDNA is passed down by a mother to all of her children (both sons and daughters), but only daughters then pass it on in turn to their children. The same strand of mtDNA, therefore, perpetuates until the interjection of a male breaks the mtDNA line as his children will inherit the mtDNA of their mother.

To test your mother’s mtDNA you can test yourself, but if you wish to test your father’s mtDNA you would have to test your father himself or an appropriate relative on your paternal side who shares his mtDNA, e.g. any of his full or maternal half siblings or a niece/nephew descended from a sister.
Mitochondrial DNA is also the only type of DNA for which the standard test is a full-sequence test (offered by FTDNA) – this means that the entire mitochondria (16,569 base pairs) is sequenced and not just select areas on it, as is the case with Y-DNA and atDNA. For a more detailed look at mtDNA testing see Chapter 6.
There are two sex chromosomes (X and Y) and each person inherits one from their mother and one from their father. Your sex is determined by which one you inherit from your father. The Y-chromosome is the sex chromosome that determines the male gender, so if you inherit a Y-chromosome from your father then you will be male. Only men have a Y-chromosome, so only men can pass it down.
The inheritance pattern of the Y-chromosome is very straightforward: it is passed from father to son down a direct male line and Y-DNA testing, therefore, covers only the father’s, father’s father’s direct paternal line.
Y-DNA is passed down virtually unchanged through many generations and for that reason it can, like mtDNA, reach back over 200,000 years in time.
Only men can take a Y-DNA test, so if a woman wishes to test her father’s Y-DNA line she will have to recruit an appropriate male to take the test for her. Due to the nature of Y-DNA it is possible for Y-DNA lines to die out, and this is commonly referred to as ‘daughtering out’: it occurs when a man only has daughters and, therefore, does not pass on his Y-chromosome to a son. Once he dies his individual Y-DNA line dies with him. That does not mean that his Y-DNA line is untestable, however, until all avenues have been exhausted. Due to the fact Y-DNA is slow-mutating, you could find a suitable Y-DNA donor many generations removed who shares the same Y-chromosome as the man whose line has daughtered out. For example, your father is deceased and you are his only daughter so you cannot test his Y-DNA line. He has no brothers, nephews or paternal first cousins, but by tracing his direct paternal line back to his third great-grandfather and then tracing just the male lines forward you could find a suitable male fourth cousin who shares your father’s Y-DNA.
Y-DNA testing is useful for surname studies, testing relationship hypotheses on the direct paternal line, finding cousins, solving paternal mysteries and investigating the ancient origins of your direct paternal line. For a more detailed look at Y-DNA testing, see Chapter 5.
The X-chromosome is the other sex chromosome and both men and woman inherit at least one of these. Since women have two X-chromosomes it is only possible for mothers to pass down an X to their children. If you inherit an X-chromosome from your mother and a Y-chromosome from your father you will be male. If, instead, you receive an X-chromosome from both your mother and your father then you will be female.
The unique inheritance pattern of the X-chromosome makes it very helpful when it comes to DNA cousin matching. If you discover a reasonable match with someone on the X-chromosome there are far fewer ancestors you need to look at in order to pinpoint the connection.
Here is a chart showing all of the ancestors from whom a male potentially inherits X-DNA:
https://thegeneticgenealogist.com/wp-content/uploads/2008/12/MaleChart350dpiVersion2-1.jpg © Blaine Bettinger
Since men do not inherit any X-DNA from their fathers, the entire paternal side of their tree can be discounted when trying to identify common ancestors with an X-DNA match. Any other male-to-male line can also be discounted, as there will never be any X-DNA inheritance between a father and son – e.g. the maternal grandfather’s paternal line can always be ruled out. Another key point is that just because you could potentially have received some X-DNA from a certain ancestor, it does not follow that you definitely did. For an X-DNA match a man only has to look at eight out of his thirty-two third great-grandparental lines thus narrowing the connection down significantly.
Here is a chart showing all of the ancestors from whom a female potentially inherits X-DNA:
https://thegeneticgenealogist.com/wp-content/uploads/2008/12/FemaleChart350dpiVersion2.jpg © Blaine Bettinger
Women have double the X-DNA men do, so they also tend to have far more X-DNA matches and more lines that need to be included in any investigation. It is not possible to narrow a female X-DNA match down to a side in the way you can with a male X-DNA match, but there are still many lines that can be ruled out. In particular the paternal grandfather line can be instantly discounted, thus removing an entire quarter from the equation and leaving women with only thirteen of their thirty-two third great-grandparents’ lines to investigate.
If you wish to test your X-DNA you cannot purchase a standalone test like you can for Y-DNA – instead X-DNA is generally incorporated into autosomal DNA tests. All autosomal testing companies include the X-chromosome during testing, but not all report on it. 23andMe and FTDNA do report on it, but Ancestry and MyHeritage currently do not.
For a more detailed look at using X-DNA for genealogy, see Chapter 4.
Autosomal DNA is the all-rounder of the DNA testing world since it covers all of your ancestral lines as opposed to just one like Y-DNA and mtDNA. For instance you have sixteen second great-grandparents, but when you test Y-DNA or mtDNA you are exploring only one of those sixteen lines, whereas autosomal testing covers them all. These tests can be taken by both men and women and matches can be related to you on any of your lines.
You inherit 50% of your autosomal DNA from your mother and 50% from your father. Parent-child, however, is the only relationship that shares such an exact percentage of atDNA; all other percentages are approximate.
Autosomal DNA is structured by chromosomes (chr for short) – these are organized packages of DNA found within the nucleus of every human cell. There are forty-six chromosomes in total: twenty-two pairs of autosomal chromosomes (known as autosomes), and one pair of sex chromosomes (chr 23/X and Y).
The most important concept to take on board here is the fact that chromosomes come in pairs – twenty-three pairs overall. This means you have two copies of chromosome one, two copies of chromosome two and so on. Technically only chromosomes 1 to 22 make up autosomal DNA as the final pair (chromosome 23) are the sex chromosomes. Chromosomes are numbered from largest to smallest with chromosome 1 being the largest and chromosomes 21 and 22 being the smallest. The Y-chromosome is also reasonably small and the X chromosome is three times larger than it.
