 Confirming and supporting your family tree research
Confirming and supporting your family tree researchNow that we have covered the inheritance patterns of autosomal DNA, it is time to delve into how to work with autosomal DNA test results. This chapter will provide a practical guide on how to get the most out of these results for genealogy purposes.
Before we begin it is important to understand that you may discover something unexpected about your ancestry when you take an atDNA test. DNA is the one record set that does not and cannot lie or make mistakes, whereas human beings can and sometimes do. While those who receive surprising results are exceptions, you have to be aware of and prepared for this possibility when you choose to test. Additionally, while DNA cannot deliberately mislead, it can be misinterpreted and occasionally false conclusions are drawn from it. Look upon DNA as an additional evidence source that requires understanding, interpretation and most of all amalgamation with other research-based evidence.
Here is a summary of what you can use autosomal DNA testing for:
 Confirming and supporting your family tree research
Confirming and supporting your family tree research
 Breaking down brick walls (both direct and collateral)
Breaking down brick walls (both direct and collateral)
 Adding new branches to your tree
Adding new branches to your tree
 Discovering and connecting with new cousins
Discovering and connecting with new cousins
 Testing hypotheses about relationships and ancestors
Testing hypotheses about relationships and ancestors
 Solving adoption, unknown parentage and other unknown ancestor mysteries
Solving adoption, unknown parentage and other unknown ancestor mysteries
 Mapping your DNA back to your ancestors
Mapping your DNA back to your ancestors
 Learning about your ethnic make-up
Learning about your ethnic make-up
There are two distinct aspects to the results that you receive with an autosomal DNA test:
1. DNA match lists
2. Admixture estimates
When you take an autosomal test at AncestryDNA, Family Tree DNA (FTDNA), 23andMe, MyHeritage or Living DNA you are provided with a DNA match list that displays the names of others who have tested at the same company and share segments of DNA with you in accordance with the thresholds used by that specific company.
The DNA match list is by far the most important and genealogically useful part of an autosomal DNA test. Admixture information is initially interesting and exciting to look at, but it is via working with your DNA matches that you will get the most out of your autosomal DNA test results. You can continue to work with your DNA match lists indefinitely as they grow over time, while you will probably only look at your admixture estimates a handful of times.
No matter which company you have chosen to test with, the best way to begin working with your DNA match list is to start at the top, where your closest matches are located, and work down towards your more distant matches. Your closest matches share the most DNA with you, so theoretically they should be the easiest to work out and in general they will be, but there is always a chance either you or your match has an error on your tree or within your known ancestry that increases the difficulty level e.g. a research error or an NPE in a recent generation (NPE stands for non-paternity event, but I prefer ‘not the parent expected’, which is a phrase coined by genetic genealogist Emily Aulicino.)
Most of us will have so many distant matches that it is impossible to ever look at them all, and with databases expanding so rapidly more and more new matches are being added to our lists on a daily basis. In order to get the most out of the new matches, it is imperative to direct your efforts in the right areas and, in general, that means working with the highest matches possible.
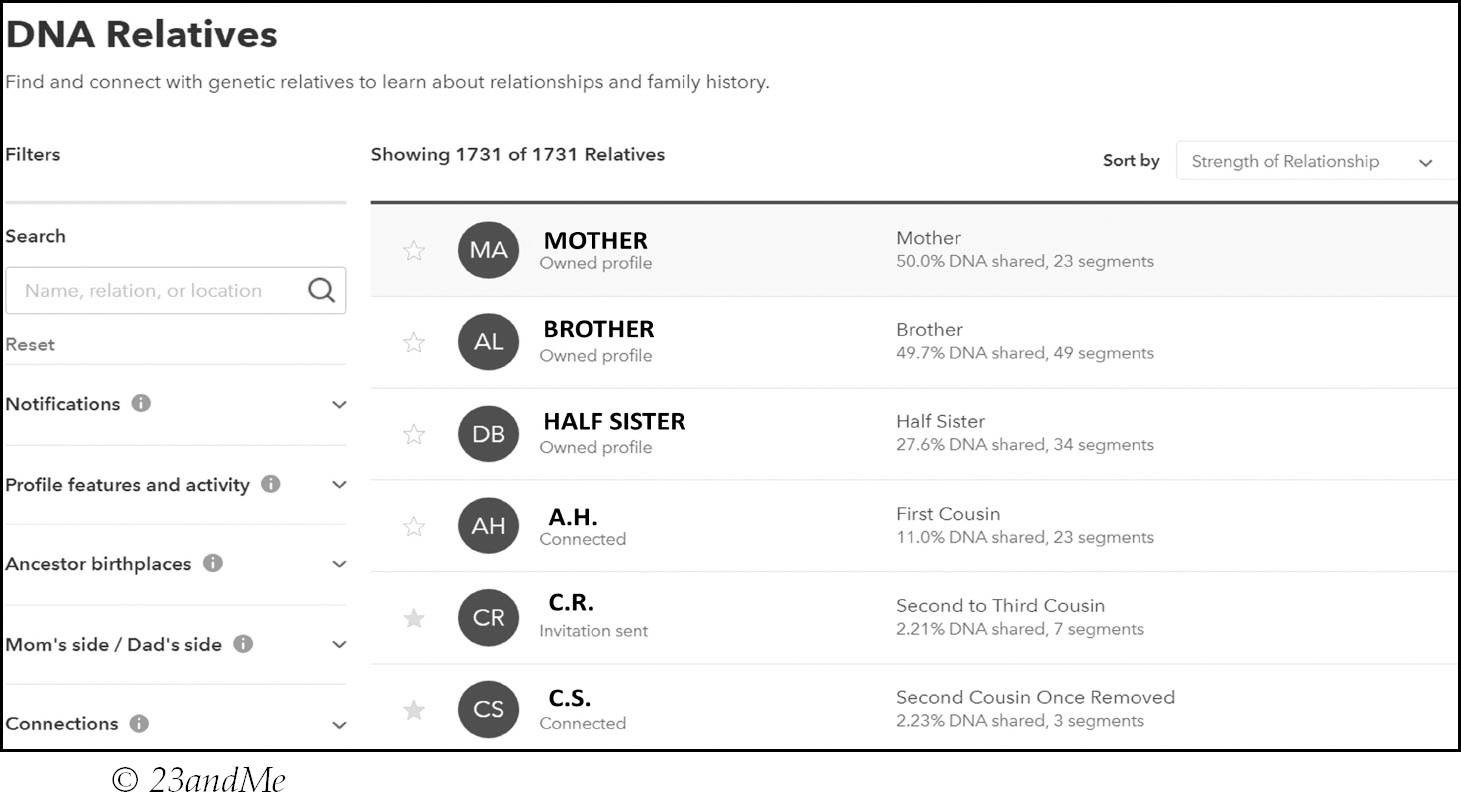
After checking out your DNA homepage, the next thing you want to look at is your DNA match list page, but what does a DNA match list look like? The answer is that it depends on the company you have tested with as they all have different features, tools and ways of displaying similar information.
Here is the top of my 23andMe match list as an example:
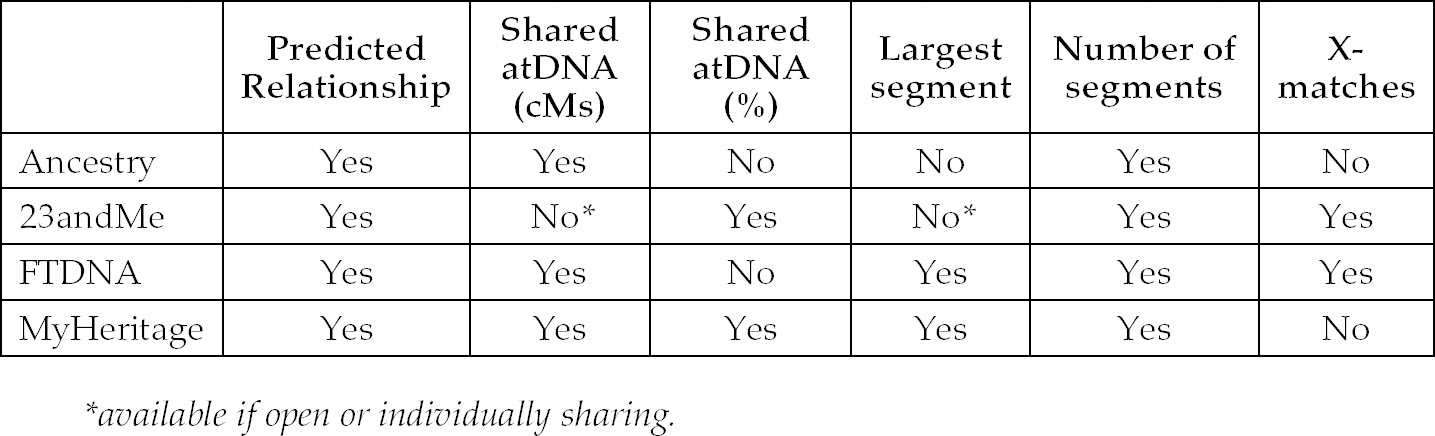
The following table gives a summary of the basic data which the various companies report on:
Relationship Predictions
Each of the testing companies has their own relationship prediction categories and ranges e.g. first–second cousin, fourth–sixth cousin, fifth– eighth cousin. These are a rough guide to the expected level of relationship between the test-taker and the match. No matter where you have tested, however, I would recommend consulting the extremely useful DNA Painter Shared cM Project Tool to get the most accurate picture of possible relationships and probabilities.
The Shared cM Project is one of the best resources available for understanding ranges of shared cMs and relationship probabilities. It is a collaborative project, run by American genetic genealogist Dr Blaine Bettinger, which collates data from real matches and displays the averages and ranges for a wide variety of relationships. There have been over 25,000 submissions to the project so far. I recommend submitting your own data from confirmed relationships using the data submission portal here: https://docs.google.com/forms/d/e/1FAIpQLSc5a0SIHIeiwLl5Wxn4sLqgnRV-su2klK2W_YzIJc9xq2i4zw/viewform
The most exciting tool to emerge out of the Shared cM Project in recent years is ‘The Shared cM Project 3.0 tool’ created by DNA Painter developer Jonny Perl. This automates the investigation process for you and can help enormously with relationship predictions and probabilities. When you look at the number of cMs you share with a DNA match and want to identify the possible and probable relationships for that level of sharing, go to the tool, either key in the cM or percentage total (percentage is the default at 23andMe) and it will generate a full list of probabilities, percentages and relationships. It is incredibly helpful and simple to use.
My first cousin Ann shares 634cM with me and is predicted to be a second cousin at Ancestry, but if she shared just 6cM more the prediction would be first cousin. It is best to investigate the number of cMs as opposed to the relationship estimate, so take the cM number and plug it into the Shared cM Project Tool.
The result shows that there is only a 0.9% probability that Ann is a second cousin, but there are many possible relationships at higher probabilities. Always look at the highest probability relationships first, but do not dismiss the lower probabilities and beware of confirmation bias: try to disprove a preferred outcome as opposed to trying to prove it. In this case probability suggests my known 1C (first cousin) is more likely to be a half 1C or 1C1R, so I had to consider that possibility. Due to the much higher amounts of DNA she and I both share with many other close relatives, however, it is certain she is a slightly low-sharing 1C and the relationship falls within the second highest probability grouping at 38.9%.
