Covariance and precision matrix estimation and inference pervade every aspect of statistics and machine learning. They appear in the Fisher’s dis-criminant and optimal portfolio choice, in Gaussian graphical models where edge represents conditional dependence, and in statistical inference such as Hotelling T-test and false discovery rate controls. They have also widely been applied to various disciplines such as economics, finance, genomics, genetics, psychology, and computational social sciences.
Covariance learning, consisting of unknown elements of square order of dimensionality, is ultra-high dimension in nature even when the dimension of data is moderately high. It is necessary to impose some structure assumptions in covariance learning such as sparsity or bandability. Factor models offer another useful approach to covariance learning from high-dimensional data. We can infer the latent factors that drive the dependence of observed variables. These latent factors can then be used to predict the outcome of responses, reducing the dimensionality. They can also be used to adjust the dependence in model selection, achieving better model selection consistency and reducing spurious correlation. They can further be used to adjust the dependence in large scale inference, achieving better false discovery controls and enhancing high statistical power. These applications will be outlined in Chapter 11.
This chapter introduces theory and methods for estimating large covariance and precision matrices. We will also introduce robust covariance inputs to be further regularized. In Chapter 10, we will focus on estimating a covariance matrix induced by factor models. We will show there how to extract latent factors, factor loadings and idiosyncratic components. They will be further applied to high-dimensional statistics and statistical machine learning problems in Chapter 11.
9.1 Basic Facts about Matrices
We here collect some basic facts and introduce some notations. They will be used throughout this chapter.
For a symmetric m x m matrix A, we have its eigen-decomposition as follows
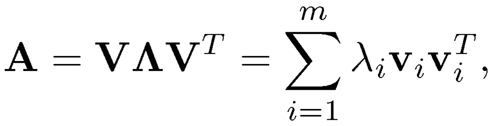 (9.1)
(9.1)
where λi is the eigenvalue of A and vi is its associated eigenvector. For a m x n matrix, it admits the singular value decomposition
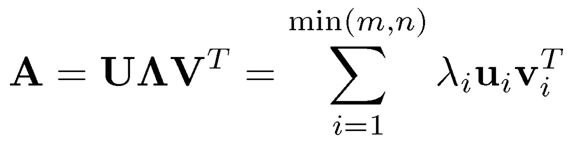 (9.2)
(9.2)
where U is an m x m unitary (othornormal) matrix, ∧ is an m x n rectangular diagonal matrix, and V is an n x n unitary matrix. The diagonal entries λi of ∧ are called the singular values of A. The columns of U and the columns of V are known as the left-singular vectors and right-singular vectors of A, respectively:
• the left-singular vectors of A are orthonormal eigenvectors of AAT;
• the right-singular vectors of A are orthonormal eigenvectors of ATA;
• the non-vanishing singular values of A are the square roots of the non-vanishing eigenvalues of both ATA and AAT.
We will assume the singular values are ordered in decreasing order and use λi(A) to denote the ith largest singular value, λmax(A) = λ1(A) the largest singular value, and λmin (A) = λm(A) the minimum eigenvalue.
Definition 9.1 A norm of an n x m matrix satisfies
1. ||A||≥ 0 and ||A||= 0 if and only if A = 0;
2. ||αA||= |α|||A||
3. ||A+B|| ≤ ||A||+||B||;
4. IIAB|| ≤ ||A||||B||, if AB is properly defined.
A common method to define a matrix norm is the induced normml: ||A|| — max||x||=1 ||Ax||. In particular, ||A||p,q = max||x|p=1||Ax||q, which satisfies, by definition,
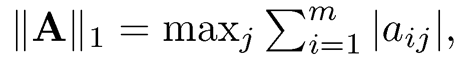
More specifically, when p = g, it becomes the matrix Lp normml:
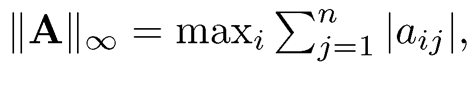
It includes, as its specific cases,
1. Operator normml: ||A||2 = λmax(ATA)1/2, which is also written as ||A||op or simply ||A||;
2. L1-normml: ||A||1= maxjΣmi=1 |aij|, maximum L1-norm of columns;
3. L∞-normml: ||A||∞ = max Σni=1, |aij|, maximum L1-norm of rows.
Definition 9.2 The Frobenius norm is defined as
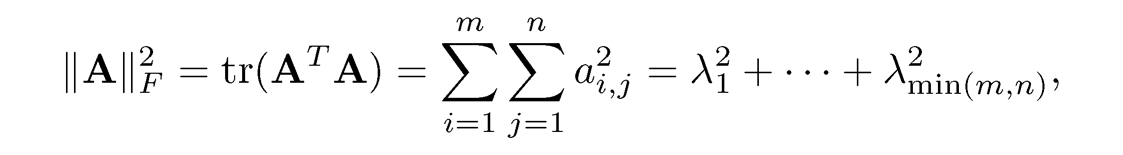
and the nuclear norm is given by
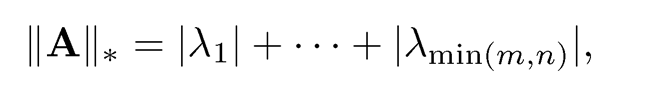
where {λi} are singular values defined in (9.2).
Operator norm is hard to manipulate, whereas ||A||∞, and ||A||1 are easier. We have the following inequalities.
Proposition 9.1 For any m x n matrix A, we have
1. ||A||2 ≤ ||A||∞ ||A||1 and ||A||2 ≤||A||1 if A is symmetric.
2. ||A||max ≤||A||2 ≤ (mn)1/2||A||max, ||A||max=maxij|aij|
3. ||A||2 ≤ ||A||F ≤ √r||A||2, r=rank(A);
4. ||A||F≤||A||*≤√r||A||F, r=rank(A)
5. n-1/2||A||∞,≤ ||A||2≤ m1/2||A||∞ and m-1/2 ||A||1 ≤||A||2 ≤n1/2||A||
Here are two basic results about matrix perturbation. For two m x m symmetric matrices A and A, let Â, let λ1 ≥ ⋯ ≥ λm and λ̂ ≥ ⋯ ≥ λ̂ m be their eigenvalues with associated eigenvectors vi, • • • , vm and îri, • • • , îrm, respectively. For two given integers r and s, write d = s — r
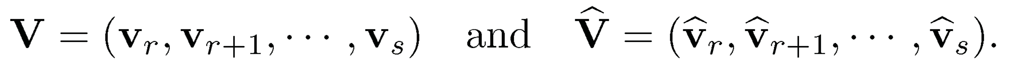
and let σ1 ≥ ⋯ ≥ σd be the singular values of V̂TV. The d principal angles between their column spaces V and V are defined as where {θj}ai=1, where θj = arccos σj. Let
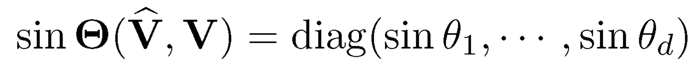
be the d x d diagonal matrix. Note that the Frobenius norm of two projection matrices
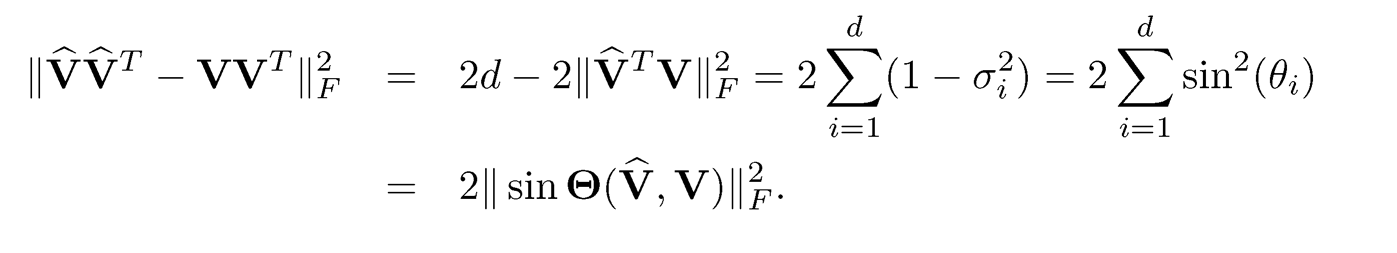 (9.3)
(9.3)
Proposition 9.2 For two m x m symmetric matrices A and Â, we have
• Weyl Theoremml: λmin(Â - A) ≤ λ̂i -λ̂i ≤ λ̂max (Â — A). Consequently, |λ̂̂i-λi|≤||Â-A||.
• Davis-Kahan sin θ Theoremml: With λ̂0 = ∞, λm+1 =-∞ and δ= min{|λ̂- λ̂| : λ̂ ∈ [λ̂s, λ̂r]λ̂̂ ∈ (∞, λ̂̂8-1] U [λ̂̂r+1, ∞)} denoting the eigen gap, we have

The result also holds for the operator norm or any other norm that is invariant under an orthogonal transformation.
The above result is due to Davis and Kahan (1970). Specifically, by taking r = s = i, we have
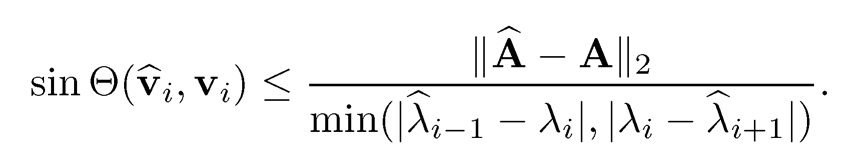
Note that

Combining the last two results, we have
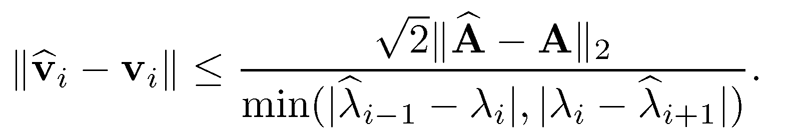
The eigen gap δ involves both sets of eigenvalues. In statistical applications, it typically replaces λ’is As by its λ’is using Weyl’s theorem. Yu, Wang and Samworth (2014) give a simple calculation and obtain
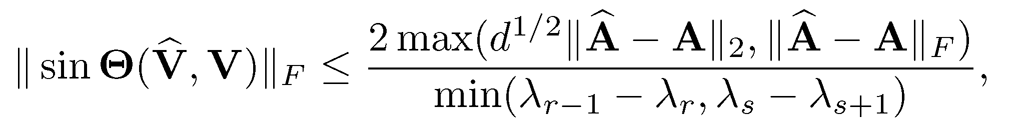 (9.4)
(9.4)
and in particular
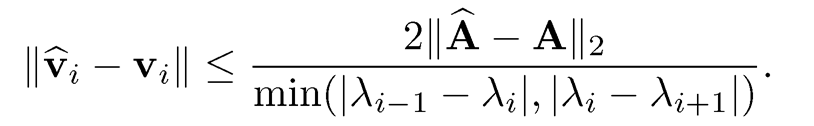 (9.5)
(9.5)
We conclude this section by introducing Sherman-Morrison-Woodbury Formula and block matrix inversion. If Σ = BΣfBT +Σu, then
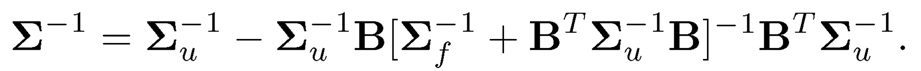 (9.6)
(9.6)
Let S = A — BD-1C be the Schur complement of D. Then
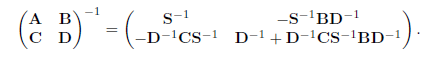
9.2 Sparse Covariance Matrix Estimation
To estimate high-dimensional covariance and precision matrices, we need to impose some covariance structure. Conditional sparsity was imposed in Fan, Fan, and Lv (2008) for estimating a high-dimensional covariance matrix and their applications to portfolio selection and risk estimation. This factor model induces a low-rank plus sparse covariance structure (Wright, Ganesh, Rao, Peng, and Ma 2009; Candés, Li, Ma and Wright, 2011), which includes (unconditional) sparsity structure imposed by Bickel and Levina (2008b) as a specific example. Let us begin with the sparse covariance matrix estimation.
9.2.1 Covariance regularization by thresholding and banding
Let us first consider the Gaussian model where we observed data {Xi}ni=1 from p-dimensional Gaussian distribution N(μ, Σ). Natural estimators for μ and Σ are the sample mean vector and sample covariance matrix
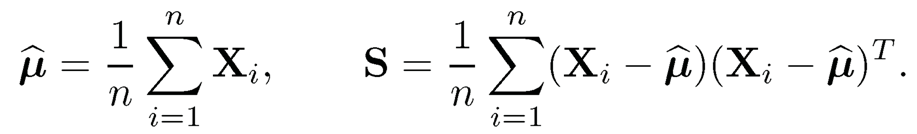 (9.7)
(9.7)
They are the maximum likelihood estimator when p ≤ n-1. However, the sample covariance is degenerate when p ≥ n. Since we estimate O(p) elements1, the noise accumulation will make the sample covariance matrix differs substantially from the population one. See Figure 9.1. It shows that the sample covariance matrix is not a good estimator, without any regularization. This is well documented in the random matrix theory (Bai and Silverstein, 2009).
To make the problem estimable in high-dimension, we need to impose a structure on the covariance matrix. The simplest structure is the sparsity of the covariance Σ. This leads to a penalized likelihood method. The log-likelihood for the observed Gaussian data is