Why do we have two copies of each chromosome?
The reason for two copies of each chromosome is because one set of twenty-three is passed down by our mother and the other set of twenty-three is passed down by our father. It is best to think of these as your maternal chromosomes and your paternal chromosomes.
Your parents also have two copies of every chromosome passed down to them by their parents, but when they create you, they can only pass half of their chromosomes down. During meiosis (cell division that occurs at the creation of an egg or sperm cell) a process called recombination takes place.
This image depicts a daughter, but the process is exactly the same for a son except for the fact dad passes down a Y-chromosome.
Let’s look closer at how it works using just one chromosome (chromosome 1 for argument’s sake, but the number is unimportant):
You can see that on her maternal chromosome one she inherited far more from her maternal grandmother than her maternal grandfather, but there’s a slightly more even distribution on her paternal copy. This could easily be reversed on some of her other maternal chromosomes – recombination is random so there is no set pattern. This is why while you inherit exactly 50% of your DNA from each parent, you only inherit approximate percentages from more distant ancestors.
You inherit progressively less of your ancestors’ DNA with each passing generation and this dropping off of DNA from more distant ancestors highlights the main limitation of atDNA – it can generally only reach back five–seven generations in time.
When you share a piece of atDNA with another person we call that piece of DNA a segment; the segment lies somewhere within the start and end locations of the chromosome in question and can come in all sorts of sizes. The way we measure these segments is via centiMorgans (usually abbreviated to cMs for short). A centiMorgan is a measure of how probable it is that a segment will recombine from one generation to the next. The most important thing to remember about centiMorgans is that the more cMs you share with a match, the closer the relationship. For that reason, it is essential to become familiar with the average number of cMs you should share with different relationship levels. Additionally, it is important to understand that there can be quite a wide range due to the random way autosomal DNA is inherited and that outliers are always possible.
Familiarise yourself with the approximate amounts of DNA you should share with different relationships:

Statistics Courtesy of the ISOGG Wiki
The only absolute is that females inherit exactly 50% and males almost exactly 50% from each parent – males actually inherit 50.8% from their mothers and 49.2% from their fathers, because the paternally inherited Y chromosome is around three times smaller than the maternally inherited X chromosome. All the rest of these percentages are approximate; for example, the 50% you inherit from a parent may be a mix of 22% from your grandmother and 28% from your grandfather. It is all down to how the cards fall during the recombination process.
Then acquaint yourself with the average number of cMs shared by different relationship levels:
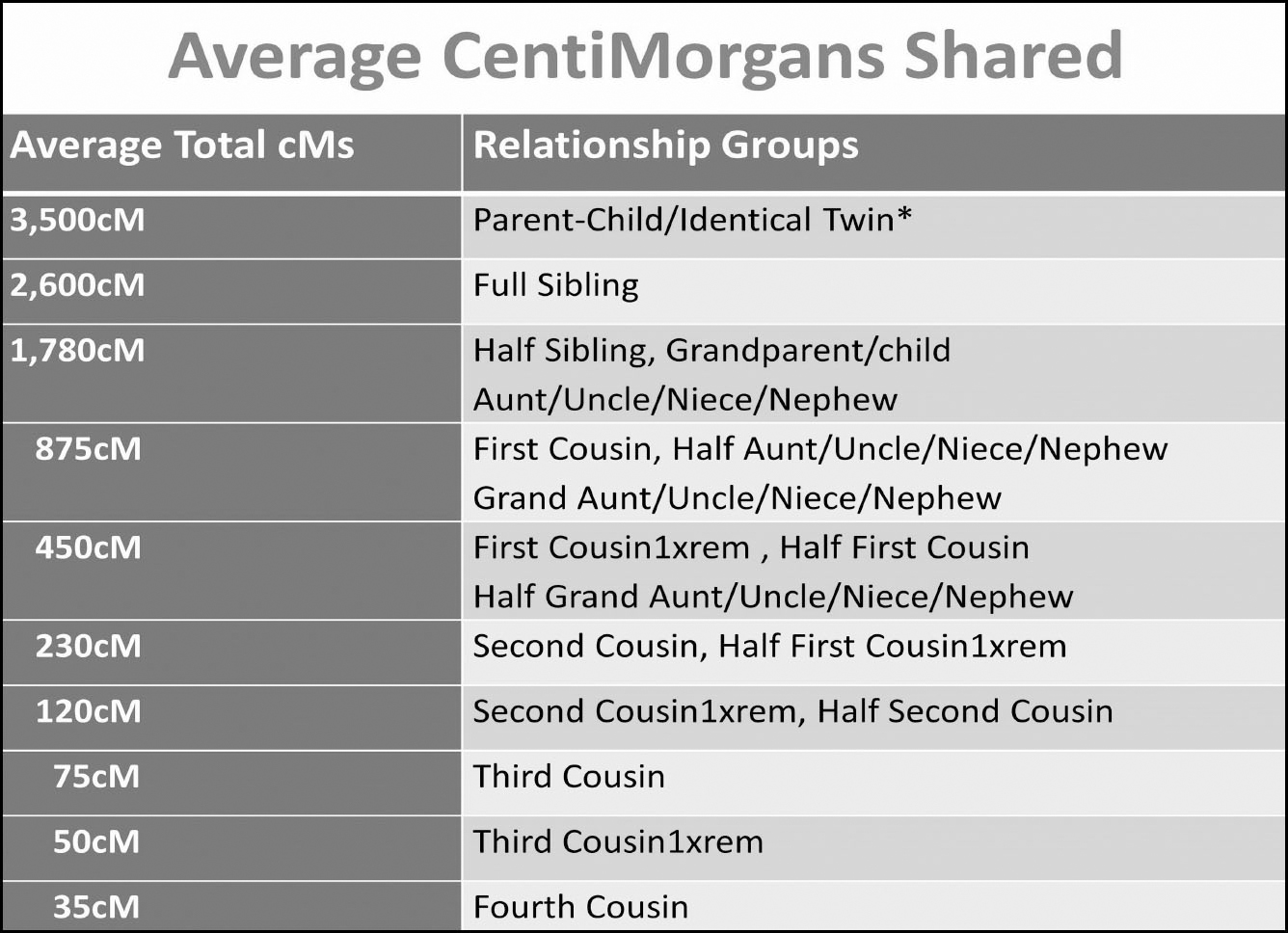
Compiled by author based on averages from Shared cM Project
*If one of your ancestors is an identical twin you will share more DNA with the descendants of the other identical twin e.g. the children of identical twins will share double the DNA of regular first cousins and will appear more like half-siblings.