This is the most up-to-date Shared cM Project chart (compiled August 2017) © Blaine Bet
https://thegeneticgenealogist.com/2017/08/26/august-2017-update-to-the-shared-cm-project.
Never take relationship predictions too literally, especially for more distant relationships, and always remember that multiple relationships could fit the same number of cMs and outliers are possible.
Bookmark this tool and use it whenever you need some help working out the potential relationships and ranges: https://dnapainter.com/tools/sharedcmv4
Shared atDNA
The total amount of shared atDNA is reported either as a percentage figure or as a number of cMs, generally on the main match page. On Ancestry this information is both on the main match page and the individual match page under the ‘i’ button. It is important to get into the habit of checking cM information when looking at new matches.
Largest Segment/Longest Block
This refers to the largest segment of DNA you share with your match. FTDNA and MyHeritage report on it and it can be seen on the DNA Comparison page on 23andMe. It is not available on Ancestry. It is often useful to filter your list by longest block on FTDNA, as opposed to shared cMs, due to the fact that segments as small as 1 cM are included in that total. As illustrated in Chapter 3, very small segments are problematic and more often than not will be false. I prefer not to count these segments so I calculate my own total, adding up just the segments over 7 cMs via the chromosome browser table view (see p76-7).
Number of Segments
The larger the number of shared segments, the closer the relationship is likely to be, but the overall amount of shared atDNA and the size of the largest segments are much more important indicators of relationship level.
X-Matches
This indicates that you share a segment on an X-chromosome with that particular match. You should note that Ancestry and MyHeritage do not report X-matches. Since X-matches can only occur on a number of specific ancestral lines, it is easier to narrow down the genealogical link to particular lines. For example, the most obvious benefit for male testtakers is that all their X-matches must be on their maternal side. FTDNA report tiny segments of X-DNA, so it is best to check the actual size of the segment on the X-chromosome via the chromosome browser tool before accepting any match highlighted as an X-match on the main match page as real.
The companies all provide differing features and tools; the following are perhaps the most useful.
Ancestral Surnames/Family Trees and Search Options
All of the companies have the facility for test-takers to list ancestral surnames and, in some cases, locations. Additionally all, except 23andMe, allow family trees to be created or uploaded, although a link to an externally hosted tree can be added to the latter. All offer surname search options, but only Ancestry and 23andMe provide location searching. It is helpful to search your entire match lists for matches with common surnames and common locations; if a match has both you have a better chance of identifying the link. Work through all of your ancestral surnames and filter by birth locations every so often in case you get any interesting hits. Keep in mind, however, that this type of filtering works better for less common surnames and locations. There are obvious benefits to uploading a tree, making it much easier to spot possible genealogical links with matches. Once you find clear indications of a link via surnames, locations or trees, you may wish to make contact with those matches. For advice on how to go about this, see p82-4.
Shared Matches/In Common With (ICW) Tools
These tools, described differently by the various companies, list people who match both you and another match you have selected. In the majority of cases the use of shared matches will be key to working out how you and your matches relate to each other. Additionally the more confirmed relatives you have in the databases, the more useful shared matches will become.
On Ancestry the ‘Shared Matches’ list is limited to test-takers who match both you and the comparison match at the fourth to sixth cousin or closer level i.e. sharing over 20 cM. You cannot, however, see how many cMs they share with each other, unless they share their results with you. FTDNA’s ‘In Common With’ list is similar in that it allows you to see which of your matches also match one specific match, but not how well they match each other. 23andMe’s ‘Relatives In Common’ and MyHeritage’s ‘Shared DNA Matches’ tools are more detailed, as they include both the amount of DNA you share with the shared match and the amount shared between them and the comparison match.
It is important to be aware that it is always possible a match could connect to you in one way and the shared match in another. Don’t assume that matches only relate to you in one way – even without endogamy it is always possible that you could have more than one connection with a given match.
Predicted Phasing
This process has already been described in Chapter 3, and although none of the companies provide full phased segment data, there are some tools for narrowing your matches to the maternal or paternal side of your family.
Ancestry adds ‘Mother’ and ‘Father’ filters if parents test, but be aware only matches in the fourth to sixth cousin or closer categories will appear on these lists.
The Family matching feature on FTDNA offers ‘Maternal’, ‘Paternal’, and ‘Both’ buckets that populate if you attach a family tree and link entries of known relatives on that tree to their test results via the complimentary Linked Relationship tool. This tool uses uploaded results for relatives from parents to third cousins. The ‘Both’ bucket will only contain full siblings and cousins who match on both sides of your tree.
If you have a parent tested at 23andMe it is automatically noted on your match list if matches are on that side, e.g. ‘Mother’s side’. If the other parent is not tested a very useful ‘Not Mother’s/Father’s Side’ tool is also available to separate out paternal or maternal matches.
Shared Ancestor Hints (SAHs)/Smart Matches
These hints, provided by Ancestry and MyHeritage respectively, are generated when the same ancestor or ancestral couple is found in both your linked family tree and that of a match. Remember that, although often accurate, errors are always possible and these are only ‘hints’ – they do not prove that you and your match inherited the DNA you share from the common ancestors identified.
Sharing Results
Some companies facilitate the sharing of results with your matches – sharing match lists in particular and having them share theirs with you can be very beneficial. Sharing is currently possible at Ancestry and FTDNA (via projects).
There are pros and cons to testing at each of the major testing companies, but it is important to get into as many of the different databases as you can in order to maximise your chances of detecting your best matches. I have many more matches at Ancestry than elsewhere. That will be a standard experience for many since Ancestry has the largest database, but it does not mean your best matches will also be at Ancestry, as you simply do not know where they may choose to test. The only way to be confident you are not missing out on any important matches is to get into all the databases. DNA testing has significantly reduced in price over the past few years and frequent sales make this a more affordable goal. Additionally several of the companies (FTDNA, MyHeritage and LivingDNA) accept free transfers, as does GEDMatch (Ancestry and 23andMe do not) so it is possible to get into all of the databases without buying standalone tests for each of them.
It is absolutely vital to pair traditional research with DNA analysis if you want to get the most out of DNA testing for genealogy. One of the most important things to consider both before and after you test is the building of your own family tree. The more robust and extensive your tree, the more you will be able to achieve with your DNA results and the more cousin matches you will be able to identify. Try to work as many lines forward to the present as you can, including collateral lines, since it is your distant living cousins who will show up on your DNA match lists. If one or both of you do not have your tree developed enough then neither of you will be able to identify the connection.
For example, my match Mary has a tree that only goes back to her grandparents, one of whom was Alfred Kerr from Lanarkshire, Scotland. My great-grandmother was a Kerr from Lanarkshire so I checked my extensive tree for Mary’s grandfather Alfred and he was already on it. It took very little effort to identify this connection as all the required research had been completed previously and this DNA match enabled me to make contact with a new cousin, add a recent branch to my tree and confirm a line. Verifying the accuracy of your tree via DNA evidence is one of the most important reasons to DNA test for genealogy and your goal should be to confirm as many lines as possible.
Extensive tree-building, however, is not possible for everyone, including adoptees and those with recent unknown ancestor mysteries or brick walls. While building a personal tree if you possibly can is extremely helpful, it is not essential to success as DNA is also the best tool for uncovering unknown ancestry. For those who do not have a recent unknown ancestor mystery, however, spending time building out a tree should be a top priority.
As covered in Chapter 3, it is very important to test your older generations if at all possible, starting with parents and grandparents. This will not only give you more of your ancestors’ DNA to work with, but will also help you enormously with narrowing down your results; testing just one parent can narrow your matches by 50%.
If you do not have older generations to test then test your peers (half-siblings, first cousins, second cousins etc). Full siblings are not included for narrowing purposes, but are also very important testtakers if you do not have parents to test, as they received DNA from them that you did not.
Note that half relationships are incredibly helpful for narrowing purposes: while you share less DNA with a half-relative than a full one, all of the DNA you do share can be effectively narrowed a further generation since the common ancestor is one parent, one grandparent etc.
Let’s look at how this works in practice. Since I have many close relatives tested I can significantly narrow down my connection to this match, D.H., via researching our shared match list:
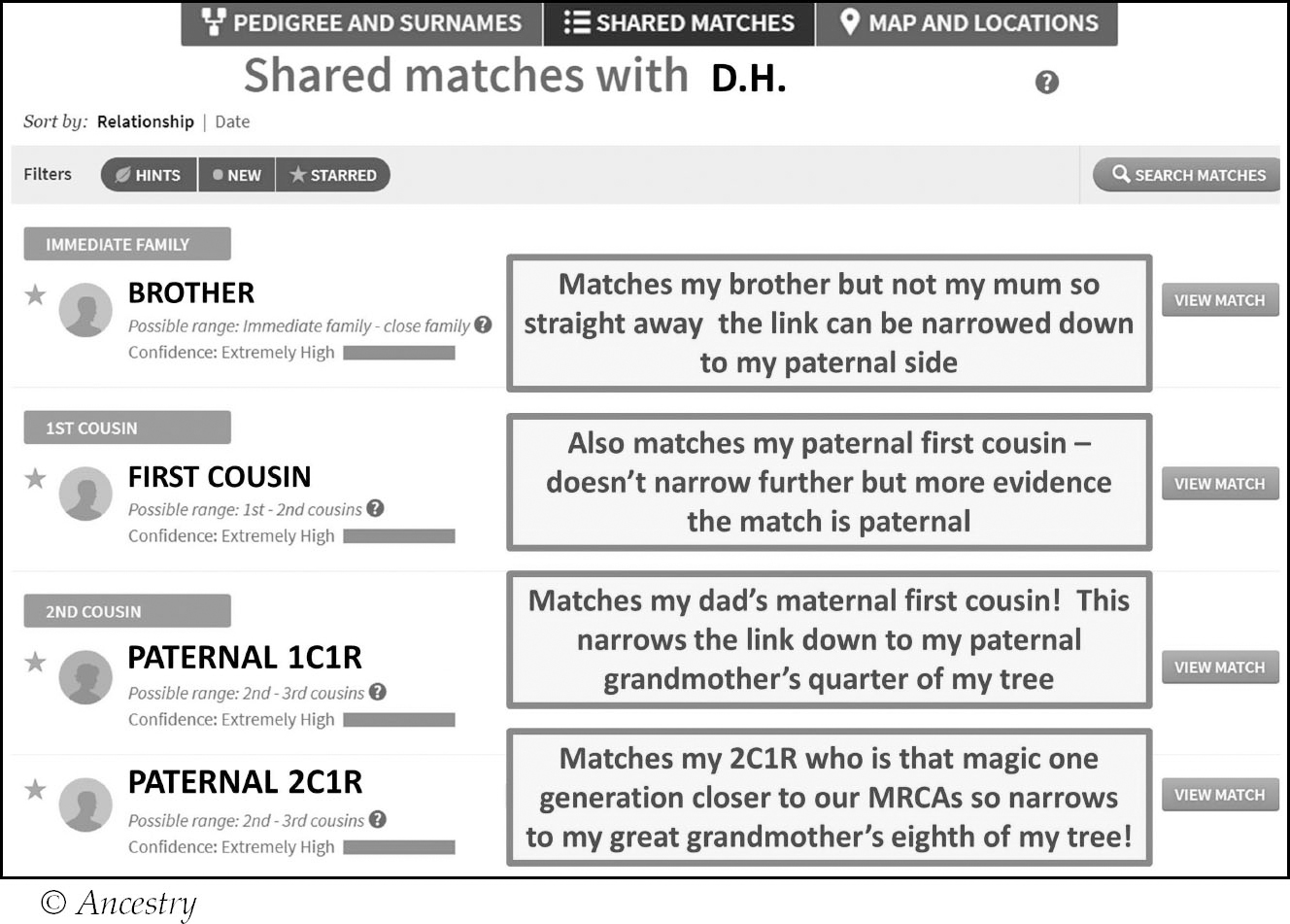
Via this analysis I have eliminated 87.5% of my tree and can focus on my great-grandmother’s eighth to find the connection. My great-grandmother, Janet Cullen, was the daughter of John Cullen and Janet Nicol, so it must lie on one of those lines. Investigating D.H.’s tree one name stood out – her grandmother Christina was a Nicol. I now have a common surname that matches with the small part of my tree D.H. narrows to, so it is worth investigating further.