Substitution μ by its MLE X̄, expressing (Xi — X̄)Σ-1(Xi, — X̄)T as tr(Σ-1(Xi — X̄)(Xi — X̄)T) where tr(A) is the trace of a matrix A, we obtain the negative profile log-likelihood
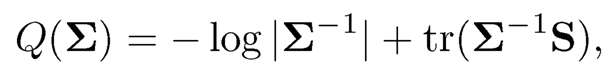 (9.8)
(9.8)
after ignoring the constant term -n/2 log(2π) and dividing the result by —n/2. Thus, the penalized likelihood estimator is to minimize
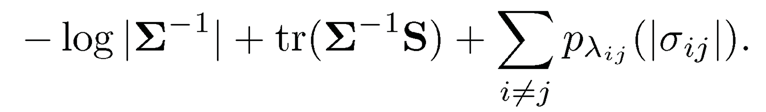 (9.9)
(9.9)
_______________
11,000 × 1,000 matrix involves over half a million elements, whereas 10,000 x 10,000 contains over 50 million elements!
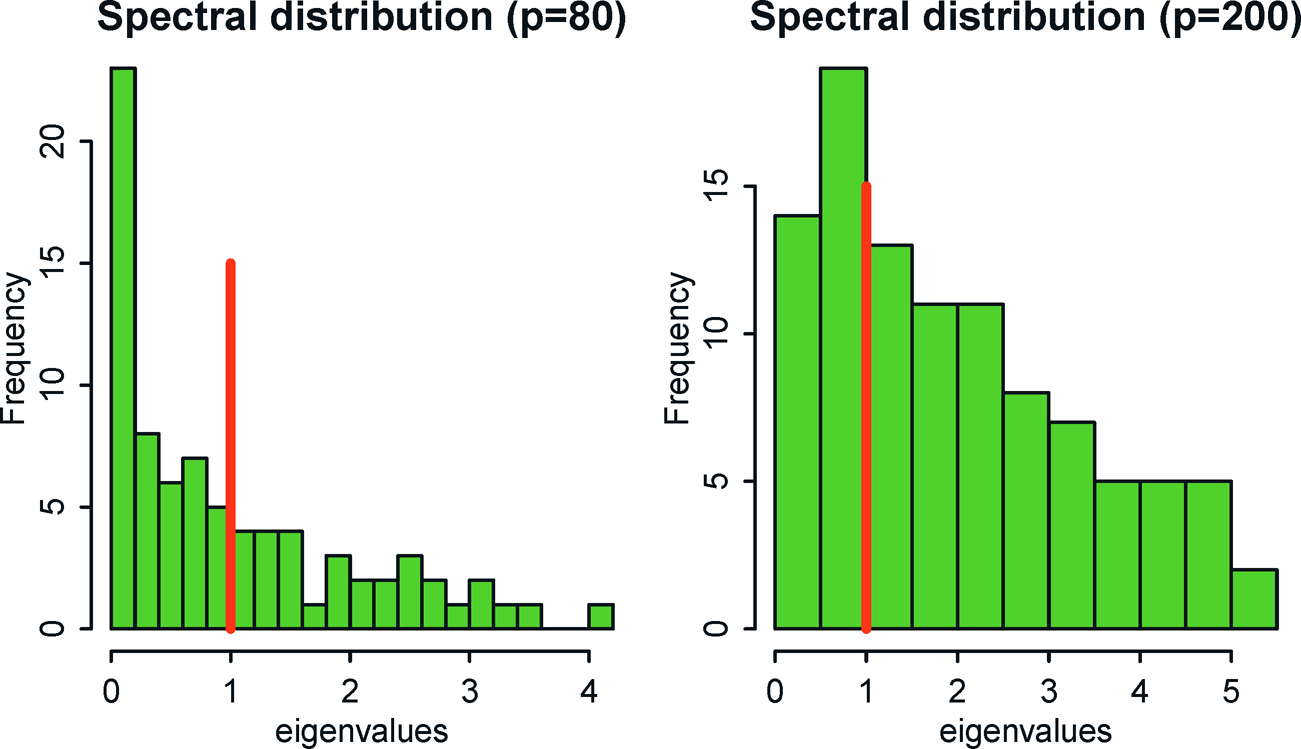
Figure 9.1: Empirical distributions of the eigenvalues of the data {Xi}ni=1 simulated from N(0, Ip) for n = 100 and p = 80 (left panel) and p = 200 (right panel). For the case p = 200, there are 100 empirical eigenvalues that are zero, which are not plotted in the figure. Note that the theoretical eigenvalues are 1 for the identity matrix Ip. Theoretical and empirical spectral distributions are very different when n and p are of the same order.
Here, we do not penalize on the diagonal elements, since they are not supposed to be sparse.
The penalized likelihood estimator is unfortunately hard to compute because (9.9) is a non-convex problem even if the penalty function is convex. Bickel and Levina (2008b) explored the sparsity by a simple thresholding operator:
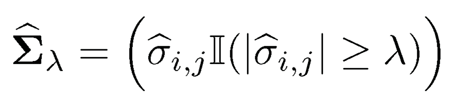 (9.10)
(9.10)
for a given threshold parameter λ, where σ̂ij =sij . In other words, we kill small entries of the sample covariance matrix, whose value is smaller than A. Again, in (9.10), we do not apply the thresholding to the diagonal elements. The thresholding estimator (9.10) can be regarded as a penalized least-squares estimator: Minimize with respect to σij
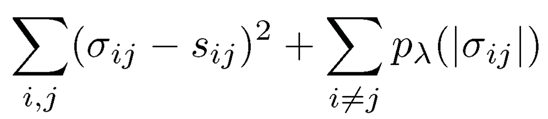 (9.11)
(9.11)
for the hard thresholding penalty function (3.15). Allowing other penalty functions in Sections 3.2.1 and 3.2.2 leads to generalized thresholding rules (Antoniadis and Fan, 2001; Rothman, Levina and Zhu, 2009).
Definition 9.3 The function τλ,(·) is called a generalized thresholding function, if it satisfies
a) a) τλ(z)| ≤ a|y| for all z, y that satisfy |z — y| ≤ λ/2 and some a > 0;
b) |τλ(z) — z| ≤λ, for all z ∈ ℝ
Note that by taking y = 0, we have τλ(z) = 0 for |z| ≤ λ/2 and hence it is a thresholding function. It is satisfied for the soft-thresholding function by taking a = 1 and the hard-thresholding function by taking a = 2. It is also satisfied by other penalty functions in Sections 3.2.2.
The simple thresholding (9.10) does not even take the varying scales of the covariance matrix into account. One way to account for this is to threshold on t-type statistics. For example, we can define the adaptive thresholding estimator (Cai and Liu, 2011) as
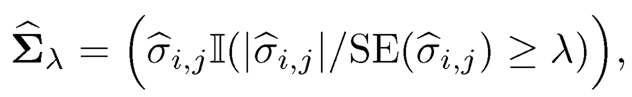
where SE(σ̂ij) is the estimated standard error of σ̂ij.
A simpler method to take the scale into account is to apply thresholding on the correlation matrix (Fan, Liao and Mincheva, 2013). Let ψ̂λ, be the thresholded correlation matrix with thresholding parameter λ ∈ [0, 1]. Then,
 (9.12)
(9.12)
is an estimator of the covariance matrix. In particular, when λ = 0, it is the sample covariance matrix, whereas when λ = 1, it is the diagonal matrix, consisting of the sample variances. The estimator (9.12) is equivalent to applying the entry dependent thresholding λij= √σ̂i,j, σ̂i,j λ to the sample covariance matrix Σ. This leads to the more general estimator
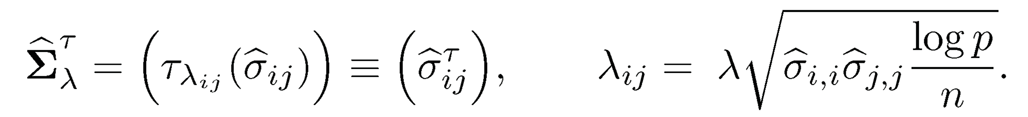 (9.13)
(9.13)
where τλij(·) is a generalized thresholding function, (σ̂ij) is a generic estimator of covariance matrix Σ, and √(log p)/n is its uniform rate of convergence, which can be more generally denoted by an (see Remark 9.1). In (9.13), we use λ\/log p/n to denote the level of thresholding to facilitate future presentations.
An advantage of thresholding is that it avoids estimating small elements so that noises do not accumulate as much. The decision of whether an element should be estimated is much easier than the attempt to estimate it accurately. Thresholding creates biases. Since the matrix Σ is sparse, the biases are also controlled. This is demonstrated in the following section.
Bickel and Levina (2008a) considered the estimation of a large covariance matrix with banded structure in which o-i3 is small when |i-j| is large. This occurs for time series measurements where observed data are weakly correlated when they are far apart. They also appear in the spatial statistics in which correlation decays with the distance where measurements are taken. They proposed a simple banded estimator as and derived the convergence rate. Cai, Zhang, and Zhou (2010) established the minimax optimal rates of convergence for estimating large banded matrices. See Bien, Bunea, and Xiao (2016) for convex banding, Yu and Bien (2017) for varying banding, and Bien (2018) for graph-guided banding. Li and Zou (2016) propose two Stein’s Unbiased Risk Estimation (SURE) criteria for selecting the banding parameter k and show that one SURE criterion is risk optimal and the other SURE criterion is selection consistent if the true covariance matrix is banded.
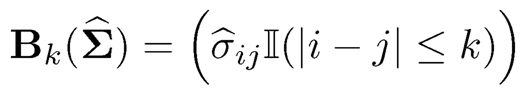
9.2.2 Asymptotic properties
To study the estimation error in the operator norm, we note, by Proposi-tion 9.1, that for any general thresholding estimator (9.13),
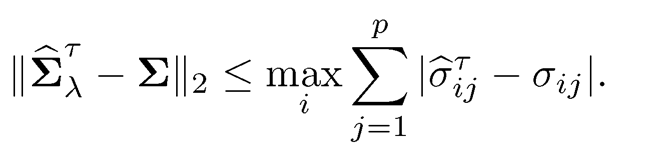 (9.14)
(9.14)
Thus, we need to control only the Li-norm in each row, whose error depends on the effective number of parameters, i.e. the maximum number of nonzero elements in each row
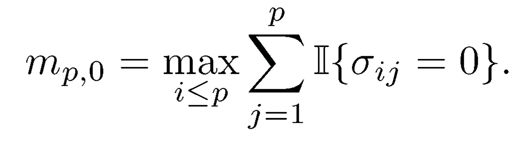
A generalization of the exact sparsity measure mo is the approximate spar-sity measure
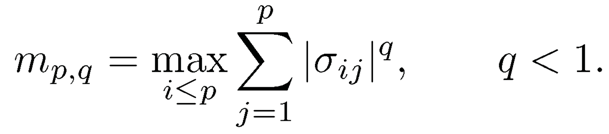 (9.15)
(9.15)
Note that q = 0 corresponds to exact sparsity mp,0. Bickel and Levina (2008b) considered the parameter (covariance) space
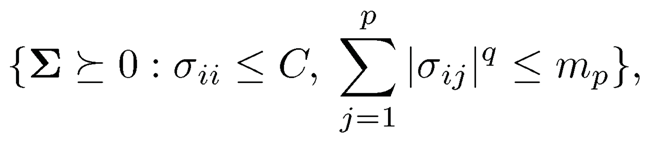
with generalized sparsity controlled at a given level mp. As a result, by using the Cauchy-Schwartz inequality |σij| ≤σiiσjj, we have
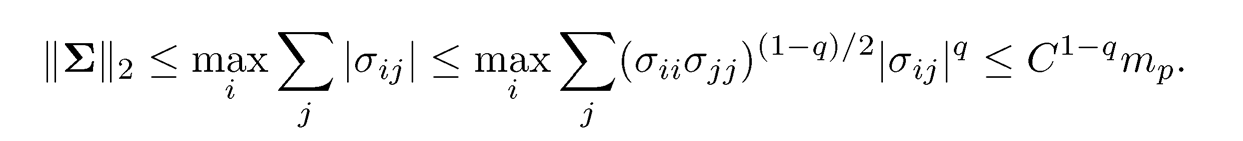
This leads to the generalization of the parameter space to (Cai and Liu, 2011)
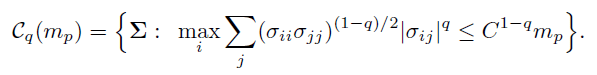 (9.16)
(9.16)
The following result is very general. It encompasses those of Bickel and Levina (2008b) and Cai and Liu (2011).
Theorem 9.1 Suppose that the pilot estimator Σ̂ of Σ satisfies
 (9.17)
(9.17)
where Cois a positive constant. If log p = o(n) and mini<p σii=γ > 0, there exists a positive constant C1 such that for sufficiently large n and constant λ, we have
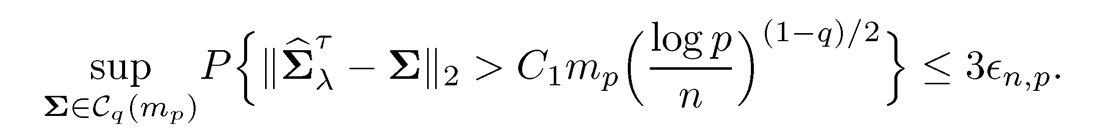
A similar result also holds over under the Frobenius normml:
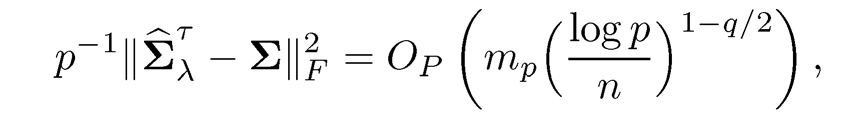 (9.18)
(9.18)
if maxi≤p σii≤ C2. In addition, if ||Σ-1|| is bounded from below, then
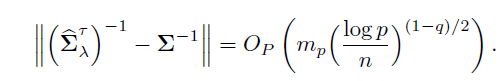
Remark 9.1 The last part of Theorem 9.1follows easily from (Exercise 9.1)
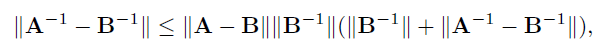
for any matrix A and B. The result of Theorem 11.1 holds more generally by replacing √(log p)/n with any rate an →0. In this case, the condition (9.17) and the threshholding are modified as
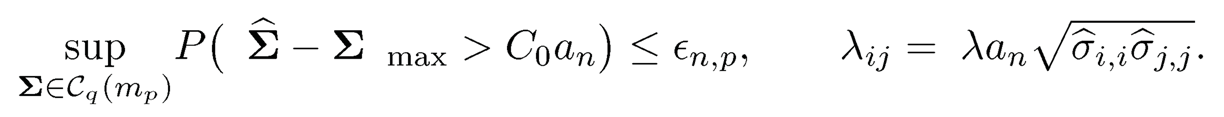
The results of Theorem 9.1hold with replacing log p/n by an, using identically the same proof.
The above Theorem and its remark are a slight generalization of that in Avella-Medina, Battey, Fan, and Li (2018), whose proof is simplified and relegated to Section 9.6.1. It shows that as long as the pilot estimator uniformly converges at a certain rate and the problem is sparse enough, we have the convergence of the estimated covariance matrix. In particular, when g = 0, the convergence rate is mp√log p/n. This matches with the upper bound (9.14): effectively we estimate mp parameters, each with rate √log p/n. Cai and Zhou (2011) showed the rate given in Theorem 9.1is minimax optimal.
If the data {Xi} are drawn independently from N(μ,Σ), and λ = M(n-1 log p)1/2 for large M, Bickel and Levina (2008b) verified the condition (9.17):
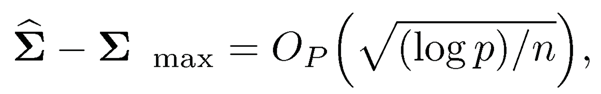 (9.19)
(9.19)
uniformly over the parameter space Cq(mp). Here, we use (9.19) to denote the meaning of (9.17). The first result in Theorem 9.1will also be more informally written as
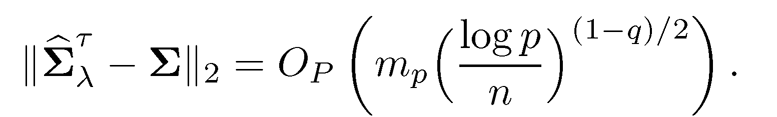 (9.20)
(9.20)
Condition (9.19) holds more generally for the data drawn from sub- Gaussian distributions (4.20) using the concentration inequality (Theo-rem 3.2). The sample covariance matrix involves the product of two random variables. If they are both sub-Gaussian, their product is sub-exponential and a concentration inequality for sub-exponential distribution is needed. The calculation is somewhat tedious. Instead, we appeal to the following theorem, which can be found in Vershynin (2010). See also Tropp (2015).
Theorem 9.2 Let Σ be the covariance matrix of Xi. Assume that {Σ-½ Xi}ni=1 lare i.i.d. sub-Gaussian random vectors, and denote by κ = sup||v||2=1 ||VTXi||ψ2, where ||X||ψ2 = inf{s > 0 : Eexp(X/s) ≤ 2} is the Orlicz norm. Then for any t ≥ 0, there exist constants C and c only depending on κ such that

where δ= C√p/n +t√n, and S is the sample covariance matrix (9.7)
The above bound depends on the ambient dimension p. It can be replaced by the effective rank of Σ, defined as tr(Σ)||Σ||2. See Koltchinskii and Lounici (2017) for such a refined result. In particular, if we take p = 2 in Theorem 9.2, we obtain a concentration inequality for the sample covariance, which is stated in Theorem 9.3.
Theorem 9.3 Let siibe the (i, j)-element of the sample covariance matrix S in (9.7) and Σij be the 2 x 2 covariance matrix of (Xi, Xj)T. Under conditions of Theorem 9.2, if {σii} are bounded, there exists a sufficiently large constant M such that when |t|| ≥ M, we have, for all i and j,
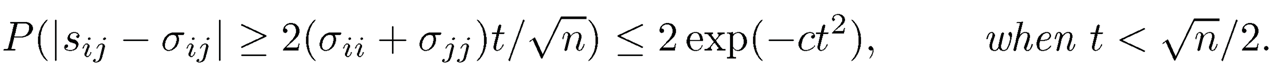
If in addition logp = o(n), we have