There are two important points to take away from this table:
1. These numbers are just the average amount shared – in many cases the reality will be different and it is best to think in terms of ranges of cMs.
2. There are different groups of relationships that could share exactly the same amount of cMs e.g. a half-sibling is very hard to distinguish from an aunt/uncle/grandparent via the number of cMs shared alone, as all of those relationships should share a similar amount.
For details of a very useful resource to consult for ranges of cMs shared and relationship probabilities, see the description of the Shared cM Project in Chapter 4.
Understanding kinship terminology is important for researching DNA matches:
Removed
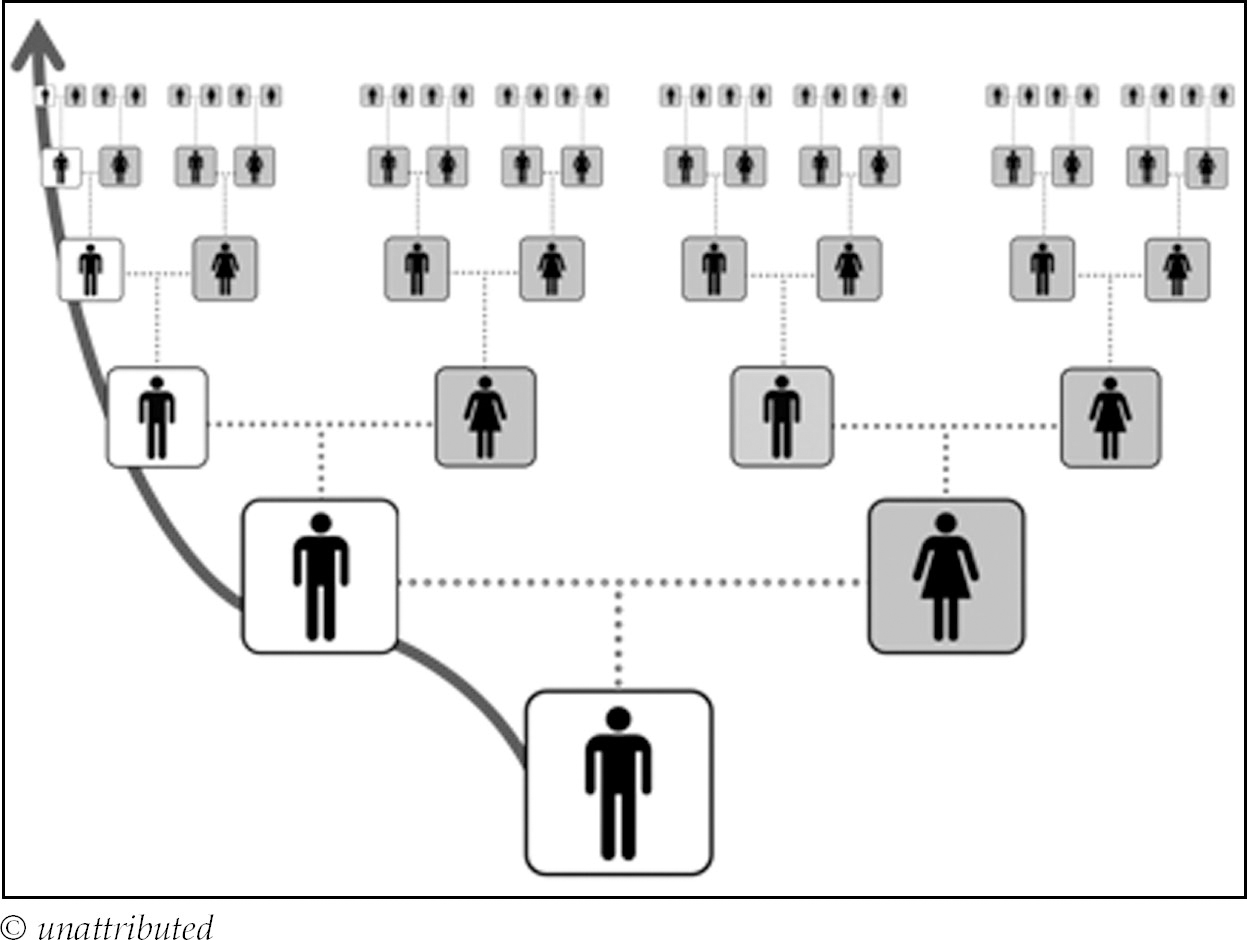
This simply means you and your cousin are not on the same genetic generation i.e. one or other of you is closer or more distant to your common ancestors. For example, second cousins share a set of great-grandparents and are both exactly three generations away from their common ancestors, so there is no ‘removed’ involved. A second cousin once removed (2C1R), however, is either one generation closer or one generation further away from the common ancestors – the great-grandparents of one must either be the grandparents or second great-grandparents of the other.
Half
Generally we do not tend to talk about half-cousins or halfaunts/uncles, but it is important to differentiate these relationships for DNA testing, since half relationship relatives will share 50% less DNA with you than their full counterparts. So what do we mean by a half first cousin? This is someone who only shares one grandparent with you as opposed to two and that happens when your parent and theirs are half-siblings. Similarly a half-aunt/uncle is your parent’s half-sibling. So when you see a complicated relationship like a half 3C2R this means that person shares one common ancestor with you at the third cousin level, but one of you is two generations closer to that common ancestor than the other. The cousin relationship is always dictated by the cousin who is closest to the common ancestors: for a half 3C2R this means the common ancestor is a second great-grandparent for one cousin and a fourth great-grandparent for the other. Whether you are the person who is closer or more distant makes no difference to the relationship calculation.