Tree investigation is a vital component of successful DNA matching and many matches can be solved if you spend time building out their trees yourself. Many will only have small trees comprising of a few names or maybe even just parents’ names, but do not dismiss these matches or trees. Do not accept the tree is too small to find the link – be proactive and build your own versions of the match’s tree to see if you can pinpoint the connection. Before deciding to spend time on this, however, you have to evaluate whether the match is worth the effort: would a 12 cM one-segment match with no clues be worth it? Probably not. Would a 90 cM six-segment match be worth it? Almost certainly.
Let’s return to D.H. as an example: her tree only reaches back to her grandparents, so traditional research was required to see if her grandmother Christina connects to my Nicol line. This research broke a collateral line brick wall for me as I found Christina’s father, William Nicol, was my second great-grandmother Janet’s brother, whom I had not been able to trace past the 1841 census. When exploring trees it is essential to remember that many contain errors. Always pursue sources and, if matches have large but unsourced trees, it is wise to build your own sourced versions. You may, in the process, uncover mistakes that help identify common ancestors.
Knowing the age of your match can be very helpful when it comes to determining how they may relate to you. The general starting point is that people of a similar age are most likely to be of the same genetic generation, but do not assume that will always be the case as there will be exceptions to the rule. Be aware that only 23andMe and MyHeritage display the ages of matches, and only if they have consented to share this information.
The most telling aspect to look out for when it comes to genetic generational anomalies is an older father skewing the generations. A perfect example of this is my mother and her 2C2R Rob who is two generations closer to their common ancestors despite being three years younger. The reason for this anomaly is that Rob’s grandfather was in his sixties when his father was born.
In order to be confident there are no generational anomalies, look closely at the trees of your matches going back several generations and if your and their parents, grandparents and great-grandparents all fit within regular generation spans (20–35 years) then the likelihood that you and your similarly aged match are part of the same genetic generation is much higher. If, however, you find a much older father or very long generations then you have to factor that into relationship prediction calculations.
A chromosome browser is a tool that provides a visual representation of the exact segments of DNA shared between a test-taker and one or more (in the case of one-to-many chromosome browsers) of their DNA matches. They provide information on the start and end locations of matching segments and the amount of cMs in those segments. FTDNA, 23andMe and MyHeritage all provide Chromosome browsers, but Ancestry does not.
All chromosome browsers display twenty-three lines to represent the twenty-three Chromosomes (twenty-two autosomes plus the X-chromosome). Remember that there are twenty-three pairs of chromosomes though, so why is there only one line on the chromosome browser to represent each pair? The reason is that it is not possible to automatically differentiate between the maternal and paternal chromosomes so they are basically merged together. Never lose sight of the fact that both exist, however, and two people matching you on the same segment on the same chromosome could be matching on either your maternal or paternal copy. It is possible they match each other but equally likely they do not, so you have to find out if they do through comparing them to each other if possible.
Here is my comparison with my brother on 23andMe:
An interesting additional feature of 23andMe’s chromosome browser is that it supplies information on whether shared segments are half or completely identical (often referred to as fully identical). If a segment is half-identical this means you share it on just one of your two copies of that particular chromosome and if it is completely identical you share it on both copies. 23andMe also makes it possible for you to compare not only yourself to a number of your matches, but also your matches to each other via their DNA comparison tool. Knowing how your matches match each other can be extremely helpful when trying to work out a connection.
FTDNA and MyHeritage also provide one-to-many chromosome browsers, but it’s important to note MyHeritage does not report on the X-chromosome. You will notice some criss-crossed greyed out areas on chromosome browsers; these indicate SNP poor regions that have not been tested, because there is not enough data available to sample them.
Chromosome mapping is fun, but it is more than just a fascinating DNA jigsaw puzzle you might be interested in working on, as it can bring real benefits to your family history research. The more DNA you can map back to your ancestors, the easier it becomes to narrow matches down and successfully pinpoint cousin connections. This is a process you can begin working on as soon as you have test result data from known relatives, and is likely to remain a work in progress while you encourage more relatives to test and attempt to identify connections with matches.
So what exactly is ‘chromosome mapping’ and how do you do it? Chromosome mapping is the process of assigning specific segments of DNA to specific ancestors with the use of confirmed cousin matches. If you share a segment of DNA with a known second cousin, it is extremely likely that piece of DNA was passed down to you and your second cousin by your shared great-grandparents (your most recent common ancestors) and therefore you can map this shared DNA back to them. You do not have to only practise chromosome mapping on your own chromosomes either – if you have parents and other relatives tested you can also map their DNA.
It is only possible to undertake chromosome mapping if you have segment information to work with, so this means it cannot be done with Ancestry matches unless they transfer their raw data somewhere that provides segment information. You can currently map matches from FTDNA, 23andMe, MyHeritage and GEDMatch.
GenomeMatePro (GMP) has an excellent chromosome mapping section and in 2017 a new tool named DNA Painter, created by developer Jonny Perl, was released. It has fast become the go-to tool for chromosome mapping. Different colours are used to represent particular ancestors and the segments that have been mapped back to them.
Chromosome mapping can also be carried out independently by recording specific segments of DNA and which ancestor they have been assigned to on, for example, a spreadsheet, but DNA Painter undoubtedly facilitates the process and produces a clear visual representation of what otherwise might seem a dry collection of numbers.
It is exciting to see your map develop and chromosomes start to fill up, but it is also extremely useful. The advantage of starting your chromosome mapping early is that whenever you get new matches, you can check the segment data for them in case they match up on any of the segments you have already mapped. Whenever they do, you can instantly narrow them down to the ancestors the DNA is already mapped to.
I would advise first spending time getting to grips with the tools at the testing sites and working your way through your top matches, but once you have a bit more experience, chromosome mapping is an excellent way to advance your DNA adventure. It can be quite addictive trying to map as much as you possibly can and see exactly which pieces of your DNA came from which of your ancestors.
As we have seen, FTDNA’s shared match list is called ‘In Common With’ but they also provide a ‘Not In Common With’ tool which can be equally useful.
It is important not to assume too much from ‘Not In Common With’ lists, as the only relationship for which you can run a ‘not in common with’ search and narrow to a side is parent/child. For all other relationships there is a chance the test-takers on the list still match you on the same side. For example, a list generated between my mum and her maternal first cousin Isobel only tells me that these test-takers match my mum but do NOT match Isobel. I cannot assume that they are paternal matches, however, as mum and Isobel only share approximately 25% of their maternal DNA, meaning there is 75% they do not share and these test-takers could match my mum on segments within that 75%. There is a way I can potentially narrow some of them down to paternal, however, and that is by comparing segment data in the chromosome browser. Let’s focus on a match named ‘Kate’ and analyse the exact segments she and Isobel share with my mum.
While my mum shares a large number of segments with her first cousin, there is only one significant segment with Kate. The interesting thing to note, though, is the fact that this segment is on chromosome 9 and clearly overlaps with a segment my mum also shares with Isobel.
The best way to investigate start and end locations is to view the data in a table, using the Detailed Segment Data link near the top of the screen.
The numbers confirm that part of the segment of DNA on chromosome 9 my mum and Kate share does indeed correspond with the same segment that my mum and Isobel share (from start location 108,966,384 to end location 124,646,012 – since these numbers are large it is generally easier to shorten them to 108.9 – 124.6). The ‘Not In Common With’ list, however, reported that Isobel and Kate do not match each other. Since Isobel matches my mum on her maternal copy of chromosome 9, we can therefore deduce that Kate must match my mum on her paternal copy of chromosome 9. Kate’s match can be narrowed down to the paternal half of my mum’s tree since it is not possible for two people to match you on the same segment but not match each other, unless they are matching you on opposite copies of the chromosome. After further research into Kate’s family tree this was proven correct as she is a 4C2R via a paternal line. While FTDNA is the only site that provides a ‘Not In Common With’ tool, this kind of inferred segment analysis can be undertaken on any of the sites that provide segment data so long as your starting point is two matches who do not match each other.
Moving a step beyond identifying ‘in common with’ matches is the process of triangulation.
Triangulation is another major term that you will often hear in the genetic genealogy community. It occurs when three or more DNA testtakers all match each other on the exact same segment of DNA. When this transpires it indicates that these people all have a common ancestor from whom they inherited their shared DNA segment.
True triangulation is not possible at Ancestry as it requires segment data which is not supplied, but it can be partially completed at FTDNA (partially as it requires the co-operation of the match to complete a full comparison) and can be fully realised at 23andMe, MyHeritage and GEDMatch.
Let’s look at an example of triangulating a segment of DNA using the chromosome browser at FTDNA. When looking at my mum’s account and comparing her to her maternal first cousin Isobel and another match named Kim, one chromosome immediately stands out – Isobel and Kim both match my mum on a segment of DNA on chromosome 7. This only tells us that they both match my mum, however, and the key to triangulation is that all three must match each other. In order to determine whether they triangulate or not we need to know if Isobel and Kim also match each other on the same segment on chr 7. Unfortunately this is not something the chromosome browser on my mum’s account can tell us, as I cannot compare Isobel to Kim. There are, however, two ways I can find out whether Kim and Isobel match each other. I can look at the ‘In Common With’ list for one of them to see if the other is on it, or I can use the matrix tool which can be accessed from the FTDNA dashboard.
The matrix confirms that Isobel and Kim do match each other, making it more likely that the segment they both share with my mum is on the same copy of chromosome 7, but it does not prove that it is, since Isobel and Kim could conceivably match each other in a different way.