This theorem can be used to verify condition (9.17). Hence (9.18) and (9.20) hold for thresholded estimator (9.13), when data are drawn from a sub-Gaussian distribution.
9.2.3 Nearest positive definite matrices
All regularized estimators are symmetric, but not necessarily semipositive definite. This calls for the projection of a generic symmetric matrix Σλ into the space of semipositive definite matrices.
The simplest method is probably the L2-projection. As in (9.2), let us decompose
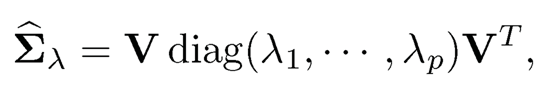 (9.21)
(9.21)
and compute its L2-projected matrix
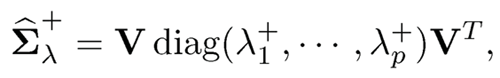
where λ+i =max(λi, 0) is the positive part of λi. This L2-projection is the solution to the least-squares problems: Minimize, with respect to X ≥0 the problemml:

Another simple method is to compute
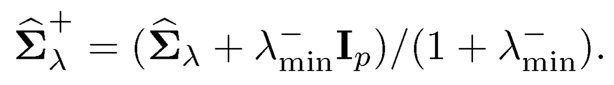
where λ-min is the negative part of the minimum eigenvalue of Σ̂λ, which equals —λmin when λmin < 0 and 0 otherwise. Both projections do not alter eigen-vectors, using the same matrix V as the original decomposition (9.21). When applied to the correlation matrix, the second method still yields a correlation matrix.
The above projection gives a simple and analytic solution. However, it is not necessarily optimal. For example, Qi and Sun (2006) introduced an algorithm for computing the nearest correlation matrix: For a given symmetric matrix A, find a correlation matrix R to minimize
 (9.22)
(9.22)
To apply the algorithm (9.22) to our thresholding estimator Σ̂λ, we compute the standardized input
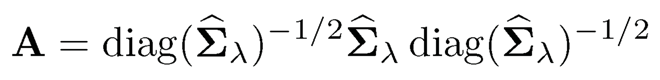
first and then apply (9.22) to obtain R̂λ, and transform back to the covariance matrix as
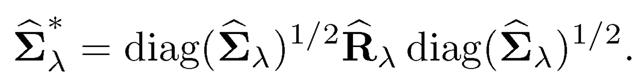
Of course, if the thresholding is applied to the correlation matrix as in (9.12), we can then take the input A = ψ̂λ. After the nearest correlation matrix projection, we transform the estimate back.
There is a R-package called nearPD that computes (9.22). The function is also called nearPD.
As a generalization of (9.22), one can also solve the following optimization
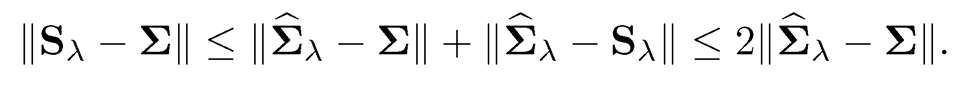
for a given δ ≥ 0. Namely, we wish to find a correlation matrix R with minimum eigenvalue δ such that it is closest to A. When δ= 0, it reduces to problem (9.22).
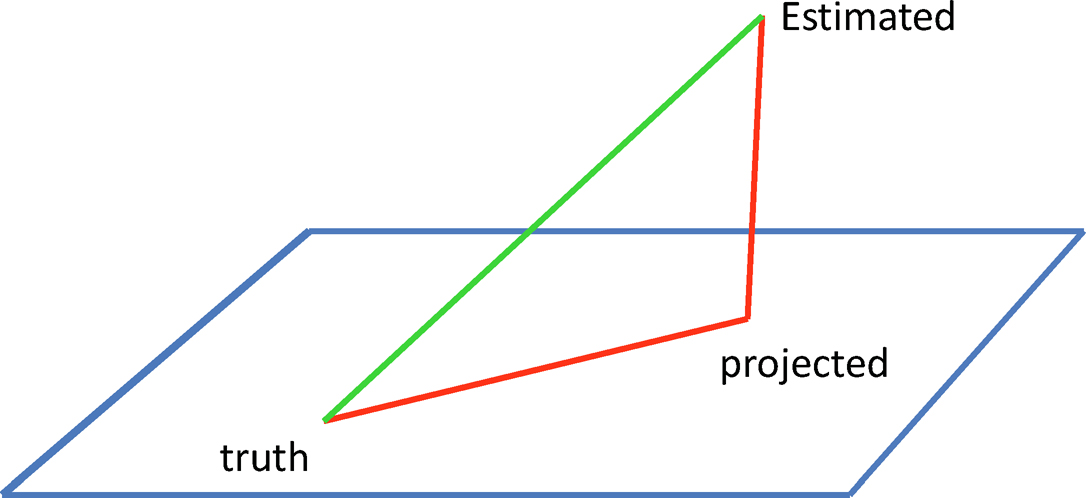
Figure 9.2: The projected matrix cannot be twice worse than the unprojected one by using the triangular inequality. In the case of a rotationally invariant norm, the projected matrix is indeed closer to the truth.
More generally, we can cast the following projection problemml:
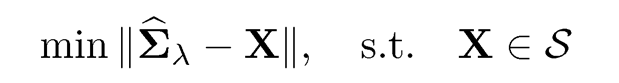 (9.23)
(9.23)
where S is a set of matrices such that Σ ∈ S is a feasible solution. Let Sλ be a solution. Then, we have

Figure 9.2 gives an illustration. In other words, the projection cannot make the estimator much worse than the original one. For example, if we take the elementwise L∞-norm ||A||=|| A||max, then the projected estimator Sλ can not be twice as worse as the original Σ̂ λ in terms of elementwise L∞-norm. This minimization problem can be solved by using cvx solver in MATLAB (Grant and Boyd, 2014) for the constraint λmin(X) ≥ δ.
Another way of enforcing semi-positive definiteness is to impose a constraint on the covariance regularization problem. For example, in minimizing (9.11), we can impose the constraint λmin(Σ) ≥ τ. Xue, Ma and Zou (2012) propose this approach for enforcing the positive definiteness in thresholding covariance estimator:
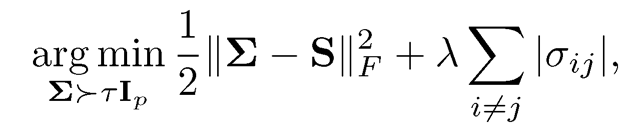
and the above eigenvalue constrained sparse covariance estimator can be solved efficiently via an alternating direction method of multiplier algorithm. A similar estimator was considered by Liu, Wang, and Zhao (2014).
9.3 Robust Covariance Inputs
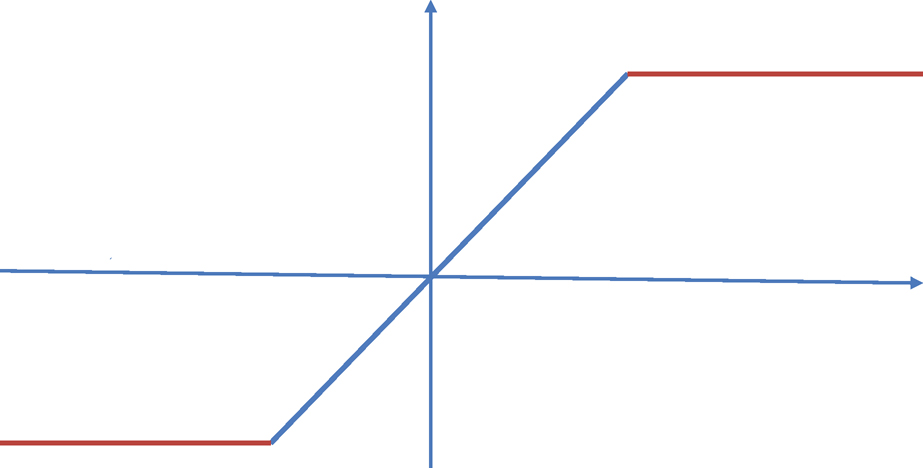
Figure 9.3: Winsorization or truncation of data.
The sub-Gaussian assumption is too strong for high-dimensional statistics. Requiring tens of thousands of variables to have all light tails is a mathematical fiction rather than reality. This is documented in Fan, Wang and Zhu (2016). Catoni (2012), Fan, Wang, Zhu (2016) and Fan, Li, and Wang (2017) tackle the problem by first considering estimation of a univariate mean /..t from a sequence of i.i.d random variables , Y1, ⋯Yn with variance σ. In this case, the sample mean Y provides an estimator but without exponential concentration. Indeed, by the Markov inequality, we have
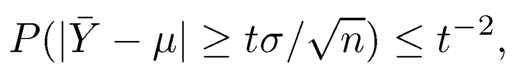
which is tight in general and has a Cauchy tail (in terms of t). On the other hand, if we truncate the data Ỹi =sign(Yi) min (|Yi|, τ) with τ ≍ σ√n (see Figure 9.3) and compute the mean of the truncated data (usually called Win-sorized data), then we have (Fan, Wang and Zhu, 2016)
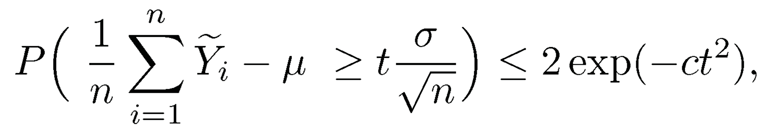 (9.24)
(9.24)
for a universal constant c > 0 (see also Theorem 3.2). In other words, the mean of truncated data with only a finite second moment behaves very much the same as the sample mean from the Gaussian data: both estimators have Gaussian tails (in terms of t). This sub-Gaussian concentration is fundamental in high-dimensional statistics as the sample mean is computed tens of thousands or even millions of times, and with sub-Gaussianity, the maximum error will not accumulate to be too large.
As an example, estimating the high-dimensional covariance matrix Σ — (σij) involves O(p) univariate mean estimation, since the covariance can be expressed as σij= E(XiXj) — E(Xi) E(Xj). Estimating each component by the truncated mean yields an elementwise robust covariance estimator
 (9.25)
(9.25)
where a is the method of estimating the univariate mean. When a = T, we refer to the truncated sample mean as in (9.24) and when a = H, we refer to the adaptive Huber estimator in Theorem 3.2. For example, for the Winsorized estimator (a = T), we have
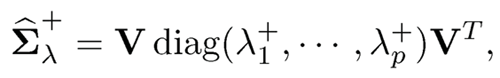
and E(Xi)T^ = n-1Σnk=1 sign(Xki)min (|Xki|τ) in (9.25). We would expect this method performs similarly to the sample covariance matrix based on the truncated data:
 (9.26)
(9.26)
where X̃i is the elementwise truncated data at level τ.
Assuming the fourth moment is bounded (as the covariance itself involves second moments), by using the union bound and the above concentration inequality (9.24), we can easily obtain (see the proof in Section 9.6.3)
 (9.27)
(9.27)
and condition (9.17) is met. In other words, with robustification, when the data have merely bounded fourth moments, we can achieve the same estimation rate as the sample covariance matrix under the Gaussian data, including the regularization result in Theorem 9.1. Formally, we have
Theorem 9.4 Let u =maxi,j SD(XiXi) and v = maxi SD(Xi) and σ max(u, y) be bounded. Assume that log p = o(n).
1. For the elementwise truncated estimator ΣT when σ ≍ σ√n, we have
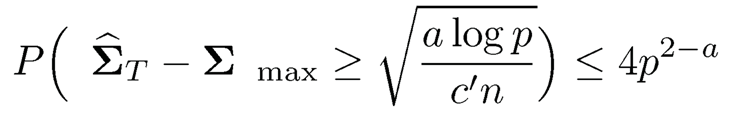
for any a > 0 and a constant c’ > O.
2. For the elementwise adaptive Huber estimator ΣH, when τ = √n I (alog p)σ, we have
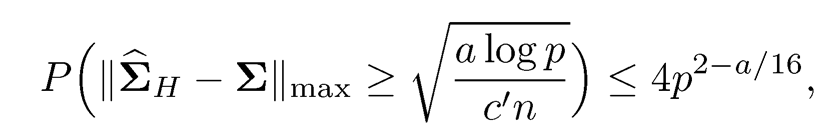
for any a > 0 and a constant c’ > O.
Instead of using elementwise truncation, we can also apply the global truncation. Note that
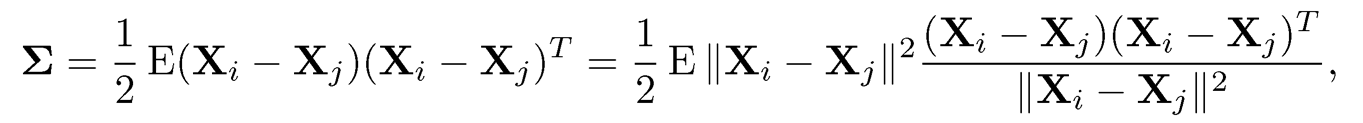
which avoids the use of mean. Truncating the norm ||Xi -Xj|at τ leads to the robust U -covariance
 (9.28)
(9.28)
which does not involve estimation of the mean due to symmetrization of Xi -Xj. Fan, Ke, Sun and Zhou (2018+) prove the follow result.
Theorem 9.5 Let v =½||E{X-μ){X-μ)T} +tr(Σ)Σ+2Σ||2. For any t>0, when τ≥ vn½/(2t), we have
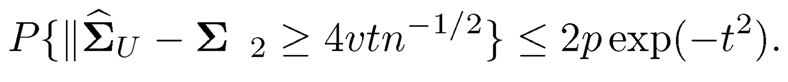
As in the proof of Theorem 9.3, the result in Theorem 9.5also implies the sharp result in elementwise L∞-norm II·||max, namely condition (9.17).
Before the introduction of the robust U-covariance matrix estimator, Fan, Wang and Zhu (2016) and Minsker (2016) independently propose shrinkage variants of the sample covariance using fourth moments. Specifically, when EX = 0, they propose the following estimator:
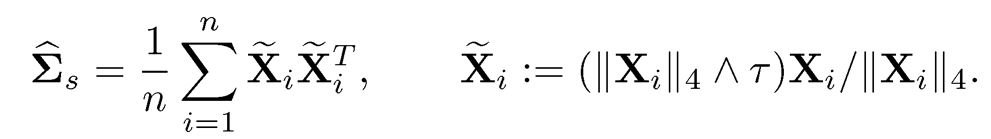 (9.29)
(9.29)
It admits the sub-Gaussian behavior under the spectral norm, as long as the fourth moments of X are finite.
For an elliptical family of distributions, Liu, Han, Yuan, Lafferty, and Wasserman (2012) and Xue and Zou (2012) used rank correlation such as the Kendall τ correlation (9.46) or Spearman correlation and (9.49) (see Section 9.5) to obtain a robust estimation of the correlation matrix. After estimating the correlation matrix, we can use Catoni’s (2012) method to obtain robust estimation of individual variances. Such constructions also possess the uniform elementwise convergence as in Theorem 9.4.
9.4 Sparse Precision Matrix and Graphical Models
The inverse of a covariance matrix, also called a precision matrix, is prominently featured in many statistical problems. Examples include Fisher dis-criminant, Hotelling T-test, and the Markowitz optimal portfolio allocation vector. It plays an important role in Gaussian graphical models (Lauritzen, 1996, Whittaker, 2009). This section considers estimation of the sparse precision matrix and its applications to graphical models.
9.4.1 Gaussian graphical models
Let X ~ N(μ, Σ) and Ω= Σ-1. Then, it follows that the conditional distribution of a block of elements given the other block of variables is also normally distributed. In particular, if we partition