Once you know who the common ancestors are with a match, use a chart such as this to identify the actual cousin relationship:
Another key concept to understand is that overlap and outlier matches exist and it is always possible that any given match could fall into these categories. An overlap match shares in between two relationship levels, for example 875cM is the average shared between first cousins and 440cM between 1C1R or half first cousins, but what if someone shares 650cM with you? That is in the overlap zone between those two relationship levels and could just as easily be a first cousin as a 1C1R. Matches in the overlap zone are outliers: an outlier is a match that shares either significantly more or less cMs than average. There are a wide range of outliers from small to extreme.
Small outlier
A small outlier could be first cousins sharing 700cM on the low side or 1,000cM on the high side.
Extreme outlier
An extreme outlier could be first cousins sharing 570cM on the very low side or 1200cM on the very high side.
Don is a confirmed 2C1R match sharing 295 cM across fourteen DNA segments.
Mary is another confirmed 2C1R match sharing 13 cM across one DNA segment.
Don and Mary share exactly the same relationship with me, but the difference in the amount of cMs is enormous! These are extreme outliers at both ends of the scale and they demonstrate the need to be careful with relationship predictions.
How can I be so sure my low-sharing with Mary is an outlier, though, and there is not a problem with the relationship? There could be an error such as a misattributed parentage event. Equally perhaps the high-sharing with Don is indicative of a secondary relationship between us. The only reason I can be sure these scenarios are incorrect is because I have tested many other close relatives and the majority of them match both of these test-takers well within the average sharing range for the paper trail relationships.
My half-sister and, to a lesser extent, my brother and one first cousin also share far less than normal with Mary, but the majority of my first and second cousins share exactly in the correct range with both me and Mary, so I can classify this as an outlier situation. Being able to determine overlaps and outliers is just one of several excellent reasons for testing as many other family members as you can.
If a close relative (anyone up to second cousin level) does not match you then there is almost certainly a problem that needs to be addressed, as 100% of close relatives up to the second cousin level should match you and there have been, to date, no known instances of genuine second cousin or closer relations not matching each other. From third cousin onwards, however, there is an ever-increasing chance that genuine paper trail cousins may not share any DNA at all:
The reason for this takes us back to how autosomal DNA is inherited and the fact that we inherit progressively less from our ancestors the further back in time we go:
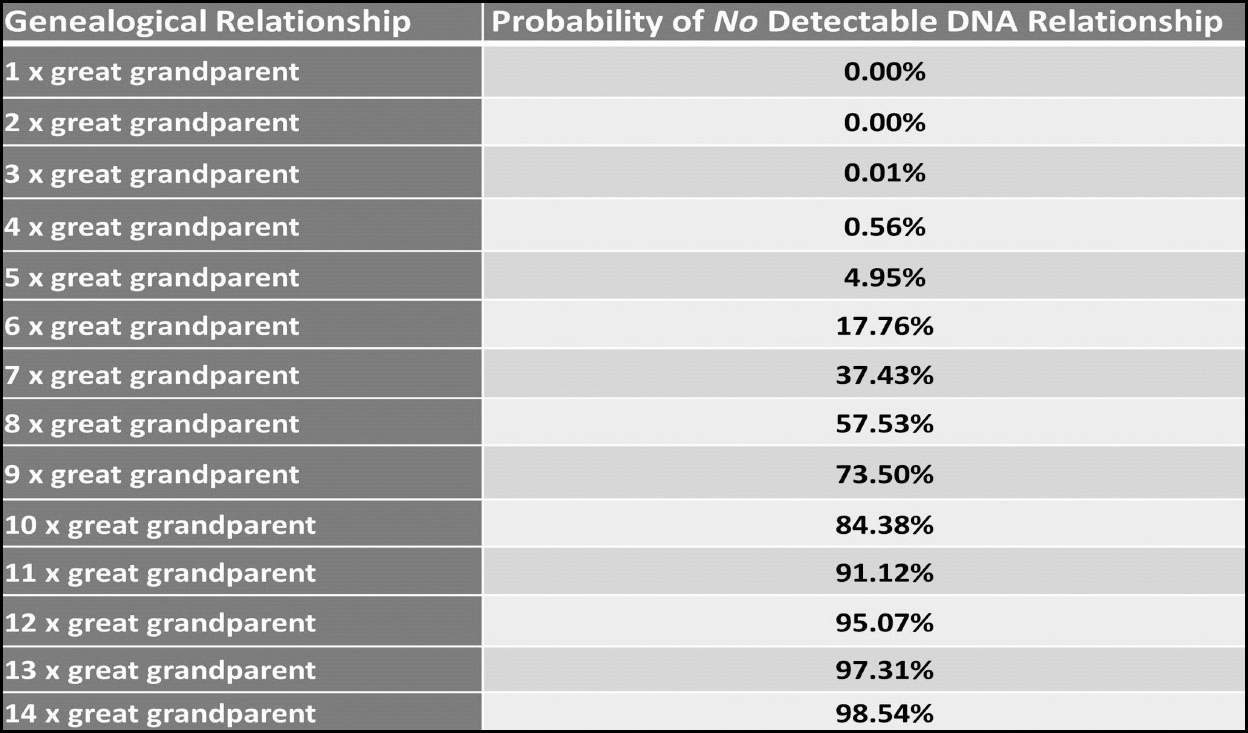
https://isogg.org/wiki/Cousin_statistics Probability of sharing DNA. © Jonathan Hamm
Past the point of fifth great-grandparent level, the probability of no detectable DNA increases exponentially. For example, we each have 128 fifth great-grandparents, and it is almost certain that we will have inherited no DNA from several of these. By the time we reach back to our 1,024 eighth great-grandparents we will have inherited DNA from less than half of them. This means we will have paper trail ancestors that are not genetic ancestors. These more distant ancestors still belong on our family trees – we just did not inherit any segments of DNA from them, so they are not part of our genetic make-up. The further back you can take your tree, the more ancestors who did not contribute to your DNA will be on it. In fact it has been calculated that almost half of our DNA comes from just 200 of our twentieth generation ancestors, the other half coming from around 1,200 ancestors.