In order to obtain proof I would either need to have access to one of their match lists, or they would have to do the comparison on their own accounts. Thankfully, in this case, I manage Isobel’s test so I have the required access to compare them:
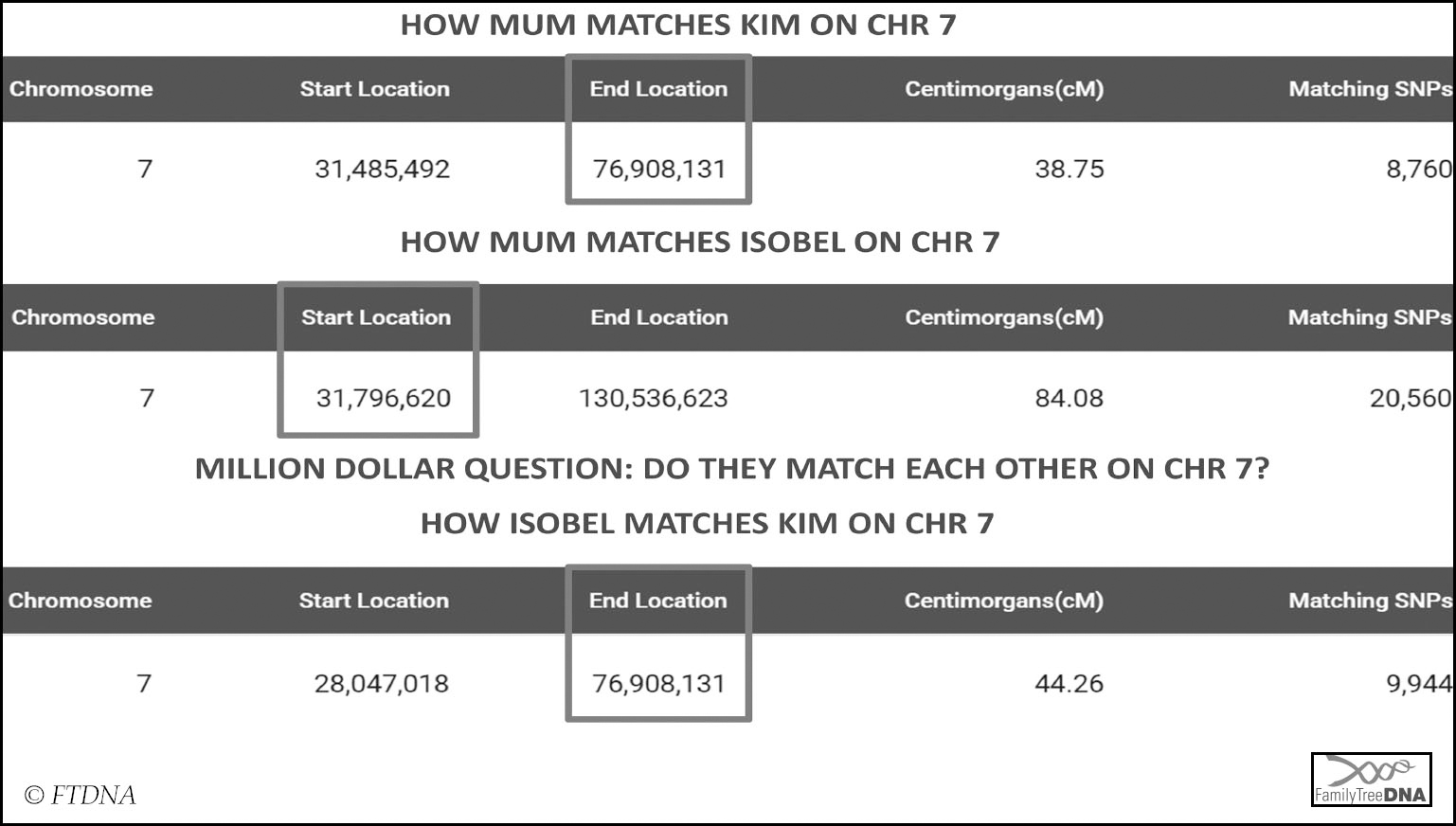
We have triangulated! Mum, Isobel and Kim all match each other on the same segment of DNA on the same copy of chromosome 7, so they inherited this piece of DNA from the same common ancestor. In this case it is my mother’s maternal chromosome 7 as Isobel is her known maternal first cousin. We cannot say that the full segments they share with each other are triangulated though – we have to count the triangulated segment from the highest start location between the three of them to the lowest end location since that is the exact segment they all share. In this case that would be 31.7 – 76.9.
The chromosome browser at MyHeritage automates this process for you.
If triangulated segments have been detected between you, your comparison match and the shared match, a triangulation symbol is displayed. In order to see the details in the one-to-many chromosome browser, click on this symbol.
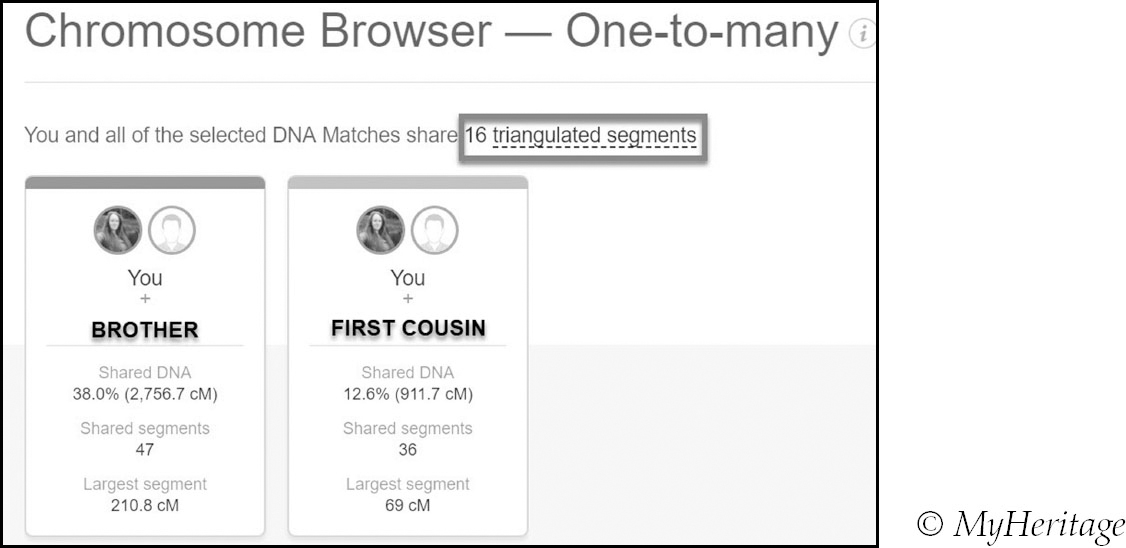
The site highlights all of the triangulated segments and provides tables with full details of the triangulated and regular segments.
23andMe also provide automated triangulation information if a match is open or individually sharing with you. Any match with a ‘Yes’ in the ‘Shared DNA’ column on the ‘Relatives In Common’ list shares a triangulated segment and any with a ‘No’ shares DNA with both you and your match but not on the same segment.
Triangulation can be useful for narrowing down matches and identifying groups of people who all share common ancestors, but it also has to be remembered that triangulation between cousins at more distant levels is a rare occurrence and attempting to triangulate small segments is not generally useful. Triangulation is a helpful technique, but the majority of DNA puzzles can be solved without it.
There is a great deal of frustration in the genetic genealogy community about lack of trees hampering success. Many feel discouraged by the fact that a large number of test-takers do not upload trees and often ignore these matches. While this frustration is understandable, and it would be ideal if every testtaker uploaded a robust, well-documented tree, it is unrealistic to expect this.
While there are many matches who do not have a tree, there are many others who do but just have not linked their tree to their DNA results. Ancestry highlights when matches have unlinked trees, however, so you can investigate them, but always bear in mind that the home person of the tree will not necessarily be the test-taker. Linking your tree to your results is one of the most important steps to take when they first arrive.
I conducted a study into how many trees are available on Ancestry for my top 200 matches (excluding those I have tested myself as they could skew the data):
| Tree Designation | Number | Percentage |
| Linked Public Tree | 79 | 39.5 |
| Linked Private Tree | 15 | 7.5 |
| Unlinked Tree | 47 | 23.5 |
| No Tree | 59 | 29.5 |
| Total Public Trees | 126 | 63.0 |
| Total Trees | 141 | 71.0 |
The fact that 71% of my top 200 have a tree of some kind and 63% of those are public belies the idea that the majority of test-takers have no tree. While this is an individual study, several other genetic genealogists have conducted similar ones and ended up with comparable results. Additionally, out of the fifty-nine test-takers with no trees, many provided family history information upon contact that helped me work out the common ancestors in eighteen cases, so do not bypass those without a tree. We also have to take into account the quality of the trees and many will not be helpful, but even a tiny tree can often be enough to solve a match if you are prepared to work with it.
Also never forget that some people will have no tree or private speculative trees because they are adopted or have unknown parentage. What if there really is no tree though? The obvious step is to attempt to make contact.
Once you have familiarised yourself with the various processes and tools described above, you may feel it is time to take the plunge and contact some selected matches. One of the keys to successfully using your DNA test results for genealogy purposes is contacting matches and eliciting successful responses. Connecting with your matches can provide the breakthroughs needed to identify common ancestors.
At Ancestry, 23andMe and MyHeritage you can only contact matches via their proprietary messaging systems. This can be hit and miss at times, but do not be discouraged from trying. FTDNA and GEDmatch supply email addresses, which are often a more fruitful contact method. Also, do not avoid messaging those with no tree or private trees – some of my most helpful contacts have emerged from those circumstances.
Personalise your profiles to let people see you are friendly and interested in contact and they may be more willing to reply.
Before sending an opening message to a match do some preliminary research. Investigate their tree (if they have one) and shared matches to try to work out the connection. Can you pinpoint any shared surnames or locations? Can you narrow the link down via matches to other confirmed cousins?
In the subject title and first line of your message always state who matches whom. Never write ‘I match you’ or ‘you match me’. You have to make it as easy as possible for your contact to find and evaluate the match. Many people manage multiple tests, so if you do not identify the match properly they will not know which test to check. Write something along these lines: ‘My test under the name ML matches the test you manage under the name JS’. If the match is at GEDmatch include the kit numbers involved. If the match is at FTDNA, however, it is not useful to provide FTDNA kit numbers because it is not possible to search the FTDNA database by kit number.
Your initial message should be short, friendly, informative and nontechnical. It should aim to grab your match’s attention and, if possible, offer an incentive to reply: I call this ‘carrot dangling’ or, more aptly, ‘record and photo dangling’! If you do not have such an incentive to offer, try to find one by doing some research.
Tailor contact to the match and situation. Tell matches you would love to work with them to figure out your common ancestors. Do not be vague, but also do not give everything you know away at once either, as too much information can overwhelm or be a disincentive to replying. Give them the chance to be involved in the discovery process and ask you questions too.
Offer family history information and ask for some in return. Include a simple question or two but no more than that – you do not want to bombard them or make replying anything other than quick and easy. I would caution against asking for invites to private trees; private tree owners may offer invites themselves but, equally, they may be reticent to reply if you ask for one outright. Instead ask for basic information which they may feel more comfortable providing. If your match does not have a tree, ask if they could supply some simple details about their four grandparents.
Do not assume that your match understands genealogy. DNA testing has become the new gateway to genealogy, so it is very possible the reason there is no tree is because your match has never compiled one. Asking for a tree, therefore, may not be the best idea as they could feel daunted if they do not have one to share. Additionally, try not to befuddle them with genealogical or DNA terminology like pedigree chart, MRCA, centiMorgans (cMs), segments, chromosome mapping etc. This can come later once you have established a dialogue.
It is also important not to assume your matches can see everything you can on subscription sites. On Ancestry, for instance, those without a subscription cannot see your tree, so you have to offer them an alternative viewing method; you could send an invite or – my personal preference – offer to email a copy. I recommend sending a pdf pedigree chart for the pertinent side or line of your tree as this focuses the match’s attention on the correct ancestors and is easier for them to assess.
Always try to migrate the contact from the testing company’s messaging platform to email as that is better for long-term communication and you can attach images and photographs, which is currently not possible on most of the messaging platforms.