then the conditional distribution of
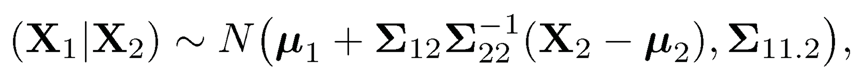 (9.30)
(9.30)
where Σ11.2 = Σ11 — Σ12Σ-22Σ21 is the Schur completement of Σ22.
Let Ω = Σ-1 be the precision matrix, and , Ω11, Ω12, Ω21 and Ω22 be its partition in the same way as Σ. Then, it follows from the block matrix inversion formula at the end of Section 9.1, we have Ω11 = Σ-111.2. In particular, when X1 = (X1, X2)T and X2 = (X3, ⋯ , Xp)T, we can compute Σ-111.2 analytically for the 2 x 2 matrix Σ11.2. It then follows easily that
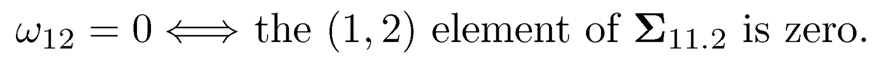
In other words, ω12 = 0 if and only if X1 and X2 are independent given X3, ⋯ , Xp. We summarize the result as follows.
Proposition 9.3 For a Gaussian random vector X with precision matrix Ω=(ωij), (ωij) = 0 if and only if Xi and Xi are conditionally independent, given the rest of the other random variables.
This result will be extended to the Gaussian copula in Section 9.5 and serves as the basis of the graphical model. The latter summarizes the conditional dependence network among the random variables {Xj}Pi=1 via a graph. The vertices of the graph represent variables X1, ⋯, Xp and the edges represent the conditional dependence. The edge between i and j exists if and only if ωij # 0. Figure 9.4 gives such an example for p = 6.

Figure 9.4: The left panel is a precision matrix and the right panel is its graphical representation. Note that only the zero pattern of the precision matrix is used in the graphical representation.
9.4.2 Penalized likelihood and M-estimation
For given data from a Gaussian distribution, its likelihood is given by (9.8). By (9.8), reparameterizing Σ by Ω = (ωij) and adding the penalty term to the likelihood leads to the following penalized likelihood:
 (9.31)
(9.31)
The optimization problem (9.31) is challenging due to the singularity of the penalty function and the positive definite constraint on Ω. When the penalty is the Lasso penalty, Li and Gui (2006) used a threshold gradient descent procedure to approximately solve (9.31). Their method was motivated by the close connection between 6-boosting and Lasso. Various algorithms have been proposed to solve (9.31) with the Lasso penalty. Yuan and Lin (2007) used the maxdet algorithm but that algorithm is very slow for high-dimensional data. A more efficient clockwise descent algorithm was proposed in Baner-jee, El Ghaoui and d’Aspremont (2008) and Friedman, Hastie and Tibshirani (2008). Duchi et al. (2008) proposed a projected gradient method and Lu (2009) proposed a method by applying Nesterov’s smooth optimization technique. Scheinberg et al. (2010) developed an alternating direction method of multiplier for solving the Lasso penalized Gaussian likelihood estimator. Fan, Feng and Wu (2009) studied (9.31) with the adaptive Lasso penalty. Roth-man, Bickel, Levina and Zhu (2008) and Lam and Fan (2009) derived the asymptotic properties of the estimator. They showed that
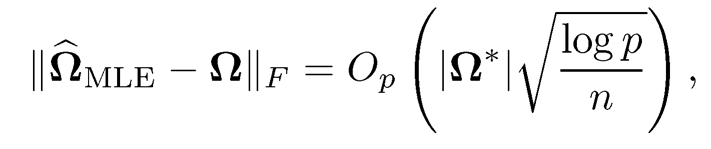
and the consistency of sparsity selection for folded concave penalty function, where |Ω*| is the number of non-zero elements of true Ω*. Under an irrep-resentable condition, Ravikumar et al. (2011) established the selection consistency and rates of convergence of the Lasso penalized Gaussian likelihood estimator.
Zhang and Zou (2014) introduced an empirical loss minimization framework to the precision matrix estimation problemml:
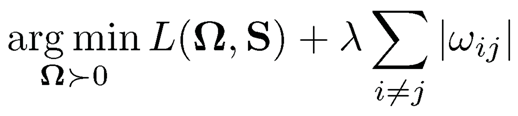 (9.32)
(9.32)
where L is a proper loss function that satisfies two conditions:
(i) L(Ω, Σ) is a smooth convex function of Ω;
(ii) the unique minimizer of L(Ω, Σ*) occurs at the true Ω* = (Σ*)-1.
Condition (i) is required for computational reasons and condition (ii) is needed to justify (9.32). In (9.31) the negative log-Gaussian likelihood is identical to the loss function Lg(Ω, Σ) = (Ω, Σ)- log det(Ω). Zhang and Zou (2014) further proposed a new loss function named D- Traceloss:
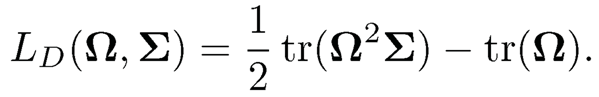 (9.33)
(9.33)
One can easily verify that LD (Ω, Σ) satisfies conditions (i) and (ii). In particular, the D-Trace loss has a constant Hessian matrix of the form (Σ⊗I+I⊗Σ)/2, where ⊗ denotes the Kronecker product. For the empirical D-Trace loss LD(Ω, S) its Hessian matrix is (S ⊗ I + I ⊗ S)/2. With the D-Trace loss, (9.32) is solved very efficiently by an alternating direction method of multiplier. Selection consistency and rates of convergence of the Lasso penalized D-Trace estimator are established under an irrepresentable condition. It is interesting to see that (9.32) provides a good estimator of Σ-1 without the normality assumption on the distribution of X. The penalized D-Trace estimator can even outperform the penalized Gaussian likelihood estimator when the data distribution is Gaussian. See Zhang and Zou (2014) for numerical examples.
9.4.3 Penalized least-squares
The aforementioned methods directly estimate the precision matrix Ω. If we relax the positive definiteness condition for Ω, a computational easy alternative is to estimate Ω via column-by-column methods. Without loss of generality, we assume μ = 0 for simplicity of presentation.
For a given j, let X-j be the subvector of X without using Xj. Correspondingly, let Σj be the (p —1) x (p — 1) matrix with jth row and jth column deleted, and σj be the jth column of Σ with the ith element deleted. For example, when j = 1, the introduced notations are in the partition:
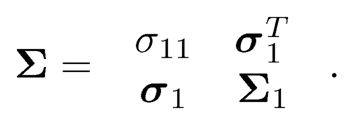
Set τj= σjj— σTj Σ-1jσj. By (9.30), we have
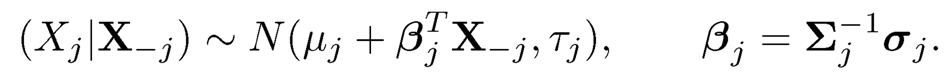 (9.34)
(9.34)
Using the similar notations to those for Σ, we define similarly Ωj and ωj based on Ω By the block inversion formula at the end of Section 9.1, we have, for example,
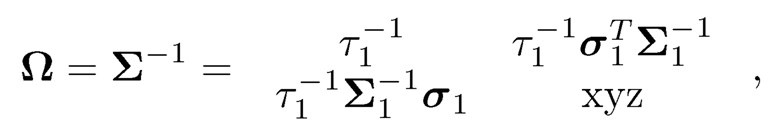
where “xyz“ is a quantity that does not matter in our calculation. Therefore, ω11= τ-11 and ω1 = β1/τ1 with β1 given by (9.34). Note that β1 is the regression coefficient of X1 given X-1 and τ1 is its residual variance. The sparsity of β1 inherits from that of ω1. Therefore, the first column can be recovered from the of X1 the rest of the variables.
The above derivation holds for every j. It also holds for non-normal distributions, as all the properties are the results of linear algebra, rather than those of the normal distributions. We summarize the result as follows.
Proposition 9.4 Let α*j and β*j; be the solution to the least-squares problemml:
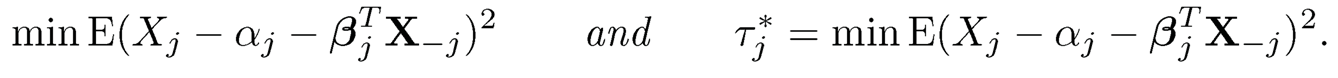
Then, the true parameters Ω* are given column-by-column as follows:

Proposition 9.4 shows the relation between the matrix inversion and the least-squares regression. It allows us to estimate the precision matrix S2 column-by-column. It prompts us to estimate the jth column via the penalized least-squares
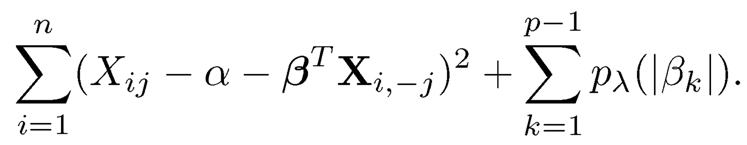 (9.35)
(9.35)
This is the key idea behind Meinshausen and Biihlmann (2006) who considered the L1-penalty. Note that each regression has different residual variance τj and therefore the penalty parameter should depend on j. To make this less dependence sensitive, Liu and Wang (2017) propose to use the square-root Lasso of Belloni, Chernozhukov, and Wang (2011), which minimizes
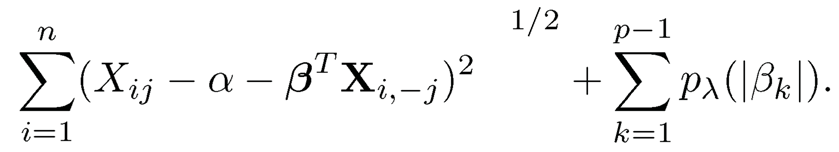
Instead of using the Lasso, one can use the Dantzig selector:
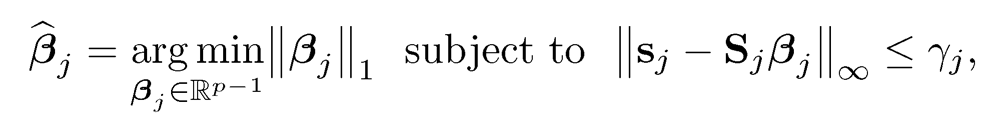 (9.36)
(9.36)
for a regularization parameter γj, where Sj and sj is the partition of the sample covariance matrix S in a similar manner to that of Σ. This method is proposed by Yuan
Sun and Zhang (2013) propose to estimate βj and τj by a sealed-Lasso: Find b̂j and σ̂j to minimize (to be explained below)
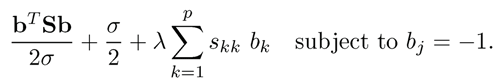 (9.37)
(9.37)
Once b̂j and σ̂j are obtained, we get β̂j = (b̂1,...,b̂j-1, b̂j+1, ..., b̂p)T and τ̂j=σ̂j. Thus, we obtain a column-by-column estimator using Proposition 9.4. They provide the spectral-norm rate of convergence of the obtained precision matrix estimator.
Note that bTSb in (9.37) is a compact way of writing the quadratic loss in (9.35), after minimizing with respect Œ3 (Exercise 9.9), also called profile least- squares. It also shows innately the relationship between covariance estimation and least-squares regression. Indeed,
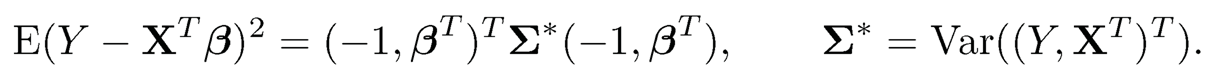 (9.38)
(9.38)
We can replace Σ* by any of its robust covariance estimators in Section 9.3. This yields a robust least-squares estimator, requiring only bounded fourth moments in high-dimensional problems. In such a kind of least-squares problem, it is essential to use a semi-positive definite covariance estimate. Projections in Section 9.2.3 are essential before plugging it into (9.38).
The column-by-column estimator Ω̂ is not necessarily symmetric. A simple fix is to do projection:
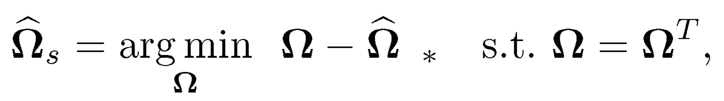
where ||·||* can be the operator, Frobenius, or elementwise max norm. For both the Frobenius and elementwise max norms, the solution is

