From DNA to Phenotypes
Michael C. Whitlock
OUTLINE
1. What is a gene?
2. Descriptions of genetic variation
3. A multiplicity of forms of inheritance
Genetic information is passed between generations in most organisms by DNA. Variation among individuals of the sequence of their DNA is the raw material of evolution. DNA codes for phenotype by sequences specifying proteins and RNAs and by regulatory elements that control when and by how much of each is made. Variation in DNA sequence can translate into variation in phenotype, which may cause fitness differences among individuals on which natural selection can act. This chapter describes the basics of how phenotypes are created from the instructions coded in DNA and introduces some of the descriptions of genetic variation used in evolutionary biology.
GLOSSARY
Allele. One of possibly many versions of DNA sequences at a given locus.
Autosome. A chromosome that is not a sex chromosome.
DNA. Deoxyribonucleic acid; the molecule used by most of life on earth to encode genetic information and to transfer that information from parent to offspring.
Gene. A region of genetic material that encodes a functional unit, like a protein or RNA.
Genetic Variance. Variance among individuals in some quantity measured on their genotypes. Genetic variance can refer to variance among individuals in the number of copies of a particular allele or to variance in the effects of those alleles on phenotypes, depending on context.
Genotype. The genetic code of an organism. Genotype can refer to the whole genome or to the alleles at a specific locus or loci.
Hardy-Weinberg Equilibrium (HWE). The frequencies of genotypes at a locus assuming independent pairing of alleles from maternal and paternal copies.
Heritability. The fraction of phenotypic variance for a trait that can be attributed to genetic effects inherited by offspring from their parents.
Heterozygote. An individual that has two different copies of DNA from each parent at a locus.
Heterozygosity. The frequency of individuals in a population at a locus that are heterozygotes. Often used to refer to the expected heterozygosity, which is the frequency of heterozygotes expected at Hardy-Weinberg equilibrium for the allele frequencies in that population.
Homozygote. An individual that has two identical copies of DNA from each parent at a locus.
Linkage Disequilibrium. An association between alleles at different loci, where the particular alleles appear together in gametes either more or less often than expected by chance.
Locus. Often synonymous with “gene,” a location in the DNA sequence.
Messenger RNA (mRNA). An RNA made by copying from DNA, used as a template in translation to produce a protein.
Phenotype. Any observable characteristic of an organism, including, for example, its morphology, behavior, and physiological or developmental processes.
Pleiotropic. Describing a locus that has effects on more than one phenotypic trait.
Protein. A covalently linked series of amino acids. Proteins are responsible for most biological functions.
Sex-Linked Locus. A locus that is located on one of the sex chromosomes.
Deer mice in the species Peromyscus polionotus are not always the same color. Some are very light brown, and some are almost black. On the beaches of Florida, where the sand is almost white in color, most mice have very light brown fur, whereas in the nearby fields the soil is dark and loamy, and the mice are darker to match. These differences in color allow the mice to escape predation by visual predators (like coyotes and hawks), because the mice are harder to spot when they are colored like their background.
These color differences are largely controlled by genetic differences between the mice. A large part of what makes a beach mouse pale is that it has a mutation at one particular place in its DNA. This region is responsible for coding the information necessary to make a protein called the melanocortin 1 receptor, abbreviated to Mc1r. This protein in the cell membrane can be activated to signal pigment-producing cells to produce the dark-brown pigment called eumelanin. Mice with more eumelanin have darker fur; mice with less have lighter brown fur. The difference can determine whether they live or die.
This chapter discusses how individuals of the same species differ from one another in their DNA, their proteins, their morphology, their physiology, their behavior, and ultimately, in fitness. We begin by reviewing, very briefly, how the phenotype of an individual is affected by its DNA.
1. WHAT IS A GENE?
The word gene refers to a region of DNA that codes for the instructions for a particular protein or RNA. (RNA is another nucleic acid, and various RNAs have important functions in biology. For example, messenger RNA [mRNA] is a copy of the information from the coding region of a protein-encoding gene, while ribosomal and transfer RNAs both help translate the instructions in mRNA into the correct sequence of amino acids to make a particular protein.) DNA is composed of just four units called nucleotides: thymine (T), adenine (A), cytosine (C), and guanine (G). The order in which these four nucleotides occur in the DNA determines its meaning.