One of the most common things many want to ask Ancestry matches to do is to transfer their results into FTDNA, MyHeritage or GEDMatch for access to segment data. This is not something I advise broaching in a first message. New contacts will generally be more receptive to transfer requests once you have built up a good rapport with them, so leave this for several messages down the correspondence line.
It is important to keep on top of your match lists and contact any new high matches as soon as you can after they appear – there is a better chance of replies when people have just received their results as they are more likely to log in at that time. That does not mean rush the contact, but have basic templates ready that can be tweaked depending on the match and situation.
Finally try to be patient! Do not send several messages in a flurry. At the end of the day many will not reply no matter how well-written your message is, but a thoughtful missive will elicit more replies than a poor one. Focus on the goal of getting any initial reply and then try to make progress from there.
Putting a system in place to organise your DNA matches is crucial and one of the most important organisational aspects is note-taking. Each of the major companies has a notes facility and it is a good idea to familiarise yourself with where they are located and get in the habit of writing notes on each match you examine. Work out what the most important information to you is and add it in the order you feel is most helpful.
My notes system:
1. cMs/segments in shorthand so 305 cMs over 9 segments written as 305x9
2. Paternal or Maternal (if it can be narrowed that far)
3. Confirmed relationship in shorthand if known e.g. second cousin once removed = 2C1R
4. Names of the most recent common ancestors (MRCAs) if known or the line I believe the match to be on if not known – failing those any shared surnames/locations
5. Date match appeared 6. Whether I have contacted the match and if so when and where I sent the message
7. If the match has responded and we are in contact (also when this occurred)
8. If the match has an unlinked or private tree
9. If the match is on any other testing sites or GEDMatch (if on GEDMatch I add the kit number)
Another way you can organise information and notes on your matches is via a custom master spreadsheet and many prefer to do this instead of or in addition to writing notes on the different websites. The advantage of having an external system is that people can delete tests from the sites and you could lose information and notes.
Genome Mate Pro (GMP), a free third-party utility, is another excellent tool for match organisation. You can add your matches from all of the different companies and combine them together, although Ancestry matches are of less use as GMP is based around the use of segment data. Matches are grouped by chromosome, which makes it easy to quickly assess things. Within each chromosome all matches are listed from the lowest to highest start location by default, but there is a great deal of filtering choice in how you wish to view your lists. Each match has a profile page on which you can write notes, so if you prefer you can use GMP as your master spreadsheet and add all the information you have gleaned about a match there. Setting up GMP is a fairly time-consuming task with a steep learning curve, but it is an extremely useful tool once you get the hang of it. There is a comprehensive set-up guide and dedicated Facebook group.
Before affordable DNA testing for genealogical purposes burst onto the scene in recent years, many family history mysteries were simply filed away in the unsolvable drawer. DNA testing has opened up a whole new world of possibility for adoptees and those with unknown parentage mysteries, and by unknown parentage mysteries I am not only referring to an unknown parent, but also perhaps an unknown grandparent or great-grandparent. DNA testing has become the logical next step for those faced with brick walls such as a grandfather’s birth certificate with a blank next to the father’s name. It is a very powerful tool that can be used to help reconnect biological family members in the here and now, as well as unlock secrets from the past, which we thought had died with those who kept them.
An important recommendation for those with an adoption or unknown parentage mystery is to get into all of the major autosomal databases. It is like going on a fishing trip and not knowing which pond your big fish might be swimming in. If you are fishing in them all then you will not miss out on landing your personal big fish.
The starting point is the same as it is for those with known ancestry – investigate your top matches and examine or compile family trees for them. I recommend building the trees of matches on one ‘Master’ tree as opposed to many separate trees; this can make it easier to spot connections between matches and, if everything is on the same tree, when links have been identified it is a simple task to merge common ancestor entries and trees.
Many cases will require contacting matches and hoping they will be willing to provide information. This can be daunting and there may well be disappointments and frustrations if close matches do not respond. Try not to get discouraged. Screenshot close match information, including trees, in case they decide to delete or go private and you lose that information.
When you do not have a tree of your own to compare to the trees of your matches, it is essential to investigate the shared matches of your highest matches and try to work out how they match each other! If you can narrow their connection down to a particular ancestor or ancestral couple it is likely they, or a set of their ancestors, are also your ancestors. Once you have identified a likely common ancestor or ancestral couple you need to build their lines forward to identify potential grandparents and ultimately birth parents via age and location. Who was in the right place at the right time? There may, at this point, be several birth parent candidates that only further targeted DNA testing can separate out. If you end up with different hypotheses to consider I recommend using the ‘What Are The Odds?’ DNA probability tool: https://dnapainter.com/tools/probability.
Target testing will involve the tricky task of contacting people on the line you have identified and asking them to test.
You need to think carefully about how to initiate this contact and have a support network on hand. It is worth reading the contact advice at www.dnaadoption.com.
Be aware that how successful you will be is totally dependent on whether or not close enough relatives test. By close enough I mean second to fourth cousins as opposed to extremely close matches such as parent, half-sibling or first cousin, which would fall under what I call ‘jackpot matches’ for unknown parentage mysteries. Some will find they do not have close enough matches to begin with, but more and more people are testing on a daily basis and match lists are constantly updating, so closer matches can come along at any point.
1. Whichever platform you test at find the shared cM/segment numbers and familiarize yourself with the relationship probabilities using the DNA Painter Shared cM Project Tool
2. Study shared matches as they may help you narrow the connection down
3. Check for shared surnames and locations
4. Investigate the match’s tree if there is one and build it out if it’s too small to be useful
5. If no tree search online in case there is one elsewhere or the match can be identified via their web presence
6. Contact the match once you have fully investigated the potential connection
7. Work with segment data if available using phasing (when possible), chromosome mapping, inferred segment matching and triangulation
This case study centres on the breaking down of a brick wall via DNA matches.
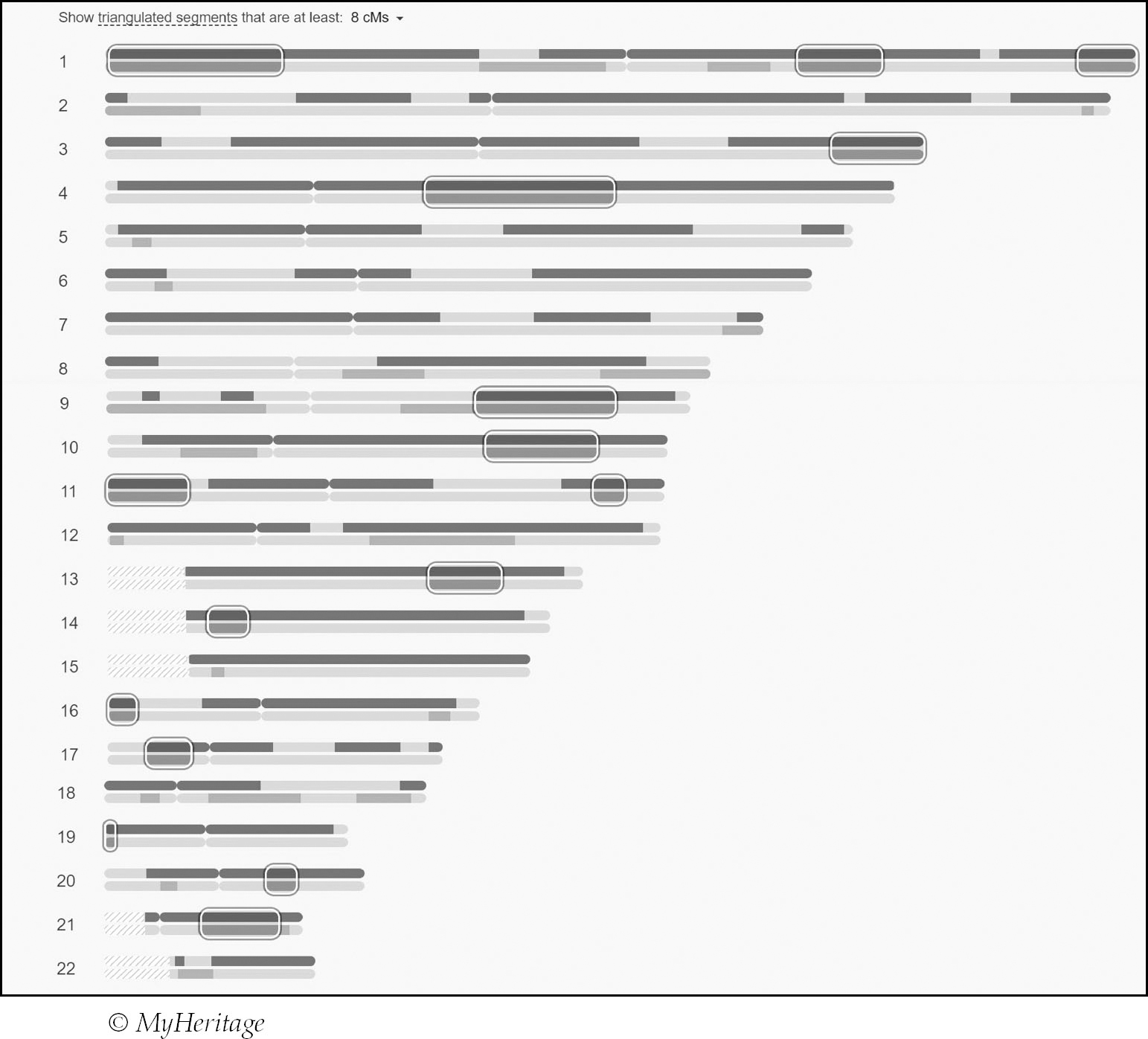
My third great-grandfather Robert Cullen represented a longstanding brick wall on my tree (see p88).
Robert died in the 1820s and there is no existing documentation containing the names of his parents. I have searched births and baptisms and there is one possibility:
Robert Cullen, born 7 June 1802, Barony, Lanark, Scotland, son of John Cullen and Elizabeth Morton.
Robert was born long before statutory registration began in Scotland in 1855 – up until this point there was no legal requirement to register a birth in Scotland so, while his baptism may have been registered, it is perfectly possible no record exists. Additionally, although he married and died in Glasgow I have no proof that he was actually born there. For these reasons there is simply no way for me to be sure whether the Robert born in Glasgow in 1802 to John Cullen and Elizabeth Morton was my Robert and, thus, it is far too speculative to add to my tree.
One of the first matches I ever confirmed a connection with was a paternal fourth cousin and upon comparing notes on our common ancestors, Robert Cullen and Margaret Whitehead, I discovered her great-grandmother (Robert’s granddaughter) was named Elizabeth Morton Johnston. I found corroboration that she called herself this on the Australian birth record of one of her children.
It is quite a coincidence that the name Elizabeth Morton was given to a potential great-granddaughter of an Elizabeth Morton so I filed this away as a decent piece of documentary evidence in support of John and Elizabeth being my Robert’s parents, but with the caveat that it is still circumstantial. The DNA match with my fourth cousin could only confirm this line as far back as our common Cullen ancestor (Robert himself) and not his parents.
Then another DNA match came along:
S.J. predicted as a fourth-sixth cousin.
Let’s look at a selection of my shared matches with S.J:
Investigating the shared matches strongly suggested that the connection is on my Cullen line and most likely at least as far back as my third great-grandparents Robert Cullen and Margaret Whitehead, since S.J. also matches the fourth cousin with whom I share those common ancestors.