Once we get back to the fourth cousin and more distant levels, the regular amount of sharing is one segment of DNA, if that. You are likely to inherit around 1.5% of your DNA from a fifth great-grandfather and a sixth cousin also descended from the same fifth great-grandfather equally will inherit approximately 1.5% of his DNA, and this means the chances that you will both have inherited the same segment are extremely small – your sixth cousin is much more likely to have inherited a piece of the 98.5% of your shared ancestor’s DNA that you did not and, therefore, will not match you.
Having said this, you will still have far more distant cousin matches than you will close ones. Here is a study undertaken by AncestryDNA on the number of cousins that the average British person is expected to have. It is unclear which parts of Britain the statistics cover, but the study was put together from information on British birth rates, census data, parliamentary research briefings and other sources on the last 200 years:
While you will only match a small proportion of your sixth cousins, far more of your matches will be sixth or more distant cousins, since there are so many more of them in existence.
Endogamy occurs when groups of people marry within the same ethnic, cultural, social or religious circles. This leads to everyone within that group descending from a much more limited gene pool. Descendants of an endogamous population end up with numerous cousin connections between them because the same ancestors appear on their family trees in a number of different locations.
The more recent the endogamy, the more it affects how you should work with your DNA matches. If you have extensive endogamy within your family tree, disregard predicted relationships and ranges of cMs, as distant cousins will share more DNA with you and the closer relationship predictions are likely to be wrong – if someone is your third cousin three times over, they are going to share more DNA with you than someone who is just your third cousin in one way. For those with endogamy, it is often necessary to increase the cM threshold at which they start investigating matches in order to identify connections e.g. a match of 50 cMs for someone from a non-endogamous background is much more likely to be solvable than for someone who has to factor endogamy into the calculation.
If you have substantial endogamy in your tree it is worth running the free ‘Are Your Parents Related?’ tool on GEDMatch. It checks your DNA for runs of homozygosity (ROHs) which are basically segments of DNA that are identical on both your maternal and paternal chromosome copies, meaning that your parents share those segments of DNA with each other and both passed them down to you.
Pedigree collapse occurs when marriages between cousins take place and some ancestors on your tree occupy more than one slot. We each have thirty-two third great-grandparents, but if your grandparents were second cousins and shared a set of great-grandparents that would mean the great-grandparents they shared belong on your tree twice. This, therefore, reduces or collapses the number of third great-grandparents you have down from thirty-two to thirty.
While there will always be pedigree collapse if endogamy is involved, not all pedigree collapse should be thought of as endogamy. Occasional cousin marriages can also affect DNA matching, but to a much lesser extent than within endogamous communities. The impact will depend on how recently the cousin match falls on your pedigree and the degree of cousin relationship between the cousins who married. For example, if a set of your grandparents were first cousins then that will affect the amount of DNA you share with cousins on that side of your tree much more significantly than if you have a set of second cousins who married at the third great-grandparent level. The latter, in fact, is unlikely to have much impact upon your DNA matches. The key difference between widespread endogamy and pedigree collapse due to occasional cousin marriage is that any effect on DNA matches is localized to the specific line on which the cousin marriage took place, while endogamy can permeate an entire tree.
Test as many of your older generation relatives as possible; they have more of your ancestors’ DNA than you do! Your parents have double and your grandparents have triple. You will share on average around 6.25% with a first cousin once removed (1C1R), but if that 1C1R is your parent’s first cousin (I call this the ‘magic’ generation closer to your common ancestors), then testing them amounts to testing around 25% of your great grandparents’ DNA while testing yourself only covers 12.5%.
There are many benefits to testing all older generation relatives, but parents are particularly useful because even if you can only test one parent, you can separate out your matches into either maternal or paternal – just be aware that very small segment matches could be false and match neither of your parents and you may also uncover matches to both.
Your parents will have a lot matches you do not, since many of them will stem from the 50% of their DNA you did not inherit. They will also regularly match people on your match list at a higher level than you do and this can bring a match to your attention that you might otherwise overlook.
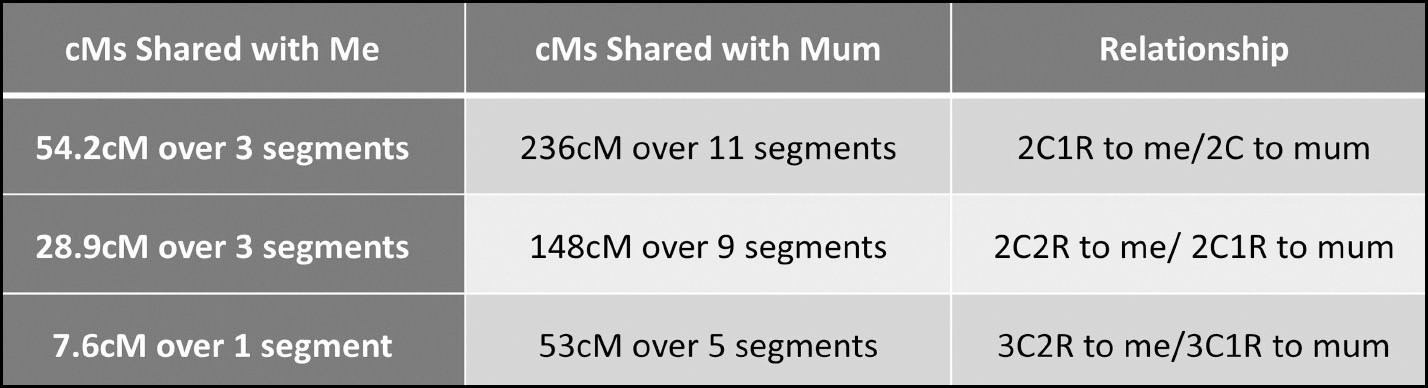
The 7.6cM match especially would not be one I would be likely to take notice of on my own list, but as soon as I see that they share over 50 cMs with my mum it suddenly becomes worth investigating.