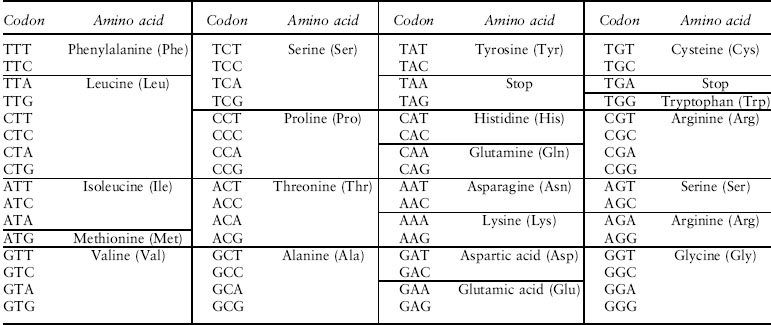
If the gene codes for a protein, it contains information describing the order of amino acids that will be linked together to make that protein. A protein is one or more polypeptides, that is, covalently linked series of amino acids. There are 20 amino acids commonly used by living organisms. Each amino acid has different biochemical properties that, together with the other amino acids in the protein and other proteins in the organism, determine the biochemical functionality of the protein. Each amino acid has one or more sequences of three nucleotides that code for it; these three-nucleotide sequences are called codons. The list of conversions between the particular codons and their associated amino acids is called the genetic code (figure 1). There are also three codons that tell the protein translation machinery to stop, marking the end of the coding sequence.

Figure 1. The genetic code
For example, the stretch of DNA that codes for the Mc1r protein is called the Mc1r gene. This gene consists of a coding region, which specifies the sequence of amino acids that make Mc1r itself, as well as a series of other DNA sequences called regulatory regions that interpret signals from the cell to determine when to create Mc1r protein and when to not. Both the coding region and the regulatory region together are called the gene.
In a diploid organism like humans or mice, each cell has two copies of most genes, one each inherited from the mother and father. Many organisms are haploid, carrying only one copy of each gene. Most microorganisms are haploid.
Every copy of a gene may not contain exactly the same DNA sequence, however. If the DNA sequence at a gene is different between two copies, we say that these are different alleles of that gene. In the beach mice, for example, one common allele for Mc1r codes for the version of the protein that causes the mice to have light brown fur, and another allele codes for Mc1r that causes mice to be darker. In this case, the two alleles differ at only one place in their DNA sequence—a difference that affects the 293rd nucleotide, changing a cytosine nucleotide in the darker allele to a thymine in the lighter allele. This DNA difference causes the 65th amino acid in the resulting Mc1r protein to be the amino acid cysteine rather than arginine (figure 2). The different biochemical properties of the proteins coded by the alleles—the arginine is large and charged, whereas the cysteine is smaller and uncharged—are what cause the fur color phenotype to differ. A difference in the DNA sequence of one nucleotide can cause a difference of one amino acid, which causes a difference in the shape or chemistry of a protein, which causes a difference in the fur phenotype, which in turn causes a difference in fitness between carriers of different alleles.
Not all changes in phenotype result from changes in the coding region. Near each coding region are sections of the DNA that regulate the expression of that gene; that is, these regulatory regions can control when, in what tissues, and in what concentration the protein is actually made. For example, the wildflower in Phlox drummondii typically has light blue flowers, as does the related species P. cuspidata. Where the two species overlap geographically, however, P. drummondii has dark-red flowers. (As a result of this difference in flower color, pollinators move less pollen between the two species, and therefore fewer unfit hybrid offspring are produced.) The dark-red flowers have much more of the pigments cyanidin and malvidin, which in part is a result of an increased expression of the transcription factor MYB. The mutation that increases the expression of MYB is not a change to its coding region, though, but a change in its gene’s regulatory element. Important phenotypic effects based on changes to gene regulation like this are very common.
Figure 2. The coat coloration in beach mice is caused by one change (C to T) in the DNA sequence of Mc1r, which causes the codon CGC to turn into TGC, changing it from coding for arginine (Arg) into coding for cysteine (Cys). The chemical structures for Arg and Cys are shown; these two amino acids have very different chemical properties that translate into divergent functions of the protein, causing differences in the coat color of the mice. This is a nonsynonymous change, because the resulting protein sequence differs between the two alleles. In contrast, a change from CGC to CGA at that same codon, shown in the bottom panel, causes no change in the amino acid sequence, because both these codons code for the same amino acid. This is a synonymous change.