Next I studied S.J.’s tree – a small, unlinked seven-person tree. It is a very interesting small tree as we have a surname in common! S.J’s grandfather was a man named Thomas Cullen who died in Glasgow. My Cullens were also from Glasgow, so I now have a common surname and location as well as confirmed cousins on my Cullen line who also match S.J. Since S.J.’s tree was not developed any further back than her grandfather I had to be proactive and build it back myself. I started by getting the 1905 marriage record for S.J.’s grandfather Thomas Cullen. The most important pieces of information I required in order to work the Cullen line backwards were the names of Thomas’ parents:
Robert CULLEN Carpet Weaver (deceased) and Margaret CULLEN M.S. GRAY
I proceeded to get Robert and Margaret’s 1881 marriage record which confirmed Robert’s parents as follows:
Thomas CULLEN Carpet Weaver and Martha CULLEN M.S. MCLEAN (deceased)
Thomas and Martha married in Glasgow in 1853, but as this was prior to statutory registration their marriage record did not contain the names of their parents. Thomas’s 1893 Glasgow death record, however, did list his parents:
William CULLEN Yarn Dresser (deceased) and Sarah CULLEN M.S. MORRISON (deceased)
I then sourced the 1879 Glasgow death record for William Cullen, which listed his parents:
John CULLEN Mason (deceased) and Elizabeth CULLEN M.S. MORTON (deceased)
The exact couple I had always thought may have been my Robert’s parents! This one match alone doesn’t prove they were his parents, but it is an excellent piece of evidence in favour of the hypothesis and I have since identified five more DNA matches that all descend from different children of John and Elizabeth and match confirmed cousins on my Cullen line. I believe the weight of evidence is now substantial enough that I can add John and Elizabeth to my tree as Robert’s parents. Every piece of extra evidence you can add to a theory via different matches gives it additional credence.
John and Elizabeth had a number of other children, so I was also able to add many new collateral lines to my tree. One of the new matches, descended from their daughter Christian, was even able to provide me with amazing early photographs from the 1850s.
Without DNA evidence I would never have been confident in John and Elizabeth being my Robert’s parents and would never have added them or their other children to my tree. I have broken down this brick wall, made new cousin connections, acquired copies of priceless family photographs and added hundreds of new collateral relatives to my tree thanks to DNA, in combination with traditional research. DNA provided the clues, but it took both DNA and traditional research to crack the case.
Remember that brick walls do not only exist on direct ancestor lines but also on collateral lines.
Many matches are difficult to solve due to lack of information, and this next case study will cover that kind of situation. The starting point is a male fourth-sixth cousin match with initials D.R., whose test is managed by a Jolene Roddin, but there is no tree, no useful shared matches and no additional information on the manager’s profile.
When a match offers such minimal information, the first thing to do is attempt contact, but unfortunately there was no reply in this case. So should I just give up and chalk it up as unsolvable? I did not and it paid dividends.
Since the manager is female and her surname starts with the same letter as the surname of the match it is likely they share that surname. This makes it probable that the match is the manager’s father, husband, brother or son. In most cases when someone manages another person’s DNA they are a close relative; I say in most cases because there are always exceptions. I manage many kits for clients and friends who are completely unrelated to me personally, for example. I am not representative of the database as a whole, however, and in this case I began by exploring the most likely option, which is that the test-taker and manager are close relatives who share a surname.
Online searching is your friend! It is always worth searching to see if you can identify the test-taker or results manager. Of course you need a bit of luck with this: if the manager was ‘Jane Smith’ as opposed to ‘Jolene Roddin’ then my chances of turning anything up via an online search would be negligible. A search for Jolene Roddin, however, led me to a US-based woman with a husband named David Roddin. This matches the initials on the test, so he quickly became the most likely candidate. Via online obituaries and traditional research methods I traced his ancestry back several generations. The majority of his lines yielded nothing until I hit upon a set of my own fourth great-grandparents! David descends from their eldest daughter, who was hitherto missing from my tree as her birth had not been registered and she was already living away from the family by the time of the first genealogically useful Scottish census (1841). Without this match I may never have known of her existence and emigration as a young woman. From nothing more than a set of initials, a manager’s name, some online sleuthing and a little work building a tree I was able to identify the most likely connection with this match and enhance my own tree even if I never receive a reply or they never upload a tree of their own.
As set out in Chapter 3, the inheritance properties of the X-chromosome are such that a match on the X can be narrowed down more extensively than a match on an autosome.
It is essential to take the gender of the test-takers being compared into account when it comes to X matches:
 An X-DNA match between two men means the connection can be narrowed down to the maternal side for both
An X-DNA match between two men means the connection can be narrowed down to the maternal side for both
 An X-DNA match between a man and a woman means the connection can be narrowed down to the man’s maternal side, but cannot be narrowed to a particular side for the woman (many lines can be discounted though, particularly her paternal grandfather’s entire quarter)
An X-DNA match between a man and a woman means the connection can be narrowed down to the man’s maternal side, but cannot be narrowed to a particular side for the woman (many lines can be discounted though, particularly her paternal grandfather’s entire quarter)
 An X-DNA match between two women is the scenario that can be narrowed the least but again there are still lines that can be discounted
An X-DNA match between two women is the scenario that can be narrowed the least but again there are still lines that can be discounted
When working with X-DNA matches make sure you fill out an XDNA table for the ancestors you could have inherited X-DNA from and consult this as it can aid you in working out the connection and will focus your search on the correct lines.
An example from FTDNA identifies Ruth as an X-match, but you have to be careful with this designation due to the fact FTDNA take into account very small segments – a match could be listed as an X-match even if only one 1 cM segment has been detected. I have outlined that small segments are problematic already, but it has been reported that there is an even higher rate of false positives on the X-chromosome, so it is best not to spend too much time on very small X-matches. A common recommendation is that in most cases only matches of 10 cMs or more should be investigated for males and 20 cMs for females.
The first thing you must do when ‘X-Match’ is listed on FTDNA is look at the match in the chromosome browser to check if the X-DNA segment is a reasonable size or should be discounted as too small. Be aware that the cMs you share on the X are not counted in the ‘Shared Centimorgans’ total on the main match page.
Even if an X-DNA segment appears to be of a reasonable size on a chromosome browser, it is always worth checking the actual numbers via the Detailed Segment Data view:
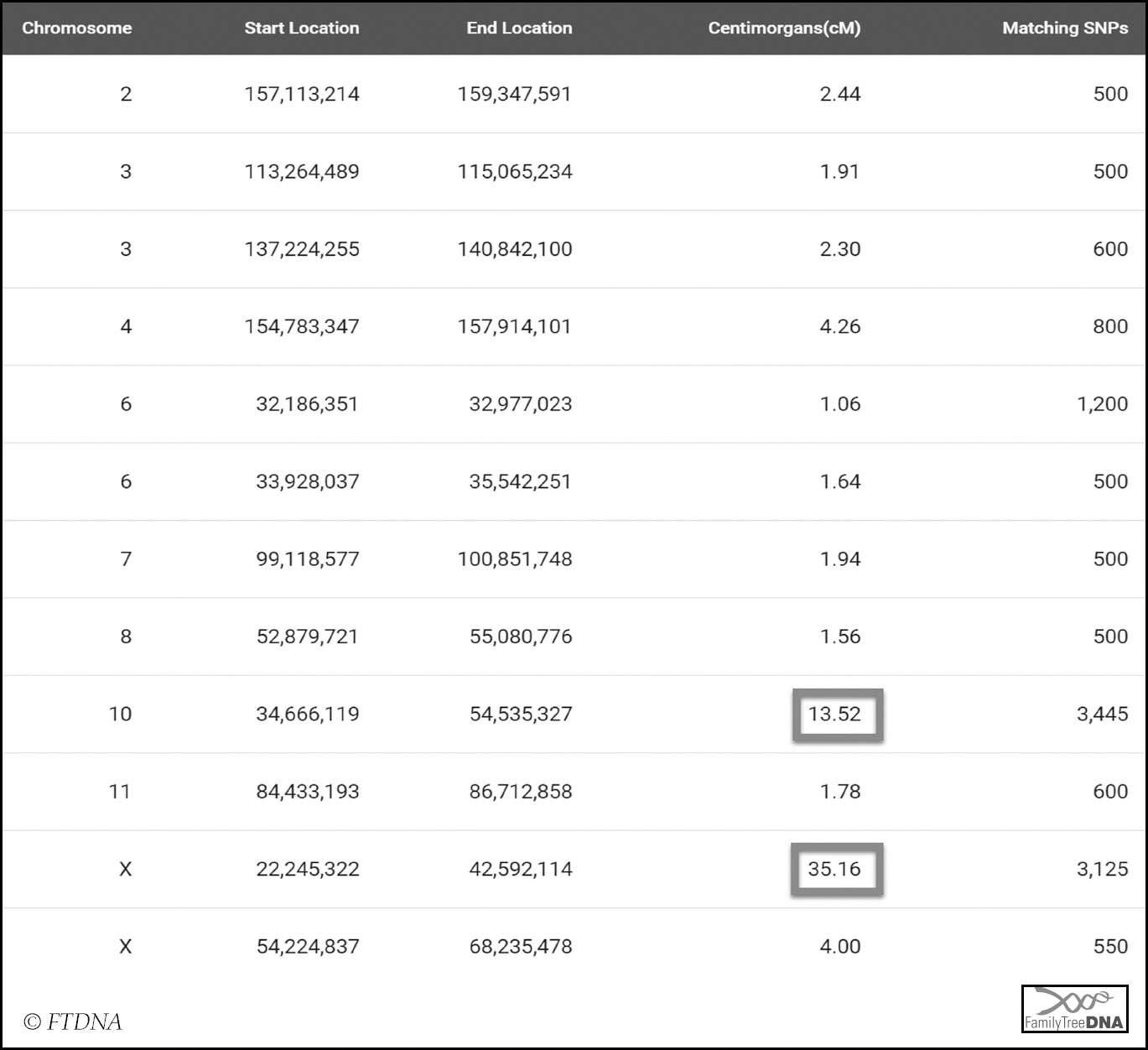
Ruth and I share a 13 cMs autosomal segment on chromosome 10, but it is clear that the largest segment shared between us is on the X-chromosome. At 35 cMs this is certainly a significant enough segment to warrant analysis.
The next step is to look at shared matches via the ‘In Common With’ tool, where Ruth matches confirmed paternal relatives. One curious detail is that my paternal half-sister, with whom I share my entire paternal X-chromosome, is not on the list. The reason for that is because my sister and Ruth do not share any autosomal DNA and you have to share a segment on an autosome at FTDNA to be considered a match, even if you share a significant X segment.