Another way of symmetrization is to perform a shrinkage and symmetrization simultaneously: Ωs, (ω̂sij) with
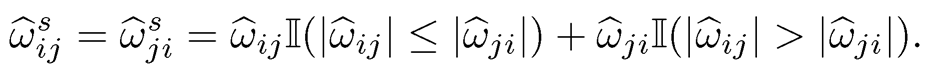 (9.39)
(9.39)
This was suggested by Cai, Liu and Luo (2011) for their CLIME estimator so that it has the property
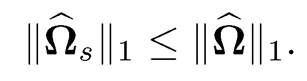 (9.40)
(9.40)
As an illustration, we use gene expression arrays from Arabidopsis thaliana in Wille et al. (2004) and construct the genomic network that involves 39genes in two isoprenoid metabolic pathways: 16 from the mevalonate (MVA) pathway are located in the cytoplasm, 18 from the plastidial (MEP) pathway are located in the chloroplast, and 5 are located in the mitochondria. While the MVA and MEP pathways generally operate independently, crosstalks can happen too. Figure 9.5 gives the conditional dependence graph using TIGER in Lin and Wang (2017). The graph has 44 edges and suggests that the connections from genes AACT1 and HMGR2 to gene MECPS indicate a primary sources of the crosstalk between the MEP and MVA pathways.
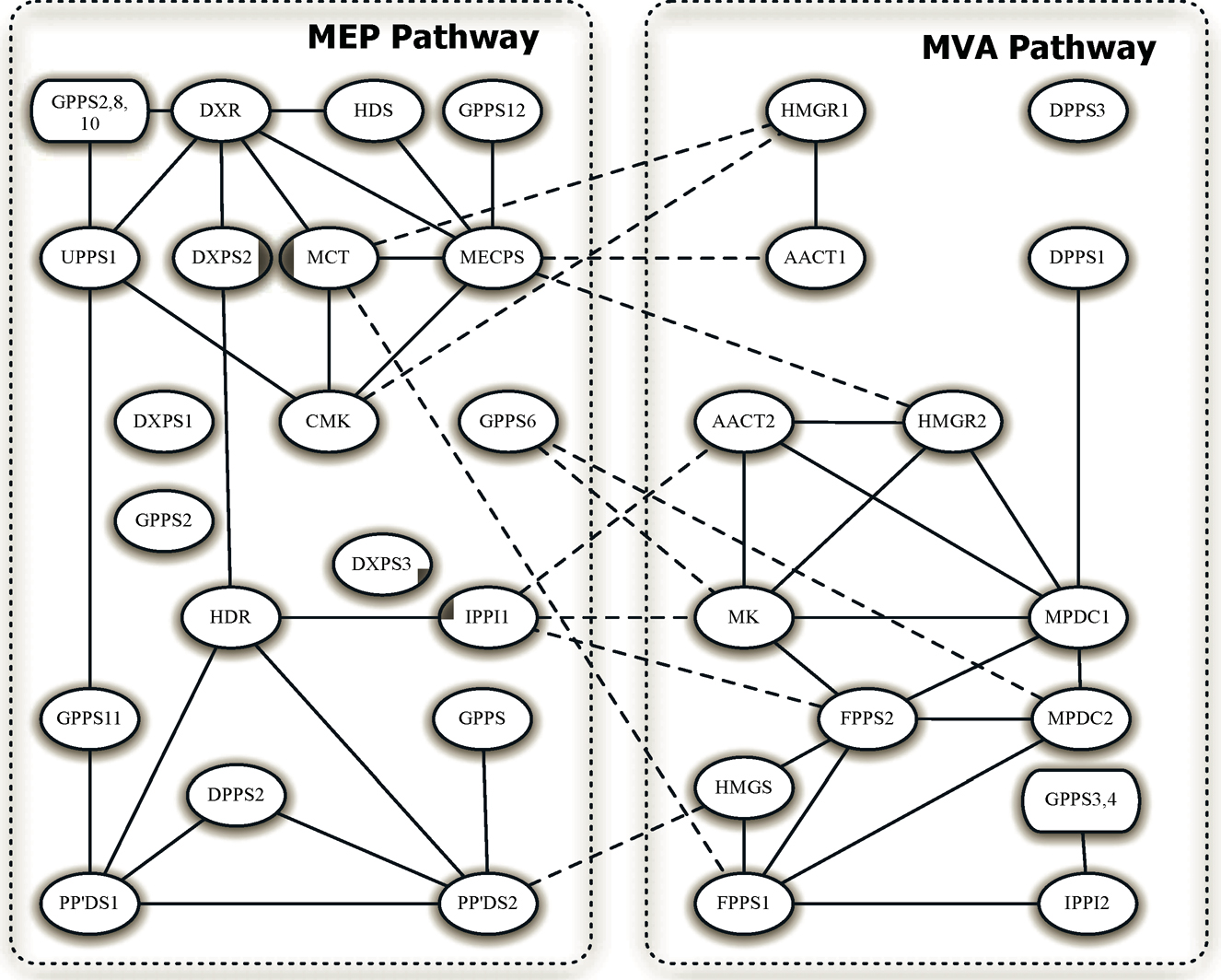
Figure 9.5: The estimated gene networks of the A ra,badopsis data. The within-pathway links are shown in solid lines and between-pathway links are presented by dashed lines. Adapted from Liu and Wang (2017).
9.4.4 CLIME and its adaptive version
For a given covariance input Σ̃, the constrained l1-minimization for inverse matrix estimation (CLIME), introduced by Cai, Liu, and Luo (2011), solves the following problemml:
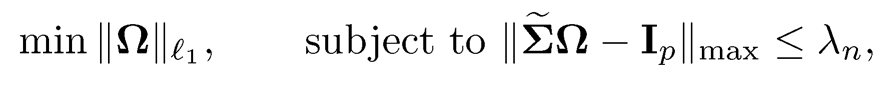 (9.41)
(9.41)
where Σ is a pilot estimator of Σ and ||Ω|| l1 = Σi,j |ωij| is the elementwise l1-norm. It is used to facilitate the computation of the solution Ω̂.
Following the Dantzig-selector argument, it is interesting to see that CLIME is more closely related to the L1-penalized D-Trace estimator in Zhang and Zou (2014). The first order optimality condition of
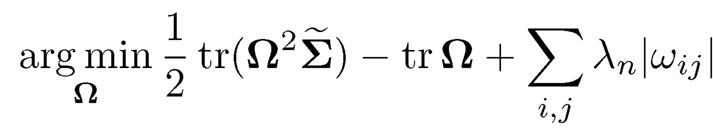
is ½(ΩΣ̃- +Σ̃-Ω) —Ip= λnZ, where Zij ∈[-1, 1]. This suggests an alternative Dantzig-selector formulation as follows:
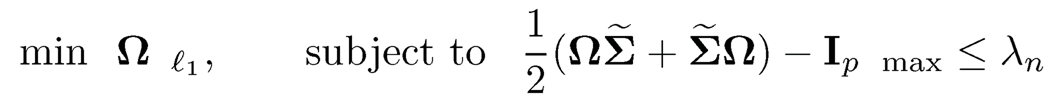
We call the above formulation the symmetrized CLIME because it is easy to show that its solution is always symmetric, while the CLIME solution is not.
The parameter λn is chosen such that
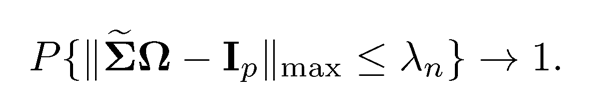 (9.42)
(9.42)
The Dantzig-type of estimator admits a very clear interpretation: in the high-confidence set {Ω : ΣΩ —Ip||max ≤ λn}, which has the coverage probability tending to one, let us find the sparsest solution gauged by the fi-norm.
Let ei be the unit vector with 1 at the jth position. Then, the problem (9.41) can be written as
 (9.43)
(9.43)
It can be casted as p separate optimization problems: find ω̂i to solve
 (9.44)
(9.44)
This problem can be solved by a linear program. Let σ̃k be the kth column of Σ. Write ω = ωj (dropping the index j in ωj) and xi =|ωi|where ωi is the ith component of ω. Then, the problem (9.44) can be expressed as
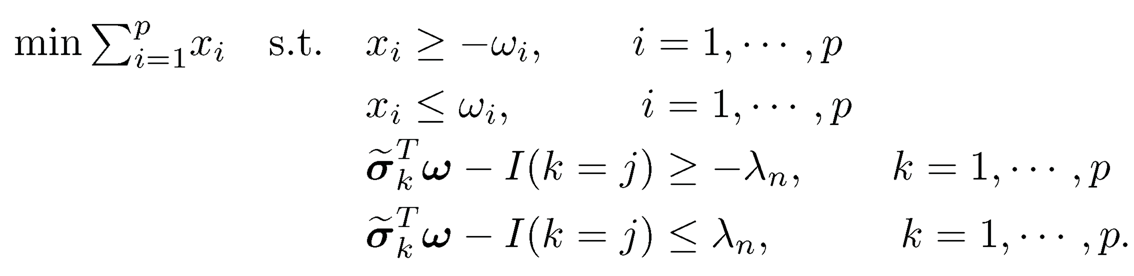
The first two constants are on the fact that xi = |ωi| and the last two constraints are on ||Σωj — ej||∞ ≤ λn. Thus, we have 2p variables x and ω and 4p linear inequality constraints.
After obtaining the column-by column estimator, we need to symmetrize the estimator as in (9.39), resulting in Ω̂s. Now consider a generalized sparsity measure that is similar to (9.16), namely,
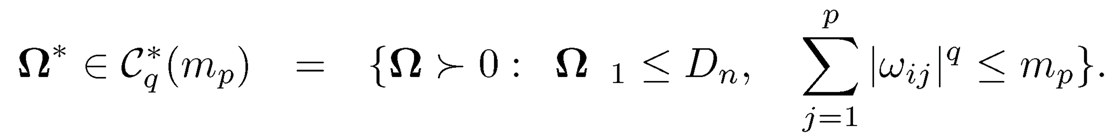
Using the sample covariance as the initial estimator, Cai, Liu and Luo (2011) established the rate of convergence for the CLIME estimator. Inspecting the proof there, the results hold more generally for any pilot estimator that satisfies certain conditions, which we state below.
Theorem 9.6 If we take λn > ‖Ω*‖ ‖Σ-Σ‖max, then uniformly over true precision matrix Ω* ∈ Cq* (mp),

In particular, if the pilot estimator Σ̅̃ satisfies
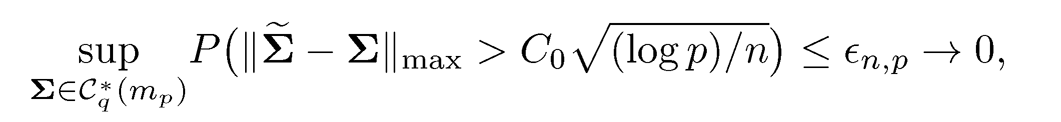 (9.45)
(9.45)
with probability at least 1 — εn,p, by taking λn = C0Dn√(log p)/n, we have
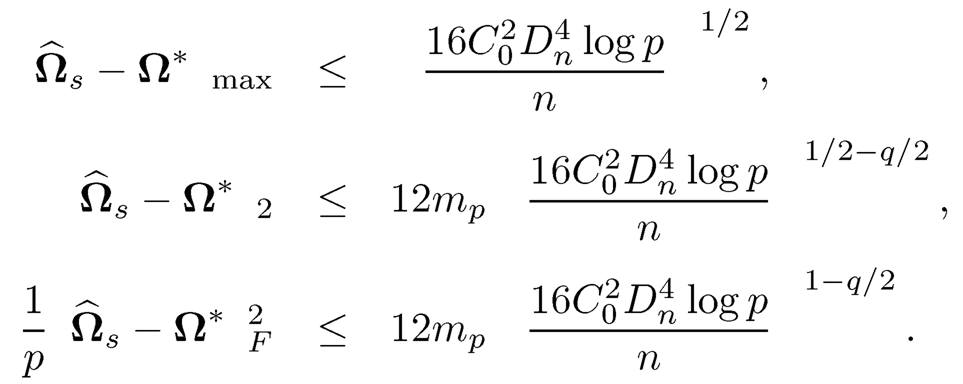
The results in Theorem 9.6 indicate that any pilot estimator that satisfies (9.45) can be used for CLIME regularization. In particular, the robust covariance inputs in Section 9.3 are applicable.
The CLIME above uses a constant regularization parameter λn, which is not ideal. Cai, Liu and Zhou (2016) introduce an adaptive version, called ACLIME. It is based on the observation that the asymptotic variance of (SΩ — Ip)ij in (9.41) is heteroscedastic and depends on σiiωjj for the sample covariance matrix S. Therefore, the regularization parameter should be of form

which depends on unknown quantities σii and ωii. These unknown quantities need to be estimated first.
Fan, Xue and Zou (2014) show that the CLIME estimator can be used as a good initial estimator for computing the SCAD penalized Gaussian likelihood estimator such that the resulting estimator enjoys the strong oracle property. Recall that the SCAD penalized Gaussian likelihood estimator of Ω is defined as
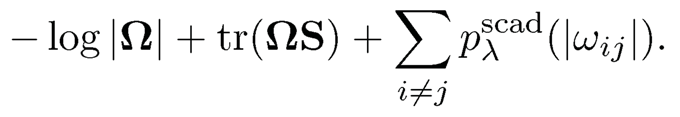
Let A denote the support of the true Ω*. The oracle estimator of Ω* is defined as

Let Ω(0) be the CLIME estimator and compute the following weighted ℓ1 penalized Gaussian likelihood estimator for k =1, 2
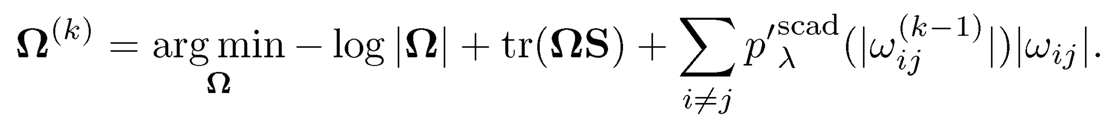
Fan, Xue and Zou (2014) show that with an overwhelming probability Ω(2) equals the oracle estimator. Simulation examples in Fan, Xue and Zou (2014) also show that Ω(2) performs better than the Lasso penalized Gaussian likelihood estimator and CLIME.
Avella-Medina, Battey, Fan and Li (2018) generalize the procedure to a general pilot estimator of covariance, which includes robust covariance inputs in Section 9.3. It adds a projection step to the original ACLIME. The method consists of the following 4 steps. The first step is to make sure that the covariance input Σ+ is positive definite. The second step is a preliminarily regularized estimator to get a consistent estimator for ωjj. It can be a non-adaptive version too. The output ωjj(1) is to make the mathematical argument valid. The third step is the implementation of the adaptive CLIME that takes into account of the aforementioned heteroscedacity. The last step is merely a symmetrization.
1. Given a covariance input Σ̃, let
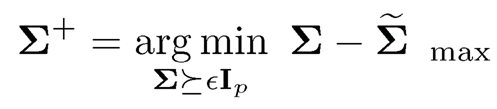
for an small ε > 0. This can be easily implemented in MATLAB using the cvx solver, as mentioned in Section 9.2.3. We then estimate σii by σii+, the ith diagonal element of the projected matrix Σ+.
2. Get an estimate of {wjj} by using CLIME with an adaptive regularization parameter via solving
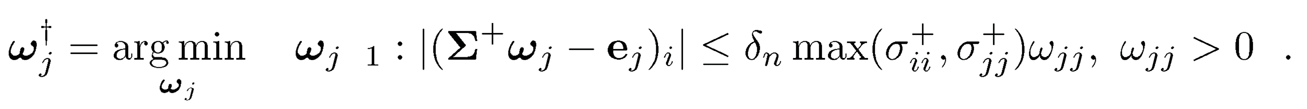
This is still a linear program. Output
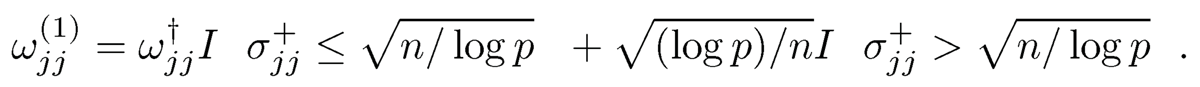
3. Run the CLIME with adaptive tuning parameter