The Central Dogma
The so-called central dogma of molecular biology is that, for most organisms, DNA is the genetic material that transmits information between generations, and mRNA is copied from genes on the DNA to carry that information to parts of the cell to produce proteins, which do most of the work of the cell. This “dogma” is largely true, but all its steps can have exceptions. Some organisms use RNA as the genetic material that is passed between generations to convey the information encoded therein. Many genes in DNA code for RNAs that themselves have physical function (and do not code for proteins at all). Some information is transmitted between generations by modifications of the DNA, such as methylation, such that genetic transmission of information to subsequent generations is not entirely composed of data in the sequence of the DNA. Parents also modify the environment of their offspring in many cases, affecting the nature of subsequent generations in nongenetic ways. Such enriching exceptions aside, however, the central dogma explains much of the nature of how information is transmitted and used by living organisms.
Variation in DNA Sequence
Differences between alleles, like those causing fur color differences in mice or flower color differences in Phlox, are the raw material of evolution. Evolution by natural selection requires heritable differences between individuals, which occur when different individuals carry alleles that produce different phenotypes. A population that contains more than one allele can have genetic variation, meaning that all individuals are not genetically identical. This variation at the DNA level can mean that there is variation at the protein level, which may translate into variation in phenotype, which can translate into variation among individuals in fitness. Only when fitness varies among individuals, and when that fitness variation has a genetic basis, can a population evolve by natural selection.
Evolutionary biologists use a variety of techniques to study this genetic variation, ranging from examination at the DNA sequence level to investigation of the genetic basis of differences in phenotypes between individuals. DNA sequencing now allows relatively inexpensive reading of the genome itself; sequencing all or part of the genome in multiple individuals within a species allows the genetic variability of that species to be measured. DNA sequences at a locus can differ in a variety of ways. Most simply, the DNA can differ among individuals at a particular nucleotide site. A single nucleotide polymorphism (SNP—pronounced “snip”) means that some individuals have one nucleotide while others have a different nucleotide at the same place in their DNA. If this difference occurs in a coding region and causes a change in the amino acid sequence of the resulting protein, it is called a replacement or nonsynonymous SNP, because the meaning of the DNA is not the same among individuals (figure 2). If a SNP occurs in a coding region but does not change the amino acid sequence, it is called a synonymous SNP. (Synonymous changes are possible because most amino acids are coded for by more than one codon—refer to the genetic code in figure 1.)
Synonymous variation is particularly useful for the study of evolution, precisely because it is unlikely to have large direct effects on phenotype. As a result, synonymous variation is often assumed to be selectively neutral, and as such often allows a “control” for the effects of selection on the genome.
DNA sequence can also vary between alleles by insertions or deletions of an extra set of nucleotides. Deletions may have little effect on phenotype, or they may remove a regulatory element or part of a coding region (or even a whole gene or multiple genes) and potentially have large phenotypic effects. Sometimes, entire genes are duplicated, potentially allowing either that protein to be made faster or for the two copies to diverge to different functions. (See chapter IV.2 for a broader description of the types of genetic variation.)
2. DESCRIPTIONS OF GENETIC VARIATION
The most basic unit in the description of the genetic variation is the allele frequency, which is the fraction of all alleles at a locus in the population of interest that have a particular sequence. (Population geneticists often, but not always, use the letters p or q to denote the allele frequency of a particular gene under study.) For example, assume that a diploid population of mice has a total of 500 individuals, which means there are 1000 copies of the Mc1r gene in that population, because each individual carries two copies of each autosomal locus. If 30 of those copies code for the light-colored allele, then the frequency of the light-colored allele in that population is p = 30/1000 = 0.03. Assuming that all other copies of Mc1r in that population are the dark-colored allele, its frequency would then be q = 1−0.03, or 0.97.
Every individual of a diploid species carries two copies of each autosomal locus, one from each of its parents. The genotype of the individual at that locus therefore must be described by keeping track of both alleles that it carries. If there are only two alleles in the population, there are three possible genotypes: individuals that carry two copies of one allele, two of the other allele, or one of each. If there are more alleles at a locus, then the number of possible genotypes is higher. If both copies at a locus are the same allele, then we say that the individual is homozygous at that locus; if the two copies differ, it is heterozygous.