Knowing that my match with Ruth is on my paternal X-chromosome immediately narrows things a further generation to just one quarter of my tree, because my paternal X-chromosome was inherited in its entirety from my paternal grandmother. Additionally I can discount even more lines using my previously compiled paternal X-DNA inheritance table:
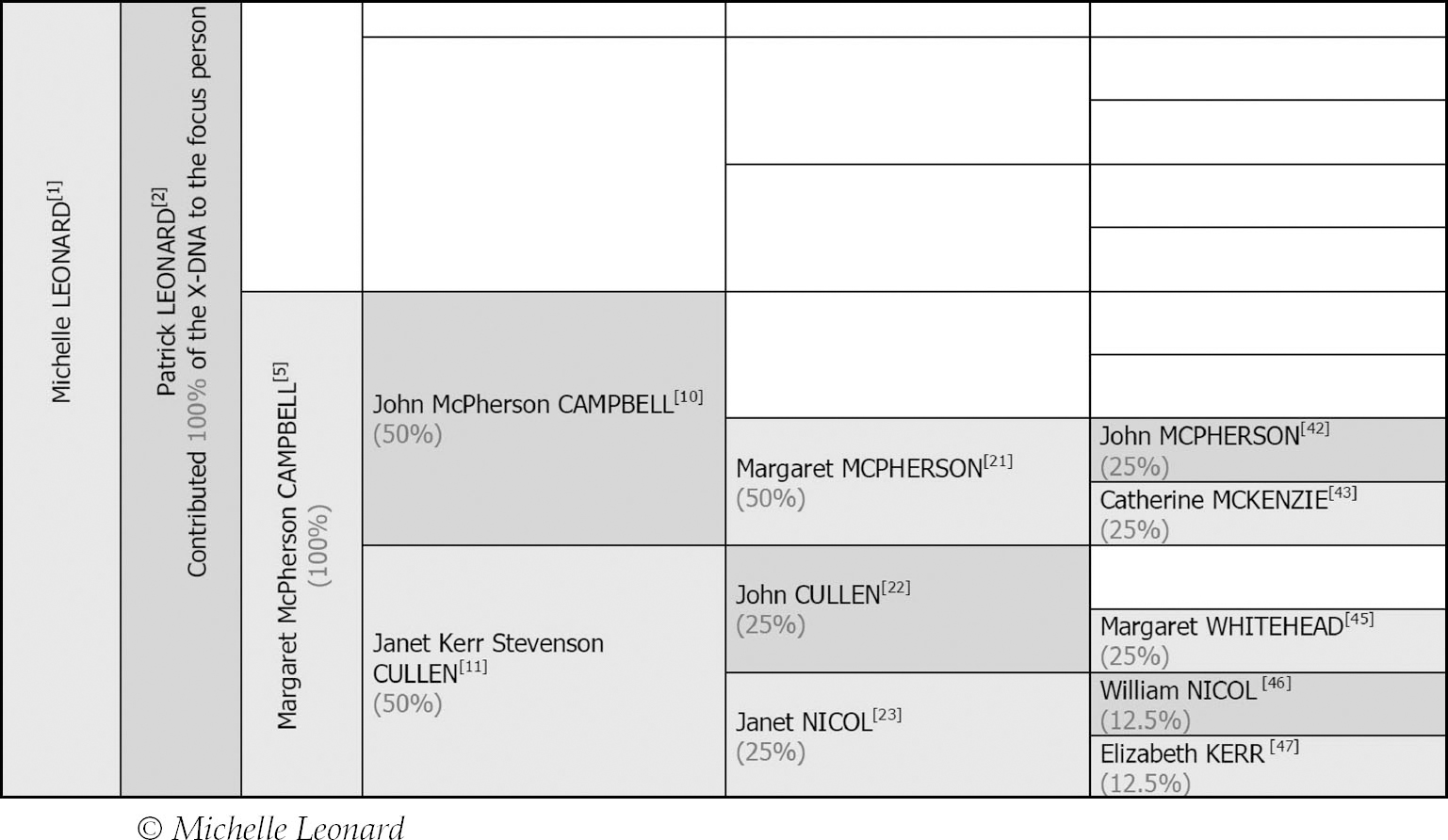
All of the white boxes represent lines I can discount since I could not have received X-DNA from those ancestors. Out of my eight paternal second great-grandparents I can only have inherited X-DNA from three of them (Margaret McPherson, John Cullen and Janet Nicol, with Margaret McPherson being statistically more likely due to the fact I may well have inherited a larger amount of X-DNA from her). Apart from the fact that 100% of my paternal X-chromosome came from my paternal grandmother, the rest of the percentages here are approximate. They are interesting as a guide and also show one of the most important differences between the X and autosomes. On an autosome the average you would inherit from a grandparent would be 25%, whereas here you could inherit 100% and the average you would inherit from a second great-grandparent is 6.25%, whereas on the X it could be 50% or even higher for a particular line. I call this the disproportionate X-DNA line: it lies on the paternal X and is the line in which a male interjects every other generation. Additionally the X-chromosome does not recombine quite as often as autosomes, so it is actually plausible that my paternal X-chromosome could be inherited entirely from my second great-grandmother Margaret McPherson: if she passed an X to my great-grandfather, who passed it whole to my grandmother, who may have passed it unrecombined to my father, who would then pass it whole to me.
Since this is an X-DNA match I only need to investigate the lines of Ruth’s tree she could have inherited X-DNA from, so I can discount a number of them and narrow things down to just a few. One in particular stands out: that of her second great-grandmother Ann McPherson.
We already know that my second great-grandmother, Margaret McPherson, could have contributed a significant proportion of my X-chromosome, so the fact Ruth also has a McPherson ancestor on an X-DNA line is a good lead to follow. It turned out that my Margaret and Ruth’s Ann were sisters. This X-DNA match gave me information on Ann’s line and enabled me to add a number of new branches stemming from her to my tree, as well as mapping a 35 cMs segment of my X-chromosome directly back to my second great-grandmother.
Let’s look at another X-DNA match, this time on 23andMe:
Linda and I do not share any autosomal DNA, but we share a whopping 43% of one of our X-chromosomes (83 cMs). Linda also matches on the same X segment as Ruth, so it seems very likely the whole 83 cMs can be mapped back to Margaret McPherson. The trouble here is that the actual connection is further back than either of us can take our trees. It would be virtually impossible, barring endogamy, to share 83 cMs with someone on an autosome and not be able to pinpoint a fairly recent common ancestor. The way the X-chromosome is inherited and the fact whole generations can be skipped, however, means matches on the X can end up being further back in time than matches on autosomes.
One thing it is important to be aware of is that Ancestry and MyHeritage do not report on the X-chromosome, so if you want to see X-Matches from those companies you have to transfer your raw data to GEDmatch or FTDNA.
The most important point to take away from these case studies is that it is essential to combine traditional genealogical research with DNA know-how in order to make the most of your results and successfully identify cousin connections.
There are a number of third-party tools that can aid you with your DNA results (we have already covered DNA Painter and GenomeMatePro), but the best known is GEDmatch.
GEDmatch is a free volunteer-run website that gives people a chance to match with those who have tested across all of the major companies and provides a suite of extremely helpful DNA analysis tools. There is also a subscription level, known as Tier 1, which provides several advanced tools for a small monthly fee.
In order to use GEDmatch you must download your raw data from your testing company and then upload it to the GEDmatch Genesis website. Instructions on how to do this are available on the website.
The most important tools you will want to get to grips with are ‘oneto-many’, ‘one-to-one’ and ‘people who match both, or one of two kits’. ‘One-to- many’ is GEDmatch’s equivalent of the match list and should be your first port of call:
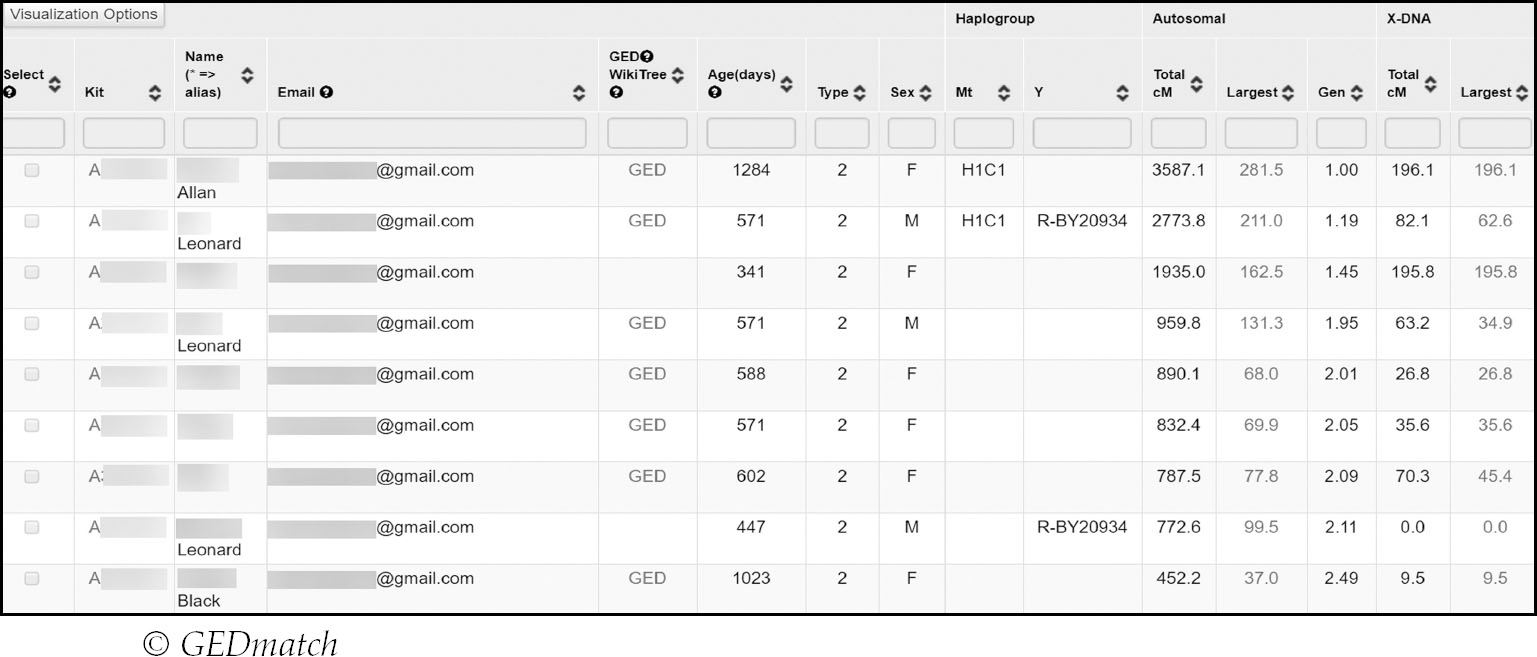
The layout is set up in such a way that it imparts a lot of information on one page – do not be daunted by all of the numbers. GEDmatch comparisons are based on kit numbers, which are generated when you upload your DNA. Each kit number migrated from the earlier classic GEDmatch website begins with a particular letter and which letter depends on the company your raw data originates from e.g. A for Ancestry or M for 23andMe. On Genesis, however, there is a new column entitled ‘Source’, which lists the company name or initial.