4. The final estimator is constructed as the symmetrization of the last estimator:

Avella-Medina et al. (2018) derive the asymptotic property for the ACLIME estimator with a general covariance input that satisfies (9.17). The results and the proofs are similar to those in Theorem 9.6.
Avella-Medina et al. (2018) conduct extensive simulation studies and show the big benefit of using robust covariance estimators as a pilot estimator, for both estimating the covariance matrix and the precision matrix. In particular, they used the elementwise adaptive Huber estimator in their simulation studies.
To assess whether their proposed robust estimator can improve inference on gene regulatory networks, they used a microarray dataset in the study by Huang et al. (2011) on the inflammation process of cardiovascular disease. Huang et al. (2011) found that the Toll-like receptor signaling (TLR) pathway plays a key role in the inflammation process. Their study involves n = 48 patients and data is available on the Gene Expression Omnibus via the accession name “GSE20129” . Avella-Medina et al. (2018) considered 95 genes from the TLR pathway and another 62 genes from the PPAR signaling pathway which is known to be unrelated to cardiovascular disease. We would expect that a good method discovers more connections for genes within each of their pathways but not across them. In this study, n = 48 and p = 95 + 62. To see whether a robust procedure gives some extra merits, they used both the adaptive Huber estimator and the sample covariance matrix as pilot estimators. We will call them robust ACLIME or “RACLIME” and ACLIME, respectively.
They first chose the tuning parameters that deliver the top 100 connections for each method. The selection results are presented in the left panel in Table 9.1 and also displayed graphically in Figure 9.6. The robust ACLIME identifies a greater number of connections within pathways and fewer connections across pathways.
They additionally took the same tuning parameter in the ACLIME optimization problem for each procedure. The results are summarized in the right panel of Table 9.1. The robust procedure detects fewer connections, but yields higher percentage (64.2% by robust ACLIME) of within-pathway connections (in comparison with 55% by ACLIME).
Table 9.1: Summary of regulatory network graphs: Number of connections detected by using robust ACLIME and regular ACLIME, with two methods of choosing regularization parameters. Adapted from Avella-Medina et al. (2018).
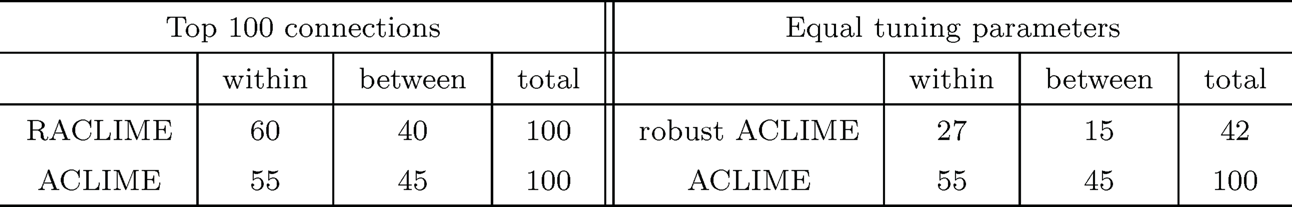
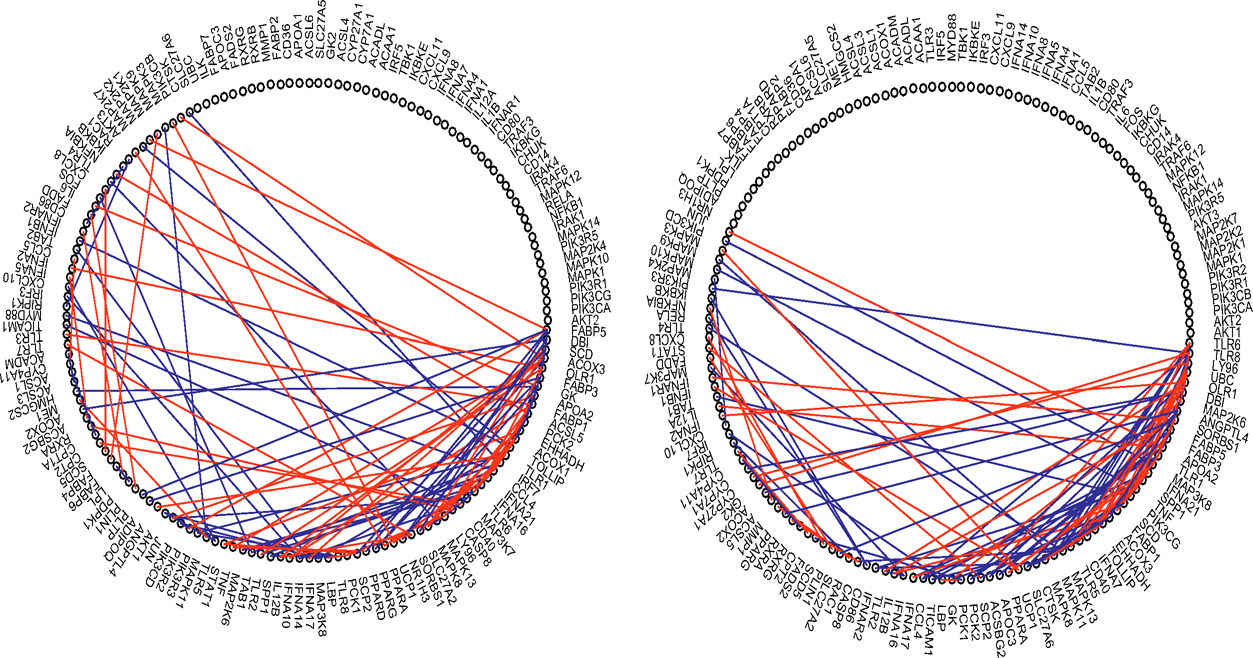
Figure 9.6: Connections estimated by ACLIME using the sample covariance pilot estimator (left) and the adaptive Huber type pilot estimator (right). Blue lines are within pathway connections and red lines are between pathway connections. Taken from Avella-Medina et al. (2018).
9.5 Latent Gaussian Graphical Models
Graphical models are useful representations of the dependence structure among multiple random variables. Yet, the normality assumption in the last two sections is restrictive. This assumption has been relaxed to the Gaussian copula model by Xue and Zou (2012) and Liu, Han, Yuan, Lafferty, and Wasserman (2012). It has been further extended by Fan, Liu, Ning, and Zou (2017) to binary data and the mix of continuous and binary types of data. In particular, the method allows one to infer about the latent graphical model based only on binary data. This section follows the development in Fan et al. (2017).
Definition 9.4 A random vector X (X1, …, Xp)Tis said to follow a Gaussian copula distribution, if there exists strictly increasing transformations g=(gj)jp=1 such that g(X) (g1(X1),…,gp(Xp))T~ N(0, Σ) with σjj=1 for any 1 ≤ j ≤ p. Write X ~ NPN(0, Σ, g), also called a nonpara-normal model.
In the above definition, we assume without loss of generality that Σ is a correlation matrix. This is due to the identifiability: the scale of functions Σ can also be taken such that Σ is a correlation matrix. Let Fj(·) be the cumulative distribution function of a continuous random variable Xj. Then, Fj(Xj) follows the standard uniform distribution and Φ−1(Fj(Xj)) ~ N(0, 1), where Φ(·) is the cdf of the standard normal distribution. Therefore,

and can easily be estimated by substitution of its empirical distribution F̂n,j(·) or its smoothed version. Therefore, Ω can be estimated as in the previous two sections using the transformed data {ĝ(Xi)}in=1.
It follows from Proposition 9.3 that gi(Xi) and gi(Xi) given the rest of the transformed variables are independent if and only if Wij= 0. Since the functions in g are strictly increasing, this is equivalent to the statement that Xi and Xj are independent given the rest of the untransformed data.
Proposition 9.5 If X ~ NPN(0, Σ, g), then Xi and Xi are independent given the rest of the variables in X if and only if Wij =0. In addition, gj(x) = Φ−1(Fi(x)).
There are several ways to model binary data. One way is through the Ising model (Ising,1925). Ravikumar, Wainwright and Lafferty (2010) proposed the neighborhood Lasso logistic regression estimator for estimating the sparse Ising model. The Lasso and SCAD penalized composite likelihood estimators were considered in Höfling and Tibshirani (2009) and Xue, Zou and Cai (2012), respectively. Another way is to dichotomize the continuous variables. This leads us to the following definition for the data generation process for the mix of binary and continuous type of data. In particular, it includes all binary data as a specific example.
Definition 9.5 Let X= (X1T, X2T)T, where X1 represents d1-dimensional binary variables and X2 represents d2-dimensional continuous variables. The random vector X is said to follow a latent Gaussian copula model, if there exists a d1-dimensional random vector Z1 (Z1, … , Zp1)T such that Z= (Z1, X2) ~ NPN(0, Σ, g) and

where II[(·) is the indicator function and C = (C1, , Cp1) is a vector of constants. Denote by X ~ LNPN(0, Σ, g, C), where Σ is the latent correlation matrix.
Based on the observed data of mixed types, we are interested in estimating Ω and hence the conditional dependence graph of latent variables Z. To obtain this, we need a covariance input that satisfies (9.17) so that the methods introduced in the previous two sections can be used. For continuous variables, its covariance can be estimated by using the estimated transformations (Liu et al., 2012; Xue and Zou, 2012). For binary data and binary-continuous data, how to estimate their covariance matrices is unclear.
Fan, Liu, Ning and Zou (2017) provide a unified method to estimate the covariance matrix for mixed data, including continuous data, using the Kendall τ correlation
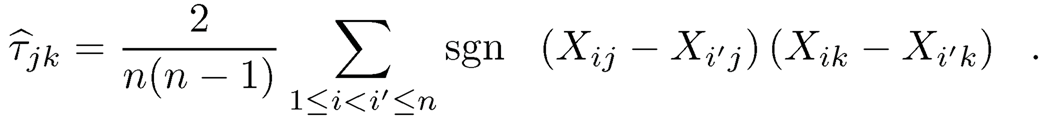 (9.46)
(9.46)
It estimates the population quantity
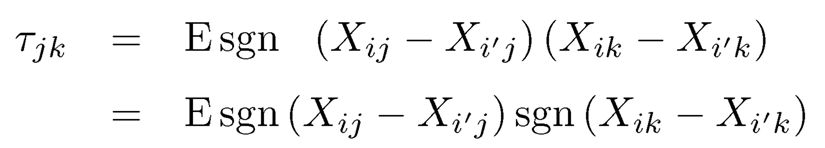 (9.47)
(9.47)
using sgn(ab) = sgn(a)sgn(b).
For binary data a and b, using sgn(a — b) = a — b, it follows from (9.47) that
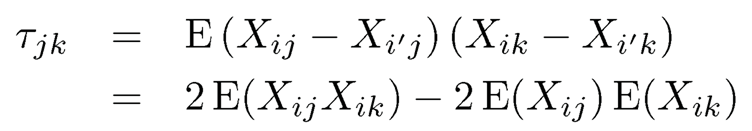
Let Δj = gj (Cj) and Φ2 (x1, x2, ρ) be the cumulative distribution function of the bivariate normal distribution with mean 0 and correlation ρ. Then, since gj (·) is strictly increasing and gj (Zij) ~ N(0, 1), we have
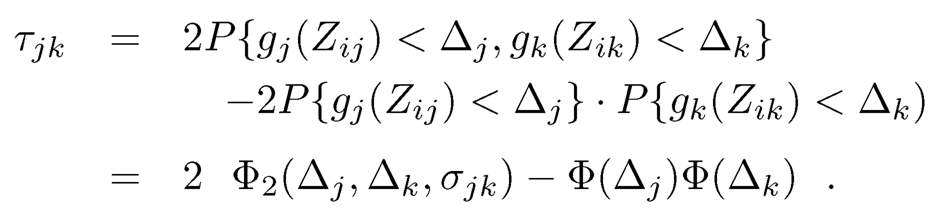 (9.48)
(9.48)
Note that P(Xj= 1) =ΦΔj. This leads to a natural estimator
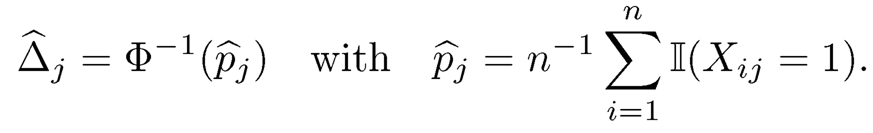
Note that Φ2 (x1, x2, ρ) is a strictly increasing function of ρ. Hence, we can estimate σij via the inversion of (9.48).
For continuous data, note that
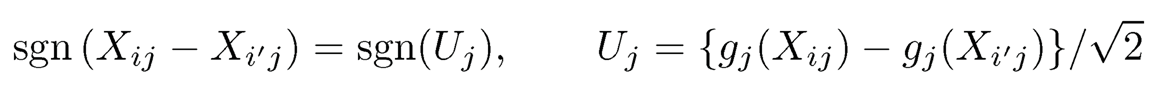
and (Uj, Uk) follow the bivariate normal distribution with mean 0 and correlation ρ. Thus, by (9.47), we have
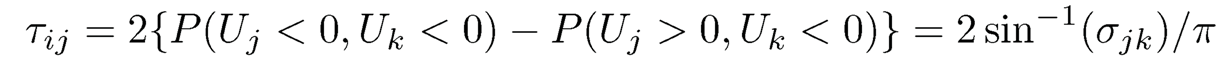 (9.49)
(9.49)
or
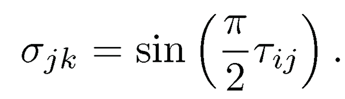
The latter was shown in Kendall (1948) that spells out the relationship between Kendall’s τ-correlation and Pearson’s correlation. This forms the basis of the rank based covariance matrix estimation in Liu et al. (2012) and Xue and Zou (2012). Note that the relation (9.49) holds for elliptical distributions or more generally the transformations of elliptical distributions (Liu, Han, and Zhang, 2012; Han and Liu, 2017). We leave this as an exercise.
Now, for the binary and continuous blocks of the covariance matrix, similar calculation shows (Exercise 9.13) that
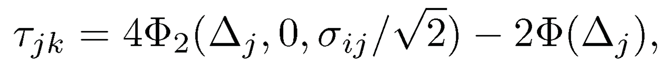 (9.50)
(9.50)
where we assume that Xj is binary and Xk is continuous.
Proposition 9.6 Suppose the mixed data {Xi}i=1n are drawn from X~ LNPN(0, Σ, g, C). Then, the relation between Kendall’s τ correlation and Pearson’s correlation are given by τij= G(σij), where the function G(·) is given respectively in (9.48), (9.49) and (9.50), for binary, continuous and binary-continuous blocks.
Now a natural initial covariance estimator is Σ in which σ̃jk= G−1(τ̂jk) for j ≠ k and σ̃jj= 1. It is easy to show that G−1(x) is a Lipschitz function of x, for |x| ≤ c, for any constant c < 1. Therefore, the concentration inequality of σ̃jk follows from that of τ̂ij , which is established in Fan, Liu, Ning and Zou (2017). The basic idea in establishing the concentration inequality is that Δj and τjk can be uniformly estimated by using the Hoeffding inequality for p̂j and the U-statistics τ̂ij. As a result, they showed that the pilot estimator satisfies condition (9.17):
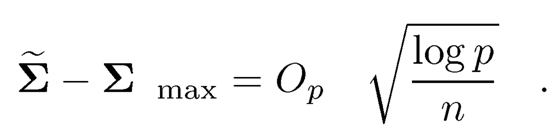
With this pilot covariance estimator, we can now estimate the precision matrix using the methods in the previous two sections and construct the corresponding conditional graph.
9.6 Technical Proofs
9.6.1 Proof of Theorem 9.1
By (9.14), we need to bound individual L1-error. Consider the event E1= {|(σ̂ij — σ̂ij| ≤ λij/2,∀i, j}. For this event, we have
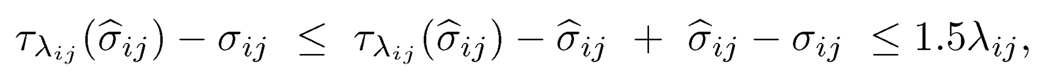
by using the property of the generalized thresholding function (see Definition 9.3b)). Thus, by considering the cases Iσ̂ijI ≥ λij and Iσ̂ijI < λij separately and using property a) of the generalized thresholding function for the second case, we have that for the event E1,
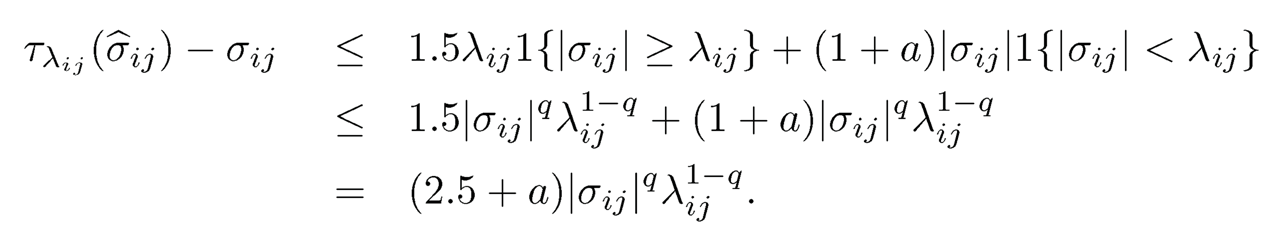 (9.51)
(9.51)
Hence, we have
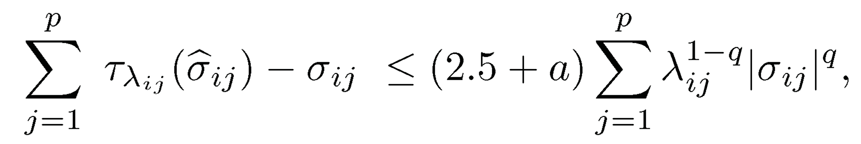
which is bounded, for the event E2 = Iσ̂̂iiσ̂̂jjI ≤ 2σ̂̂iiσ̂̂jj, ∀i, j}, further by
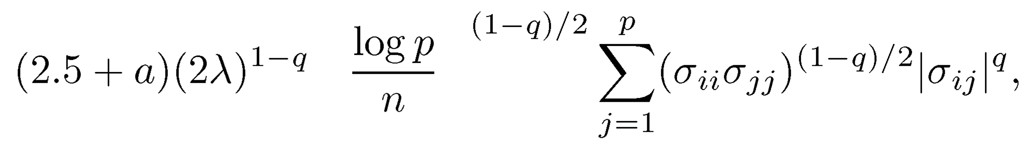
by using the definition of (9.13). Hence, for the parameter space Cq(mp), we have by using (9.14) that
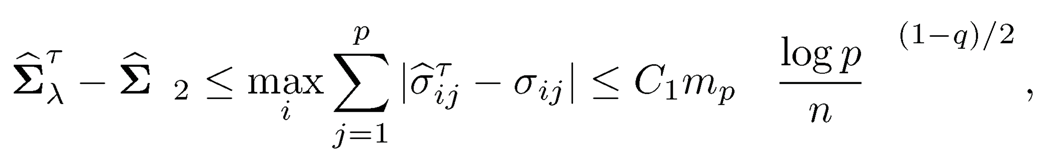
for a constant C1 (2.5 + a)(2Cλ)1−q.
It remains to lower bound the probability of the event εn ={E1 ⋂ E2}. Notice that
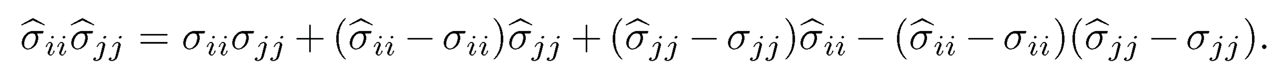 (9.52)
(9.52)
Since the last three terms tend to zero and σii ≥ γ, it therefore follows from (9.17) that, for large n
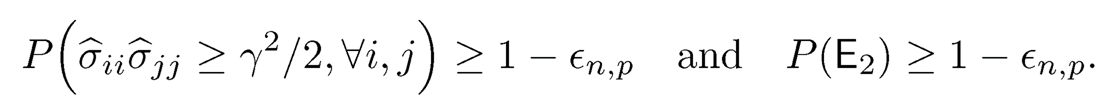
Consequently,
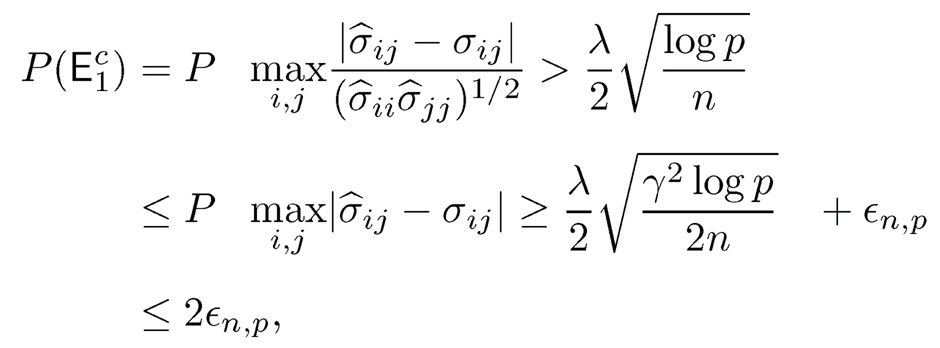
where the last one follows from (9.17) when λ is sufficiently large. Hence, we have
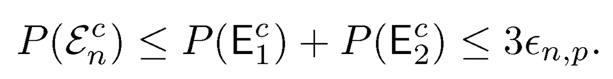
This completes the proof of the first result.
The proof of the second result follows a similar calculation. Using the same calculation as in (9.51), replacing q there by q/2, we have for the event E1