What frequencies might we expect to see of the different genotypes? Genotype frequencies can be predicted from the allele frequencies, provided a long list of evolutionary assumptions are true. If we assume that no selection, mutation, or migration affects the frequencies of alleles at a locus, that the population being studied is extremely large, and all individuals in the population are equally likely to mate with all other individuals, then the genotype frequencies can be predicted from the Hardy-Weinberg equilibrium (HWE). (In reality, the HWE is very predictive even with some selection, mutation, and migration, but its usefulness is very sensitive to that last assumption of random mating.) With HWE conditions, two alleles are paired independently to produce a diploid individual for the next generation, and as a result, the probability of a particular genotype is simply the probability that it receives one of its alleles from a random draw from the population (which will occur with a probability equal to the frequency of that allele) times the probability for its other allele. Thus, at HWE, the probability that an individual is homozygous for an allele that occurs at frequency p = 0.3 is p2 = 0.32 = 0.09. The probability that an individual is heterozygous for two alleles that occur at frequencies 0.3 and 0.6, respectively, is 2 (0.3) (0.6) = 0.36. (The 2 in that equation represents the two ways of making any given heterozygote, with a particular allele coming from either the father or mother; in either case the genotype of the offspring is the same.) If we sum the expected frequencies of all the possible heterozygotes in a population, we get the expected heterozygosity, which is a common measure of genetic diversity. Expected heterozygosity increases with larger numbers of alleles, and it is greater if the frequencies of those alleles are similar.
Genotype frequencies can deviate from HWE, especially if mating individuals are related to each other. If individuals that mate to produce offspring are more closely related than randomly chosen members of the species, they are likely to share alleles, and their offspring are more likely to be homozygous than expected by HWE.
Moreover, allele frequencies can vary substantially over space. If mating occurs locally rather than at random over the range of the species (as is in fact common), then the allele frequencies in one area may evolve to be different than in another area. Such differences can evolve either owing to chance (i.e., genetic drift; see chapters IV.1 and IV.3) or because selection may favor different alleles in different places, especially if the environment also varies spatially. For example, the Peromyscus mice described at the beginning of this chapter have very different allele frequencies of Mc1r for populations on white sand versus on dark soil.
When more than one genetic locus is examined simultaneously, we may ask whether alleles at one locus are independent of the alleles at another locus. If there is no correlation between alleles at different loci, we say those loci are in linkage equilibrium. If, however, two alleles at different genetic loci appear together more often than expected by chance, we say the loci are in linkage disequilibrium. Two loci do not have to be physically linked to be in linkage disequilibrium, but physical linkage allows disequilibrium to persist longer. Disequilibrium can be created by chance (if a particular two-locus genotype happens to leave more offspring than expected) or by selection (if two alleles at the different loci work particularly well in combination, for example). Conversely, recombination between the loci tends to reduce linkage disequilibrium.
There are many measures of linkage disequilibrium, but the most basic is D, which measures the excess of gametes in the population of a particular two-locus allele combination. If the allele frequencies of specific alleles at the first and second loci are pA and pB, respectively, then the frequency of gametes bearing both of those alleles will be pA pB +D. If the two alleles were drawn at random and independently, gametes with both would appear with frequency pA pB, that is, D would be zero. Therefore D measures the deviation in gamete frequencies from independent assortment of alleles at two loci.
Quantitative Genetic Variation
Evolution by natural selection requires that DNA differences also have phenotypic effects. If DNA differences do not affect the morphological, physiological, or behavioral phenotypes of their organisms, then long-term evolution of those phenotypes is impossible. Sometimes, we know the link between differences in DNA sequences and phenotypes (as in the beach mouse Mc1r story); far more often, we do not. Moreover, the link between genotype and phenotype is usually far more complicated than shown in the preceding examples. Nearly all traits are affected by multiple loci—sometimes interacting in complex ways—and the environment in which the organism develops almost always affects the traits. In addition, we usually do not know which genes matter for a particular trait.
If a trait is largely controlled by variation at a single locus, and if there are only two common alleles at that locus, then the phenotypes in the population may fall into two or three discrete categories (such as the light-blue or dark-red flowers in the Phlox example). If the genetic basis of the trait is more complex, or if the environment plays a stronger role, trait values are likely to vary more continuously. In such a case it becomes very difficult to determine the effects of individual genes from looking at phenotypic variation alone. For example, the height of a human being is strongly affected by that person’s genes, but no one gene controls more than about 3 percent of the variation in height. Instead, hundreds of genes and environmental factors like nutrition affect human height. In such cases, to determine how much genetic variation is available for the evolution of a particular trait, we can measure the genetic variance or heritability of that trait. These measures describe the contribution of genetics to the variance among individuals in a given trait.
The phenotypic variance of a trait is simply the variance among individuals in a population of each individual’s values of that trait. (Variance is defined in the same way as it is used in statistics, as a measure of the variation in a population.) This phenotypic variance is potentially due to both genetic differences among individuals—either at one or many genetic loci—and variation in the traits among individuals caused by differences in their environments. The relative importance of genes and the environment varies widely from trait to trait and from population to population.