This is one of the most common questions people ask when starting out on a DNA journey. Many think there is no point testing their siblings if they have tested themselves. The answer to the question, however, is ‘it depends!’ It can only actually be answered with a question in return: ‘do you have parents to test?’
If the answer to that is ‘Yes – I have both parents to test’ then, if they are willing, test both your parents. Your siblings’ DNA will not provide anything extra if you have tested your parents (unless you wish to perform visual phasing with a set of sibling results, but that is an advanced technique and not applicable to the beginner starting out – see Chapter 4). If, on the other hand, the response is ‘No’ or ‘Only one’, then the answer is a resounding ‘Yes – you should test your siblings!’. To explain why, we need to look back at how autosomal DNA is inherited:

 Siblings (including fraternal twins) share approximately 50% of their DNA
Siblings (including fraternal twins) share approximately 50% of their DNA
 Identical twins share all of their autosomal DNA so they are the exception to this rule
Identical twins share all of their autosomal DNA so they are the exception to this rule
 Using my playing card analogy this means the average siblings share around fifty DNA cards with each other
Using my playing card analogy this means the average siblings share around fifty DNA cards with each other
 It also means they each inherited around fifty DNA cards they do not share with each other
It also means they each inherited around fifty DNA cards they do not share with each other
 Two siblings, therefore, should expect to have inherited around 150 of their parents’ combined 200 DNA cards between them
Two siblings, therefore, should expect to have inherited around 150 of their parents’ combined 200 DNA cards between them
 Thus if you do not have your parents to test then testing just one sibling would get you approximately an extra 25% of your parents’ DNA to work with
Thus if you do not have your parents to test then testing just one sibling would get you approximately an extra 25% of your parents’ DNA to work with
 Their results will give you additional matches across all lines since they will match people you do not on segments of DNA from your parents that were not passed down to you personally
Their results will give you additional matches across all lines since they will match people you do not on segments of DNA from your parents that were not passed down to you personally
Here is a prime example of why testing siblings is so useful: Ben is a 23andMe match at the third to fifth cousin level: he is listed down near the bottom of my second page of matches and, according to 23andMe, we share 0.42% over two segments, which equates to 32 cMs. That is a reasonable match, but it is not going to be top of my investigation list any time soon. I have tested my brother, however, and his highest match after my mum, me and my half-sister is Ben! They share a whopping 159 cMs over seven segments in comparison to the 32 cMs shared with me. Ben now quickly shoots to the top of my investigation list. I immediately messaged him and it turns out he is my 2C1R. He shares a lot less with me than average for a 2C1R, but in the correct range with my brother. Without my brother’s test, even if I had contacted Ben and worked out the connection, I would have worried about potential issues in our line since we share so little DNA. Knowing he shares so much with my brother, however, excludes that possibility.
There are numerous terms associated with genetic genealogy which it is important to become familiar with. The following explanations should give you an understanding of many of these and their significance.
Single Nucleotide Polymorphism (SNP)
A SNP (pronounced ‘snip’) is a variation on a single DNA building block (nucleotide). Basically when a new cell is replicated and an error is made in the copying process that error is called a SNP – think of it like a spelling mistake. It is a selection of these SNPs across your entire genome that is actually tested when you take an autosomal DNA test. SNPs go hand in hand with cMs and, generally, if you share a large number of cMs with a match, you will share a large number of SNPs too. Just as there is no correlation between start and end locations and cMs, there is also no correlation between cMs and SNP density numbers. It is always best to base your research on the number of cMs.
Base Pairs (bps)
Nucleotide bases are the chemical building blocks of DNA that form the rungs on the double helix. There are four nucleotide bases – adenine (A), thymine (T), cytosine (C) and guanine (G) – and they pair up in specific ways to form base pairs, which are then used in the reporting of DNA results.
The straight letters (A and T) always pair with each other and the curved letters (C and G) do the same. This means that whenever we know which base is on one side of the helix, we automatically know the corresponding base on the other side. This is why when a sequence of DNA is reported only one side of the helix is presented, e.g. ACTGATG alone instead of ATCGTAGCATTAGC. It is estimated that there are 3.2 billion base pairs in chromosomal DNA, but only between 600,000 and 700,000 of these are covered by current autosomal DNA tests.