Hence, for the event E2, using σii, ≤ C2, we have
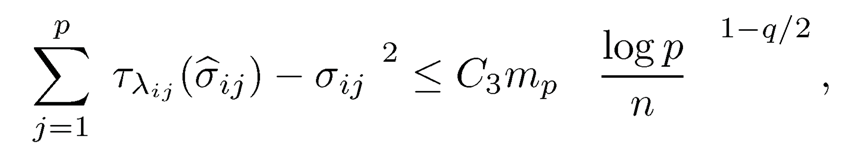
where C3 = (2.5 + a)2(2λ)2−qC2. This completes the proof.
9.6.2 Proof of Theorem 9.3
We plan to use Theorem 9.2 to prove this theorem. Consider a 2 × 2 submatrix Sij of S that involves sij. Then, the corresponding κij is bounded by κ by definition. Similarly, the corresponding
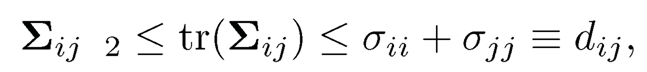
which is bounded by assumption. Hence, the constant cij is lower bounded by some c > 0 and the constant Cij is upper bounded by a constant C. Thus, for sufficiently large t that is bounded by √n/2,
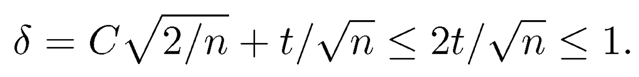
Now applying Theorem 9.2 to this 2 × 2 submatrix Sij, we obtain
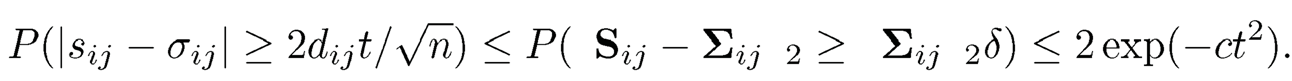
Now, take t = √a log p = 0(√n), we obtain
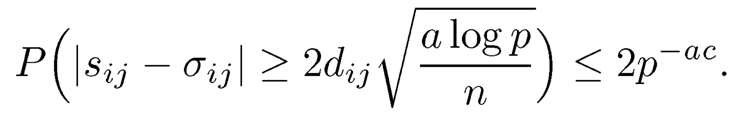
By the union bound, we have
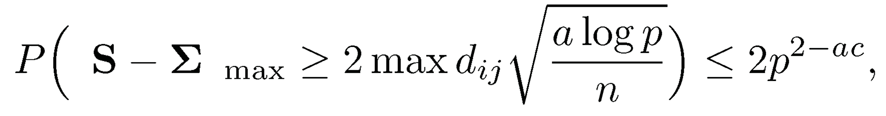
which tends to zero by taking a is a sufficiently large constant. This completes the proof.
9.6.3 Proof of Theorem 9.4
By using (9.24), for the truncated estimator, with t = √(a/c) logp, we have
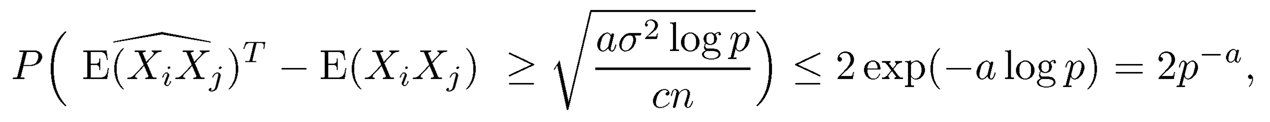
and similarly
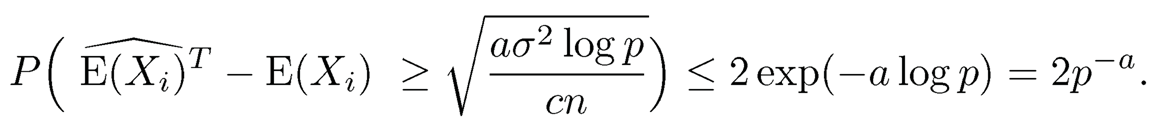
Therefore, by the union bound, we have P(E1c) < 2p2−a, where
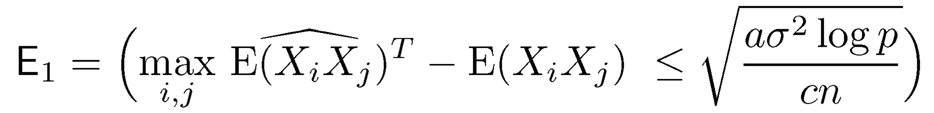
and P(E2c) ≤ 2p1−a, where
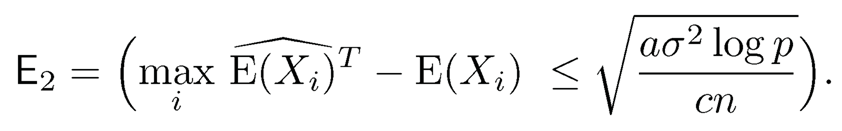
Using the decomposition (9.52) and log p = o(n), when n is sufficiently large, for the event E2, we have
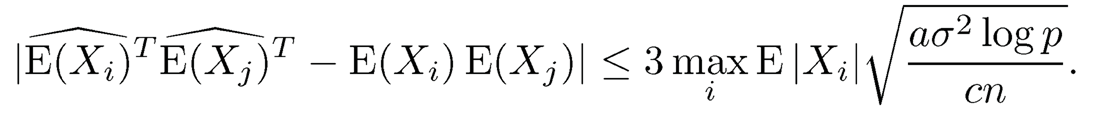
Therefore, for the event E1 ⋂ E2, we have

The result now follows from the fact that

by taking c′ = c/(1 + 3 maxi E |Xi|)2.
The same proof applies to the elementwise adaptive Huber estimator by using the concentration inequality in Theorem 3.2 d). For example, with t = √alog p, we have τ = √n/(a log p)σ and
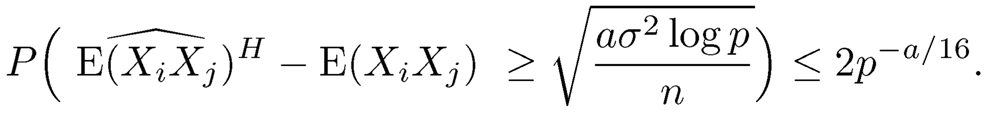
The rest follows virtually the same line of the proof.
9.6.4 Proof of Theorem 9.6
Let Ω* and Σ* be the true precision and covariance matrices, respectively.
(i) Let us first establish the result under ‖·‖max norm. Using ‖AB‖max ≤ ‖A‖max ‖B‖1, we have
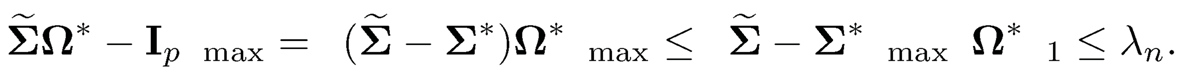
This shows that Ω* is in the feasible set of (9.41) or equivalently (9.43). Let ωj* and ω̂j* be respectively the jth column of Ω* and Ω̂. Since ωj* is a feasible solution to the problem (9.43), we have ‖ω̂j*‖1 ≤ ‖ω̂j*‖1 by the equivalent optimization problem (9.44). Hence, ‖Ω̂‖1 ≤ ‖Ωj*‖1 By (9.40), we have
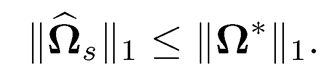 (9.53)
(9.53)
The triangular inequality entails that

where we used the fact that Ω̂s satisfies the constraint in (9.41). The inequality (9.53) implies that ‖Ωs — Ω*‖1 ≤ 2‖Ω*‖1 and it follows from the last result that
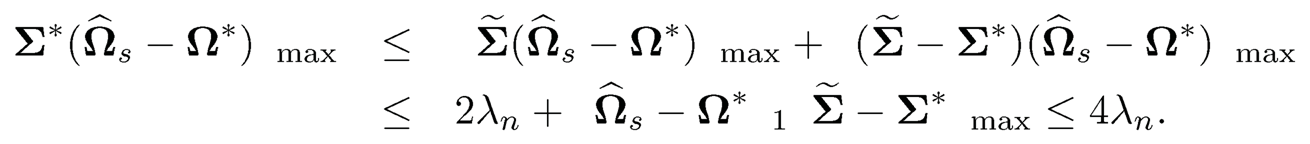
Therefore, we conclude that
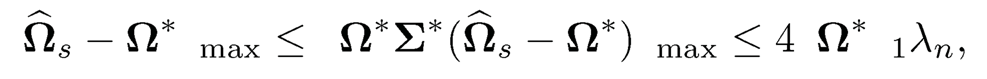
where the last inequality uses again from ‖AB‖max≤‖A‖max‖B‖1.
(ii) We next consider the loss under the operator norm. Recall that ‖Ω̂s− Ω*‖2 ≤‖Ω̂s — Ω*‖1. Thus, it suffices to bound the matrix L1-norm. Let
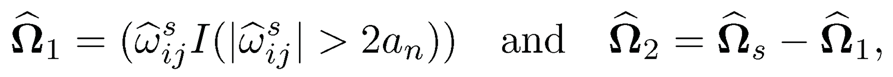
where an = ‖Ω̂s - Ω*‖max. Let ω̂̂j1, ω̂j2 and ω̂̂js be the jth column of Ω̂1, Ω̂2, Ω̂s, respectively. Then, by (9.40), we have