From an evolutionary perspective, to describe the effects of genetic variation on phenotypes, the important issue is whether the results of selection in one generation can be inherited by subsequent generations. Therefore, we wish to describe the phenotypic variation among individuals that can be inherited by their offspring. We call such heritable variation the additive genetic variance. It can be less than the phenotypic variance and, indeed, usually is.
Often, the environment in which an organism develops affects the values of its phenotypic traits as much as or more than does its genotype. The variance among individuals in a trait caused by differences in their environment is called the environmental variance for that trait. By definition, the heritability of a trait in a population is the fraction of the total phenotypic variance that is additive genetic variance. The heritability is greater if the additive genetic variance is larger, but smaller if there are many strong environmental sources of variation for that trait. For example, within North America and Europe the heritability of human height is approximately 70 to 90 percent, meaning that the majority of variation in height in these populations is caused by genetic variation. In general, the larger the heritability of a trait in a population, the faster that selection can cause an evolutionary change in that trait.
Sometimes, the environment can produce unpredictable variation in a trait, but in other cases, particular environments can cause foreseeable changes in phenotype. For example, when humans grow up with better diets, they tend to be taller. Environmentally induced changes in phenotypes can also be adaptive; for example, when conditions are dry, plants tend to have a greater proportion of their biomass as roots, which increases their ability to capture valuable water. Changes in the phenotype that result from differences in the developmental environment are called phenotypic plasticity.
3. A MULTIPLICITY OF FORMS OF INHERITANCE
There are many ways in which DNA sequence variation can affect the phenotypic variance. At one extreme, some changes to DNA sequence probably have no effect at all on the phenotype; at the other extreme, changes to a single base pair can be the difference between life and death. Some alleles have strong effects regardless of the other alleles present in the same individual, whereas other alleles strongly depend on the presence of particular alleles at other loci to produce their effects. This latter interaction between genes is called epistasis.
Some alleles produce their effects regardless of the particular alleles with which they are paired at the same locus; such alleles are dominant. Other alleles must be present in a homozygous state to produce their effects; we call these alleles recessive. Some other alleles show intermediate effects in heterozygotes; such alleles are called codominant. The most fit allele can be either dominant or recessive, depending on the locus and the environment.
Each locus may affect many traits. For example, one allele at the gene tb1 in corn, which was very important during its evolution to domestication under ancient artificial selection, causes the plant to have many branches rather than a single stalk, with more leaves, smaller leaves, smaller ears, and more tillers. When a single locus has effects on multiple traits, we say that it shows pleiotropy. Moreover, the dominance and epistasis of alleles may depend on which trait we are considering, because the interactions of alleles within and among loci may differ among traits. Genes are regulated in myriad ways, sometimes by sites close to the start of the coding sequence, sometimes by sites embedded in the introns (DNA between bits of coding sequence), and sometimes by genetic elements far away from the gene, even on different chromosomes. Regulation may depend on details such as whether a stretch of DNA is methylated, which can reflect epigenetic effects transmitted among generations.
Genetic effects can depend on the interaction of sequence differences at multiple sites, either within the same gene or between different genes. These sequence differences can be transmitted together if they are close to each other on the same chromosome, or break apart by segregation and recombination if they are farther apart. All genes are subject to mutation, and the rate of mutation and recombination can vary greatly across the genome. Nearly all genetic effects have the potential of being masked by changes in the environment.
All traits are different in the details of their genetic control. Sometimes this control is relatively straightforward, as when a single base-pair change has large and constant effects. Sometimes the factors affecting the development of a trait are very complex, as when hundreds or thousands of genes contribute small, interacting effects in addition to the multifarious effects of the environment. Sometimes the details matter to the study of their evolution, and sometimes they do not matter so much. Variation is both the fuel of evolution and its fascinating product.
FURTHER READING
Golnick, L., and M. Wheelis. 2008. Cartoon Guide to Genetics. New York: HarperCollins.
Griffiths, A. J., S. R. Wessler, R. C. Lewontin, and S. B. Carroll. 2007. Introduction to Genetic Analysis. 9th ed. New York: W. H. Freeman.
Hartl, D. L., and A. G. Clark. 2008. Principles of Population Genetics. Sunderland, MA: Sinauer.
Hoekstra, H. E. 2010. From mice to molecules: The genetic basis of color adaptation. In J. B. Losos, ed., In the Light of Evolution: Essays from the Laboratory and Field. Greenwood Village, CO: Roberts & Company.
Hopkins, R., and M. D. Rausher. 2011. Identification of two genes causing reinforcement in the Texas wildflower Phlox drummondii. Nature 469: 411–414.