If you click on a kit number, a new window will open to show the full match list for that kit – this is something you cannot easily access on the other platforms. You can also upload a Gedcom of your family tree. GEDmatch supplies full segment information (total centiMorgans/largest segment) for both autosomes and X-chromosomes. If you click on the highlighted largest cM numbers under their respective ‘Largest’ columns this will display an autosomal or X one-to-one comparison with the chosen match. A one-to-one comparison imparts even more detail as follows:
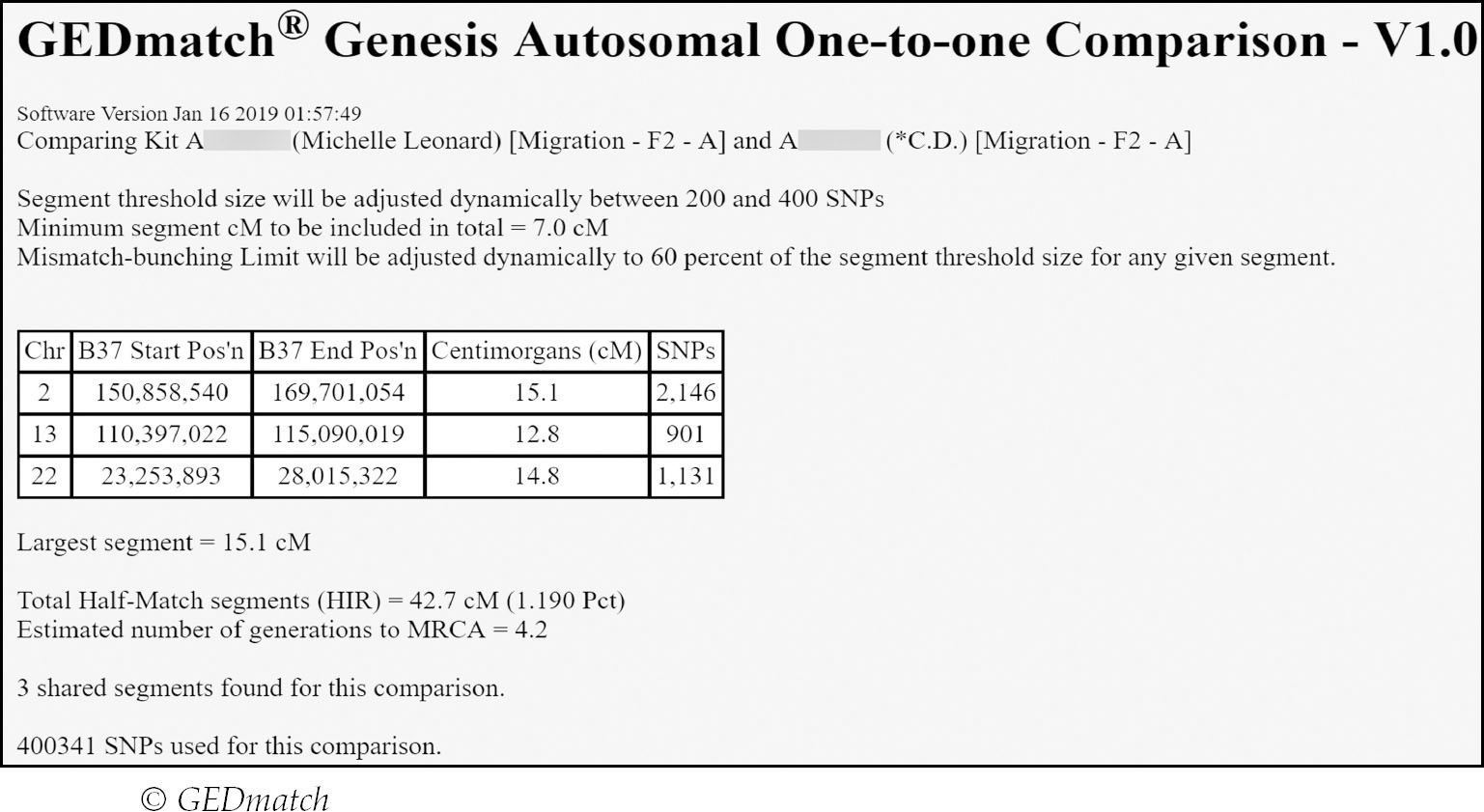
The one-to-one comparison provides the actual segment information with start/end locations and cM/SNP numbers. This is especially helpful for Ancestry matches since segment data is not reported on the testing site.
As opposed to relationship predictions, GEDmatch supplies an estimated number of generations to the MRCA under the column entitled ‘Gen’. Like other relationship prediction tools it is best not to take this number too literally and to once again use the Shared cM Project Tool instead.
GEDMatch’s equivalent of a shared match list is ‘People who match both kits or one of two kits’ (see p99).
This is very detailed and tells you exactly how much DNA the shared matches share with the two comparison kits.
It is also possible to complete full triangulation on GEDmatch both manually (via ‘one-to-one’ comparisons of three or more matches) or search via the Tier 1 ‘Segment Search’ and ‘Triangulation’ tools which automate the process.
For a comprehensive list of other third-party tools (e.g. DNAGedcom, Rootsfinder and clustering tools such as Genetic Affairs) consult the ISOGG Autosomal DNA Tools page.
Visual phasing is an advanced methodology, developed by Kathy Johnston, which can be a great aid in narrowing down matches to ancestral lines. Ideally three full siblings are required in order to undertake a visual phasing project, but it is possible with just two full and one half-sibling (as I have done) or two siblings and a number of other close relatives. When you practise visual phasing what you are doing is looking at the points on the chromosomes at which crossovers occur between siblings and using the crossover information to determine which of your four grandparents contributed which pieces of your DNA. The goal is to be able to map your DNA to your four grandparents using this method. It can be time-consuming and complex, but it is well worth the time invested.
I have developed a full visually phased reference chart for my chromosomes, which I can now use whenever I have access to segment data for a match. All I have to do is look at where we match, then consult my VP reference chart for that chromosome and I will know which of my four grandparents contributed that segment of DNA. This means I can narrow all matches down to at least one quarter of my tree. I also, however, still consult my regular chromosome map (e.g. on DNA Painter) since I have mapped many segments further back than my grandparents.
It is important to learn the technique properly before trying out visual phasing: there is a visual phasing Facebook group that provides excellent information on how to go about the learning process as well as courses and articles. Additionally, developer Steven Fox has created an incredible spreadsheet tool for visual phasing that automates much of the set-up process and is a great time saver (only available via the Facebook group).
Admixture is the most recognisable and eye-catching aspect of an autosomal DNA test. It is the feature companies highlight the most when advertising their products. For that reason many people test solely for the admixture portion of the results but, in reality, it is far less useful for genealogy than DNA match lists.
When creating admixture estimates all of the different companies use their own proprietary reference population sets and computerised algorithms. A reference population is a DNA dataset comprised of samples from individuals whose ancestry is believed to trace back to specific areas. To build these reference sets companies test people with four grandparents who all came from the same area or use academic project data along the same lines. These results are then used as a reference for the specific area concerned. There are always going to be some errors with this process, as admixture aims to predict ancestry from 500–1,000 years back in time and there is no way to know for sure that the ancestry of those tested for the reference panels genuinely does stem from the correct areas. The larger the reference populations become, however, the more accurate they should be. When a new customer takes a test, their DNA is compared to all of the reference samples and an estimate is generated via computerised comparisons. There is no universal reference set or method of calculating admixture and, therefore, results can and will differ across platforms. If you examine estimates for the same person at more than one company you will see that there are different categories, interpretations and percentages.
It is widely believed admixture results are accurate to the continental level, but more problematic and changeable when drilling down to country and regional levels. It is important not to put too much stock in very small percentages reported by any of the companies as they could be considered background ‘noise’. As DNA segments become smaller there is a greater chance of matching random and inaccurate ethnicity patterns. The science behind admixture is evolving all the time, so admixture estimates will occasionally be updated as companies improve their reference sets and algorithms.
Ancestry calls the admixture portion of their test ‘DNA Story’ and another major facet of this is the ‘Genetic Communities’ feature. Rolled out in March 2017, this has now been fully integrated into the ‘Ethnicity Estimate’ area of the results and the term ‘Genetic Communities’ has been replaced by ‘Regions’ and ‘Migrations’. The ‘Ethnicity Estimates’ are based on reference populations, but Regions do not stem from DNA analysis and are actually determined by the family trees of your matches. For example I match a large number of test-takers who have ancestors from Ulster on their tree, so the system determined that I am also likely to have ancestors from Ulster, which I do, and placed me in the Ulster Region as well as the more detailed Tyrone, Londonderry and Antrim sub-region. This is, of course, dependent upon the accuracy of the trees of your matches and will not always be correct, but it has proven very accurate for many, especially for regions with a high number of matches.
23andMe is the only company that provides a chromosome map which breaks down segments via admixture group and, if you have one or both parents tested, they also split your ethnicity by parent. This is not particularly helpful if all of your ancestry comes from similar areas, but it can become valuable when you have markedly different ancestral components. LivingDNA currently provides the most detailed regional breakdown for British and Irish ancestry.
My cousin Marion’s results are an example of how admixture analysis can generate clues to help solve unknown ancestor mysteries. Three-quarters of Marion’s ancestry is documented, but she has an unknown grandfather. In the three identified quarters there are no known Jewish ancestors, so the fact that around 25% Ashkenazi Jewish is reported in her estimates is a good clue as to the origins of her unknown grandfather.
GEDmatch also has a number of admixture calculators (mainly developed for academic purposes) that you can explore if you are interested in yet another perspective.
Admixture key points:
 Admixture estimates will vary between companies because they each use different reference populations
Admixture estimates will vary between companies because they each use different reference populations
 They are accurate to the continental level but cannot be taken too literally at the country and regional levels
They are accurate to the continental level but cannot be taken too literally at the country and regional levels
 Be especially cautious with tiny percentages as these could be erroneous
Be especially cautious with tiny percentages as these could be erroneous
 Estimates can only cover DNA you have personally inherited from your ancestors and you do not possess all of the same DNA they did
Estimates can only cover DNA you have personally inherited from your ancestors and you do not possess all of the same DNA they did
 Estimates can give you a broad idea of your origins and may provide genealogical clues if you were adopted or have a recent unknown ancestor mystery
Estimates can give you a broad idea of your origins and may provide genealogical clues if you were adopted or have a recent unknown ancestor mystery
This is an emerging science that will improve over time as more people test and reference populations increase, but be careful in reading too much into estimates for now – they are called ‘estimates’ for a reason.
 Use DNA in conjunction with traditional research as that is how to get the most out of it
Use DNA in conjunction with traditional research as that is how to get the most out of it
 Start with your largest matches and work your way down your first page
Start with your largest matches and work your way down your first page
 Test close relatives especially older generations to narrow your matches down and gain more of your ancestors’ DNA
Test close relatives especially older generations to narrow your matches down and gain more of your ancestors’ DNA
 Upload your family tree no matter how small and make sure you link it to your results
Upload your family tree no matter how small and make sure you link it to your results
 Build your tree as extensively as you can and build the trees of your matches to find links
Build your tree as extensively as you can and build the trees of your matches to find links
 Study shared matches as they may hold vital clues, especially if you have no tree of your own
Study shared matches as they may hold vital clues, especially if you have no tree of your own
 Be proactive – contact your matches! Many will not respond but keep trying and you will find new cousins to collaborate with
Be proactive – contact your matches! Many will not respond but keep trying and you will find new cousins to collaborate with
 Organise your data and take advantage of all the help out there „ Explore third party tools and techniques such as chromosome mapping, autosomal clusters and visual phasing
Organise your data and take advantage of all the help out there „ Explore third party tools and techniques such as chromosome mapping, autosomal clusters and visual phasing
 Remember there will be many matches you cannot work out, but each one you do adds to or confirms your tree
Remember there will be many matches you cannot work out, but each one you do adds to or confirms your tree
Have fun with it!
Keep in mind that genetic genealogy evolves rapidly and all of the companies will change aspects of the way they display things, so the images and exact directions given in this chapter may become out of date, but the basic premises and techniques should endure.