On the other hand,
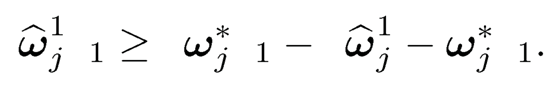
The combination of the last two displays leads to
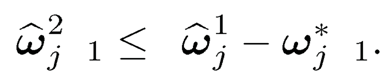
In other words, ‖Ω̂2 ‖≤ Ω̂1 — Ω*‖1.Hence,
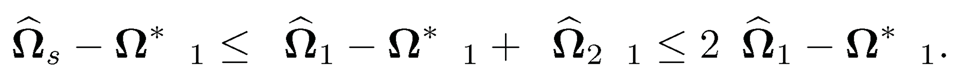
It remains to bound the estimation error ‖Ω̂2 −Ω*‖1. This is indeed a hard-thresholding estimator Ω1 for the sparse matrix Ω*. The results then follow from the proof of Theorem 9.1, which requires only sparsity and uniform consistency of the pilot estimator. More specifically, note that for the hard-thresholding function, we have a = 2. Adopting (9.51) to the current setting, we have λij = 2an there. Set dij= |ω̂ sijII ω̂ sij| ≥ 2an)-ωij*|. It follows from property b) of Definition 9.3 with a = 2 that
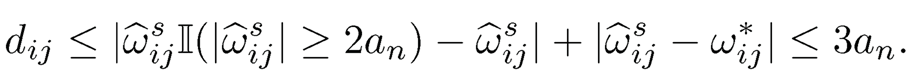
Now, separating the bounds for two cases with II{ |ωij*| ≥ an} and II{ |ωij*| < an}, using the property a) of Definition 9.3 for the second case for the hard-thresholding function, we have
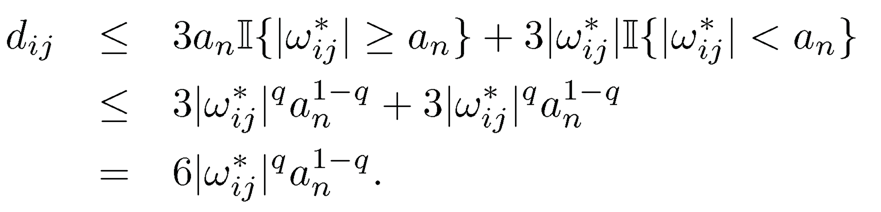
Hence,
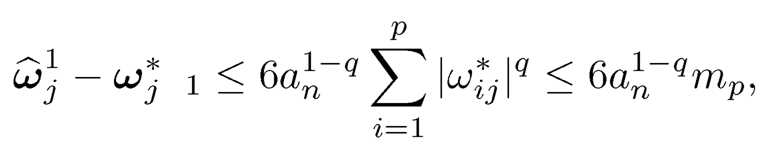
using Ω* ∈ Cq* (mp) namely, ‖Ω1 − Ω*‖ ≤ 6an1-q mp. Therefore,
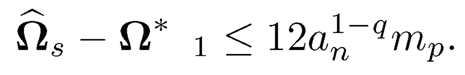
(iii) The result for the Frobenius norm follows immediately from that
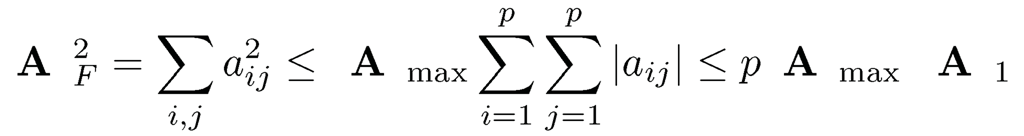
and the results in (i) and (ii).
For the event (‖Σ — Σ‖ max > C0 √(log p) /n) , the second part of the result follows directly from the choice of λn and the fact ‖Ω‖1 ≤ Dn. This completes the proof.
9.7 Bibliographical Notes
Wu and Pourahmadi (2003) gave a nonparametric estimation for large covariance matrices of longitudinal data. Estimation of a high-dimensional covariance matrix was independently studied by Bickel and Levina (2008a,b) for banded and sparse convariance and Fan, Fan, and Lv (2008) for low-rank plus diagonal matrix. Levina, Rothman and Zhu (2008) and Lam and Fan (2009) investigated various maximum likelihood estimators with different sparse structures. Xue, Ma and Zou (2012) derived an efficient convex program for estimating a sparse covariance matrix under the positive definite constraint. Xue and Zou (2014) showed an adaptive minimax optimal estimator of sparse correlation matrices of a semiparametric Gaussian copula model. The optimality of large covariance estimation was investigated by Cai, Zhang and Zhou (2010), Cai and Liu (2011), Cai and Zhou (2011, 2012a, b), Rigollet and Tsybakov (2012), Cai, Ren, and Zhou (2013), among others. Gao, Ma, Ren, and Zhou (2015) investigated optimal rates in sparse canonical correlation analysis. Cai and Yuan (2012) studied an adaptive covariance matrix estimation through block thresholding. Bien, Bunea, and Xiao (2016) proposed convex banding, Yu and Bien (2017) considered a varying banded matrix estimation problem, and Bien (2018) introduced a generalized notation of sparsity and proposed graph-guided banding of the covariance matrix. Xue and Zou (2014) proposed a ranked-based banding estimator for estimating the bandable correlation matrices. Li and Zou (2016) proposed a Stein’s Unbiased Risk Estimation (SURE) criterion for selecting the banding parameter in the banded covariance estimator. Additional references can be found in the book by Pourahmadi (2013).
There is a large literature on estimation of the precision matrix and its applications to graphical models. An early study on the high-dimensional graphical model is Meinshausen and Bühlmann (2006). Penalized likelihood is a popular approach to this problem. See for example Banerjee, El Ghaoui and d’Aspremont (2008), Yuan and Lin (2007), Friedman, Hastie and Tibshirani (2008), Levina, Rothman and Zhu (2008) and Lam and Fan (2009), Fan, Feng and Wu (2009), Yuan (2010), Shen, Pan and Zhu (2012). Ravikumar, Wainwright, Raskutti, and Yu (2011) proposed estimating the precision matrix by the penalized log-determinant divergence. Zhang and Zou (2014) proposed the penalized D-trace estimator for sparse precision matrix estimation. Cai, Liu and Luo (2011) and Cai, Liu and Zhou (2016) proposed a constrained ℓ1 minimization approach and its adaptive version to sparse precision matrix estimation. Liu and Wang (2017) proposed a tuning-insensitive approach. Ren, Sun, Zhang and Zhou (2015) established the asymptotic normality for entries of the precision matrix and obtained optimal rates of convergence for general sparsity. The results were further extended to the situation where heterogeneity should be adjusted first before building a graphical model in Chen, Ren, Zhao, and Zhou (2016).
The literature has been further expanded into robust estimation based on regularized rank-based approaches (Liu, Han, Yuan, Lafferty, and Wasserman, 2012; Xue and Zou, 2012). The related work includes, for instance, Mitra and Zhang (2014), Wegkamp and Zhao (2016), Han and Liu (2017), Fan, Liu, and Wang (2018). It requires elliptical distributions on the data generating process. Such an assumption can be removed by the truncation and adaptive Huber estimator; see Avella-Medina, Battey, Fan, and Li (2018).
9.8 Exercises
9.1 (a) Suppose that A and B are two invertible and compatible matrices. Show that for any matrix norm
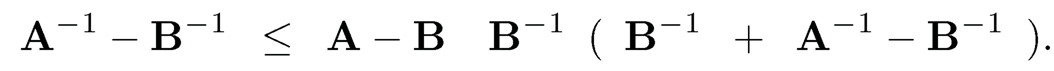
Hint: Use A−1 — B−1 = A−1(B — A)B−1.
(b) Suppose that the operator norms ‖Σ‖ and ‖Σ−1‖ are bounded. If there is an estimate Σ such that ‖Σ − Σ‖ = OP(an) for a sequence an →* 0, show that Σ̂ −1 is invertible with probability tending to 1 and that
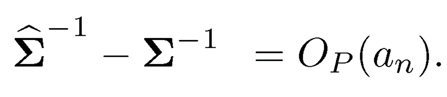
9.2 Suppose Σ is a p × p covariance matrix of a random vector X (X1, X2, … Xp)T
(a) Prove the second part of Theorem 9.1: If max1≤i≤p σii ≤ C2, uniformly over Cq(mp)
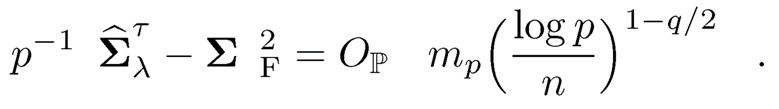
You may use the notation and results proved in the first part of Theorem 9.1.
(b) Assume that E(Xi) = 0 for all i, and σ = maxi,j SD(XiXj) is bounded. Consider the elementwise adaptive Huber estimator ΣH = (σ̂ijτ) where σ̂ijτ is the adaptive Huber estimator of the mean E(XiXj) = σij based on the data {XkiXkj}k=1n with parameter τ. Show that when τ = √n/(a log p)σ, we have
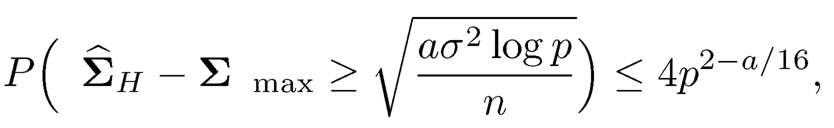
for any constant a > 0. Hint: Use Theorem 3.2(d) of the book.
9.3 Deduce Theorem 9.3 from Theorem 9.2.
Hint: Consider a 2 × 2 submatrix that involves sij and apply Theorem 9.2.
The solution is furnished in Section 9.6, but it is a good practice to carry out the proof on your own.
9.4 Complete the proof of the second part of Theorem 9.4.
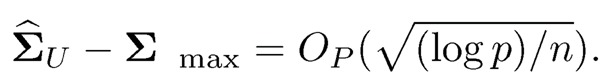
9.5 Use Theorem 9.5 to show that
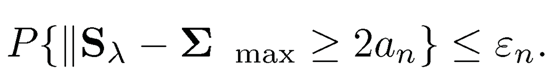
9.6 Suppose that the covariance input Σ̂ λ satisfies P{‖Σλ — Σ‖ max ≥ an} ≤ εn. Show that the projected estimator Sλ that minimizes (9.23) with ‖·‖max norm satisfies
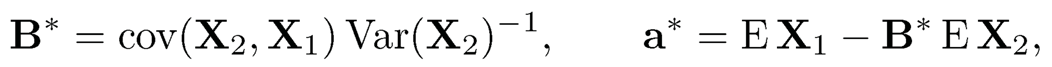
9.7 Let X ~ N(µ, Σ) and Ω = Σ-1 be the precision matrix. Let ωij be the (i j)th element of Ω. Show that Xi and Xj are independent given the rest of the variables in X if and only if ωij =0.
9.8 This exercise intends to give a least-squares interpretation of (9.30).
(a) Without the normality assumption, find the best linear prediction of X1 by a+ BX2. Namely, find a and B to minimize E ‖ X1— a — BX2 ‖2. Let a* and B* be the solution. Show that
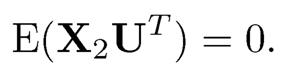
where cov(X2, X1) = E(X2—EX2)(X1—EX1)T and Var(X2) = E(X2-EX2)(X2 —EX2)T.
(b) Let U = X1 — a* — B* X2 be the residuals. Show that E U = 0 and the covariance between X2 and U is zero:
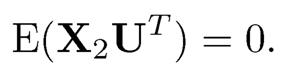
(c) Show that
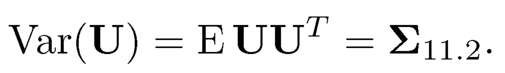
9.9 Let S be the sample covariance matrix of data {Xi}in= 1. Show that
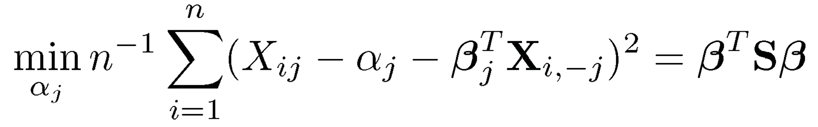
subject to the constraint that βj = —1.
9.10 Prove Proposition 9.4.
9.11 Consider the optimization problem:

For both the Frobenius norm ‖· ‖* =‖· ‖F and elementwise max norm ‖· ‖* =‖· ‖max, show that the solution is
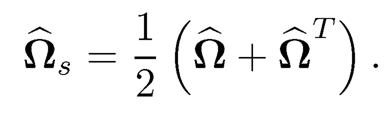
9.12 Let F(·) be the cumulative distribution function of a continuous random variable X.
(a) If g(X)~N(0, 1), show that g(·) =Φ-1(F(·)), where Φ(·) is the cumulative distribution function of the standard normal distribution.
(b) Let F̂n(x) = n−1Σi=1nII(Xi≤x) be the empirical distribution based on a random sample {Xi}i=1n and ĝn(x) = Φ−1(F̂n(x)). Show that
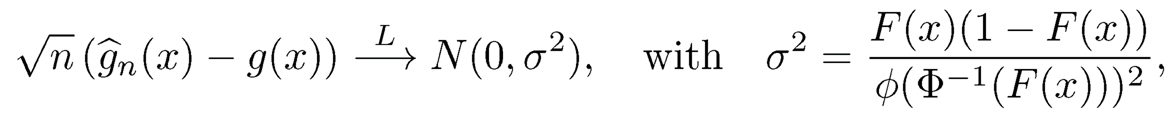
where ø(·) = Φ′(·) is the density of the standard normal distribution.
9.13 We use the notation introduced in Section 9.5.
(a) Show that the function Φ2 (x1, x1,ρ) is strictly increasing in ρ.
(b) Prove (9.49).
(c) Prove (9.50).
9.14 This is a simplified version of the next exercise.
(a) Prove the result in Exercise 9.15(b) for continuous data, rather than the mixed data.
(b) Prove the result in Exercise 9.15(b) for binary data, rather than the mixed data.
9.15 Let the function G be defined in Proposition 9.6.
(a) Show that G−1(x) satisfies
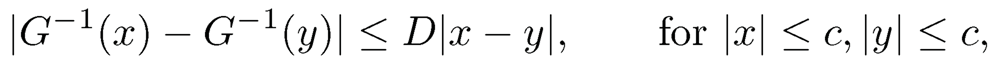
for a sufficiently large constant D, where 0 < c < 1.
(b) Let σ̃jk = G−1(τ̂jk). Prove that
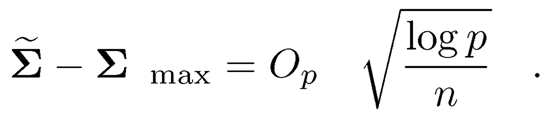
9.16 Download the mice protein expression data from UCI Machine Learning Repository:
https://archive.ics.uci.edu/ml/datasets/Mice+protein+ Expression.
There are in total 1080 measurements. Use attributes 2-78 (the expression levels of 77 proteins), as the input variables for large covariance estimation. We clean the missing values by dropping 6 columns “BAD_N”, “BCL2_N”, “pCFOS_N”, “H3AcK18_N”, “EGR1_N”, “H3MeK4_N” and then removing all rows with at least 1 missing value. For your convenience, you can also download directly the clean input data from the book website. You should have an input data of size 1047 × 71.
(a) Compute the correlation-thresholded covariance estimate with λ = 4. Report the entries of zeros and the minimum and maximum eigenvalues of the resulting estimate. Compare them with those from the input covariance matrix.
(Hint: the entry-dependence thresholds for the covariance matrix are 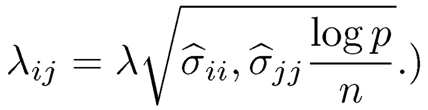
(b) Estimate the precision matrix by CLIME. (Hint: you can use the package “flare”.)
(c) Construct the protein network by using the estimated sparsity pattern of the precision matrix. (Hint: you can use the package “graph” or “ggraph”)
(d) Use the winsorized data to repeat part (a), where the winsorized parameter is taken to be 2.5 standard deviation.