Phasing
This is the process of separating out the DNA on your maternal chromosomes from the DNA on your paternal chromosomes so that you can identify which came from mum and which came from dad. Phasing can be used on both autosomes and the X-chromosome. In order to phase your DNA as fully as possible it is necessary to test your parents – testing both parents (known as trio phasing) will achieve the best results, but it is possible to undertake phasing with the results of just one parent (duo phasing). GEDMatch, in particular, offers a phasing tool that will work if you have just one parent tested. Technically, phasing identifies which values (ATCG) came from your mother and which came from your father and separates them out. Here is a look at a section of unphased DNA:
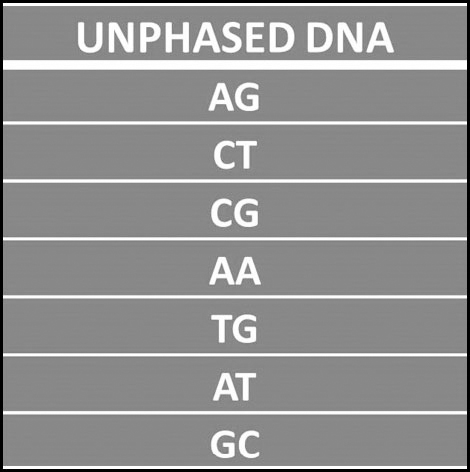
One of these values on each line came from the test-taker’s mother and the other from their father, but it is not possible to identify which is which from this result alone. Phasing the test-taker’s result against the results of their parents, however, can provide the answer:
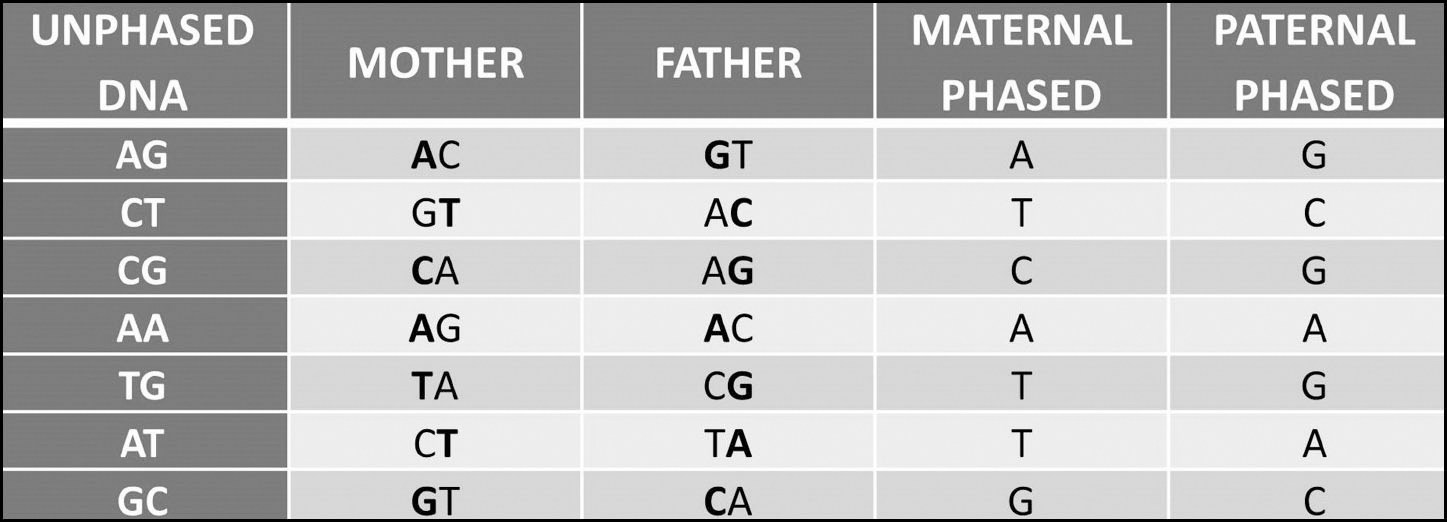
The main benefits of phasing are that it can separate your matches into maternal, paternal or both, identify false matches and aid with chromosome mapping.
Identical By Descent (IBD)
IBD segments are segments of DNA that have been passed down by a common ancestor. Research suggests 100% of segments that are 15 cMs or larger in size are IBD and the point at which 50% of segments are IBD is reached between 7 and 8 cMs. The smaller the segment, the higher the chance it is not IBD. Just the fact a segment is IBD, however, does not mean it will be easy to work out the common ancestors – for many smaller segments the connection will still be too far back to identify.
Identical by State (IBS)
IBS segments are either population segments or false segments. Since this term refers to two distinct types of segments it is problematic and can cause confusion. For this reason it is actually being phased out of genetic genealogy parlance. Two newer terms (IBP and IBC) have been created to combat this issue and it is best to use whichever of those is most appropriate for the particular segment you are investigating.
Identical by Population (IBP)
IBP segments are genuine segments that are common to a group of people from the same area. They are usually older segments that stretch beyond a genealogical timeframe, making it impossible to identify the common ancestors. Endogamous populations will also have many more of these segments.
Identical by Chance (IBC)
IBC segments are small segments that develop due to the chance mixing up of DNA values (alleles) and are, therefore, not real. In addition to IBC segments, there are a number of different terms for this phenomenon: false segments, false positives, pseudosegments, phantom segments and even frankensegments. The reason we end up with false segments is because the data is not phased. It is not possible to phase or separate out your maternal and paternal DNA segments without testing at least one parent, so in a number of your small segment matches the alleles will have been reported as a single sequence, but will actually be a mixture of alleles, some from your mother and some from your father. You, therefore, have to be alert to the fact that not all of your matches will be real cousins, as small segment matches could be IBC.
The easiest way to be 100% sure if a match is IBC or not is to test both your parents. If you are lucky enough to be able to do that and you find a match of yours does not match either of them, that raises a red flag. Many people sadly do not have that option, but it may be possible to find out that a segment is IBC by testing enough other close relatives. For example, while a match could just as easily be matching a full sibling in the same chance way they are matching you, a half-sibling could be extremely helpful. Since you only share DNA from one parent with a half-sibling, a match that you both share cannot be zigzagging back and forth between the DNA on your maternal and paternal chromosomes. This would also work with any other confirmed relatives with whom you only share one side of your tree – if the match also matches them on the same segment then you can eliminate the possibility of it being IBC.
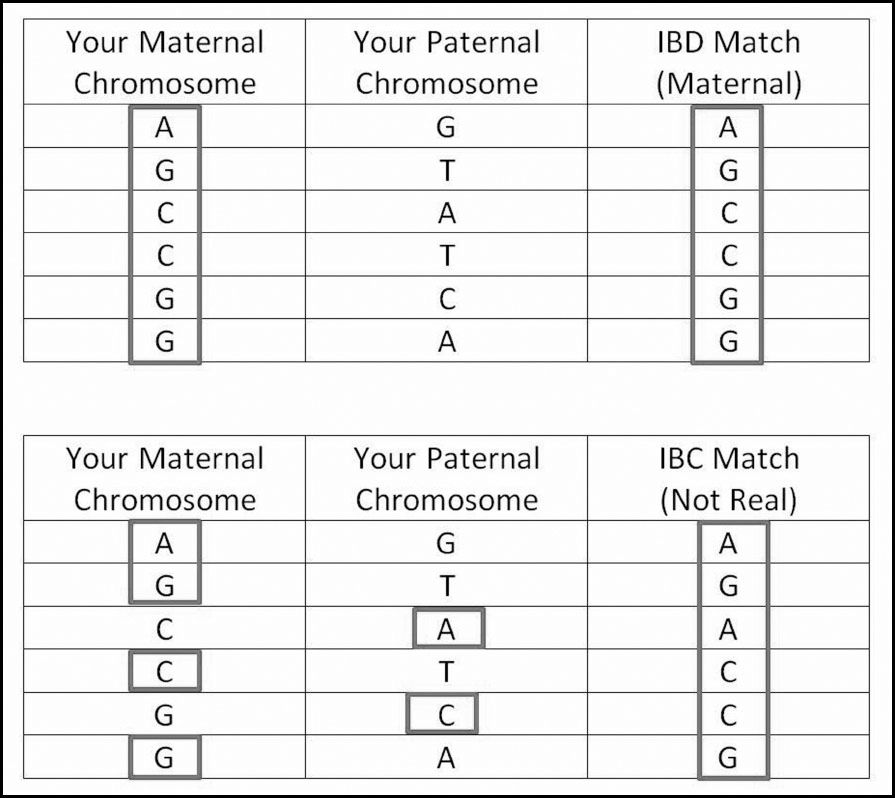
Equally, just working with larger matches generally precludes this from being a concern since the vast majority of IBC segments will be smaller than 7cMs. As a result, FTDNA’s use of small segments as small as 1cM is problematic (see Chapter 4 for how to deal with this). It is also why it is not advisable to lower the threshold below 7cMs on GEDMatch: you will share tiny segments with almost anybody if you lower the threshold far enough. This does not mean that we should completely ignore small matches (especially if common surnames/locations are present), but most people will have enough larger matches they can focus on before it is worth spending time on very small matches that could be IBC wild goose chases.
These are IBD segments of DNA that do not show up as a match due to an error in the testing process e.g. miscalls or no calls. If you have parents tested and a test-taker matches you but neither of your parents, the most likely explanation is that it is a false segment, but it is always possible there could have been an error on that particular segment during the running of the parent’s DNA and it is a genuine match after all. Only by seeing if this person also matches other close relatives who share the same segment with you and your parent can you distinguish between these likelihoods. False negatives, like false positives, will occur with smaller segments.
These occur during the testing process when a particular allele cannot be read and instead a blank value is recorded (usually as a dash or a zero).
Miscalls
These occur during the testing process when the wrong value is recorded for a particular allele.
A pile-up region (also known as excess IBD sharing) is an area of a chromosome where a large number of test-takers all match each other on the same segment of DNA but no common ancestors can be identified between them. Pile-ups are generally smaller in cM size and could be population-based segments. If you have access to segment data and find you are matching a large number of people on a particular segment, but you cannot identify common ancestors for any of the matches, then it is quite likely you are dealing with a pile-up.
An important point in relation to pile-ups and population segments is that AncestryDNA tries to exclude them from your matches. The company developed a proprietary algorithm called Timber, which attempts to eliminate segments that match large numbers of people. There has been a lot of debate since Timber’s introduction in 2014 as to how effective it is and it has been known to strip out some valid segments as well as pile-ups. If you test at different companies and find some of your Ancestry matches have also tested elsewhere, you will often notice that you appear to share more cMs with the same match on the different platform. Generally the reason for this will be that Timber has removed some segments on Ancestry.
Consult the ISOGG Wiki (https://isogg.org/wiki/Identical_by_descent) for a list of known excess IBD regions .
Number of Segments
The more segments you share with someone, the closer your relationship in general, since if you share more segments then you share more DNA overall. In the case of parents you share exactly twenty-three segments because you inherit entire chromosomes from them, whereas you may share significantly more than twenty-three segments with a sibling or aunt/uncle as segments are more broken up across the chromosomes. There is no pattern to the number of segments you share with a match, but the general rule is that if you share more than one significant segment you have a better chance of identifying a common ancestor. Be careful with the number of segments given on Ancestry, as the Timber algorithm has a tendency to break segments up and incorrectly inflate the number. When you see that you share 15 cMs over four segments with someone on Ancestry it could well be that it is actually just one segment broken up in this way.
One Segment Matches
Matches with whom you share just one segment of DNA are much more difficult to predict. For instance, a 20 cM one segment match has such a wide gamut of relationship possibilities – it could be an extreme outlier such as a 2C1R (as we have seen), but equally it could be anything from a third to a tenth or even more distant cousin, especially if a population or sticky segment is involved.
These are segments of DNA that pass down through a number of generations without recombination decreasing their size. Often you will find you and your parent share just the one segment with a match and it is the same size for you as it is for them; while many of these will not be sticky segments some of them certainly will be. These segments reach much further back in time since they have passed through several generations intact so it is extremely difficult to identify common ancestors.
Fuzzy Ends
Occasionally you will come across matches that share slightly more or less DNA with a parent than they do with a child. If the difference is only a few cMs then generally this can be attributed to a margin of error in the testing process, or what we call ‘fuzzy ends’. Every segment of DNA has a start and end location, but these can vary slightly from parent to child as reported in test results.
Non Identical Region (NIR)
This is a non-match – an area of DNA where you match someone on neither copy of your chromosomes.
Half Identical Region (HIR)
This is a segment of DNA shared on just one of your two copies of a chromosome. For instance the DNA I share with my mother is half identical, since I inherited all of my maternal chromosomes from her but none of my paternal chromosomes.
Fully Identical Region (FIR)
This is a segment of DNA that is shared on both copies of a chromosome. Outside of identical twins, who are fully identical across all chromosomes, the relatives with whom you will share the most FIRs are full siblings, since you each inherit significant amounts of DNA on both your maternal and paternal chromosomes. Double cousins and those with an endogamous background may also share more FIRs. You can view HIRs and FIRs on GEDMatch and 23andMe.
Now we are well-versed in how autosomal DNA works, Chapter 4 will explain how best to use it for genealogy.