Gene Duplication
Jianzhi Zhang
OUTLINE
1. Mechanisms of gene duplication
2. Fixation of duplicate genes
3. Pseudogenization after duplication
4. Stable retention of duplicate genes
5. Rate of gene duplication
6. Determinants of gene duplicability
7. Functional redundancy among duplicate genes
8. Functional diversification of duplicate genes
9. Future directions in the study of gene duplication
The number of genes in a genome varies by two orders of magnitude across cellular organisms. A primary mechanism underlying this variation is gene duplication, which provides raw genetic materials from which new genes and new gene functions arise. As with other types of genetic mutations, gene duplication first occurs in an individual organism, and its population genetic fate depends on its fitness effect. Even after a duplicate gene is fixed in a population, it will degenerate into a pseudogene unless its presence is beneficial to the organism. Stably retained duplicate genes are quite common in almost all eukaryotic genomes examined. These duplicates form gene families, whose members typically have similar but nonidentical functions or expression patterns. This chapter first describes the processes through which duplicate genes are generated, fixed, and stably retained. It then discusses the rate of gene duplication and factors influencing this rate. Finally, it examines the functional redundancy and divergence among duplicate genes.
GLOSSARY
Alternative Splicing. An RNA splicing process by which the exons of the RNA produced by transcription of a gene are reconnected in multiple ways. Alternative splicing leads to the production of multiple different proteins from a single gene.
Aneuploidy. Loss or gain of one to several chromosomes but not a complete set.
Concerted Evolution. An evolutionary process that explains the observation that individual members of a gene family within one species are more similar to one another than to members of the same gene family in other species, even though these members were generated prior to the divergence of the species. Concerted evolution is usually attributed to frequent gene conversions among gene family members within species.
Functional Redundancy. The condition in which two paralogous genes perform the same function.
Gene Conversion. An event in genetic recombination that converts one DNA sequence to another. It can homogenize the sequences of duplicate genes of the same species.
Neofunctionalization. Acquisition of a new function that may be qualitatively or quantitatively different from the previous function.
Paralogous Genes. Genes that are related through duplication.
Pseudogene. A dysfunctional relative of known genes that has lost its protein-coding ability or is no longer expressed.
Retroposition. Integration into the genomic DNA of a sequence derived from reverse transcription of RNA.
Subfunctionalization. Division of multiple functions of a progenitor gene into its daughter genes such that the total functions of the daughter genes are the same as those of the progenitor gene.
Unequal Crossing-over. Crossing-over between homologous chromosomes that are not precisely paired, resulting in nonreciprocal exchange of material and chromosomes of unequal length.
1. MECHANISMS OF GENE DUPLICATION
Gene duplication refers to the duplication of a segment of DNA that contains one or more genes. In 1936, Calvin Bridges reported the first case of gene duplication, observed in mutant fruit flies (Drosophila melanogaster) exhibiting extreme reduction in eye size. Through observation of the polytene chromosomes from the salivary glands of D. melanogaster larvae, Bridges showed that the mutant phenotype was caused by the doubling of a small segment of the X chromosome.
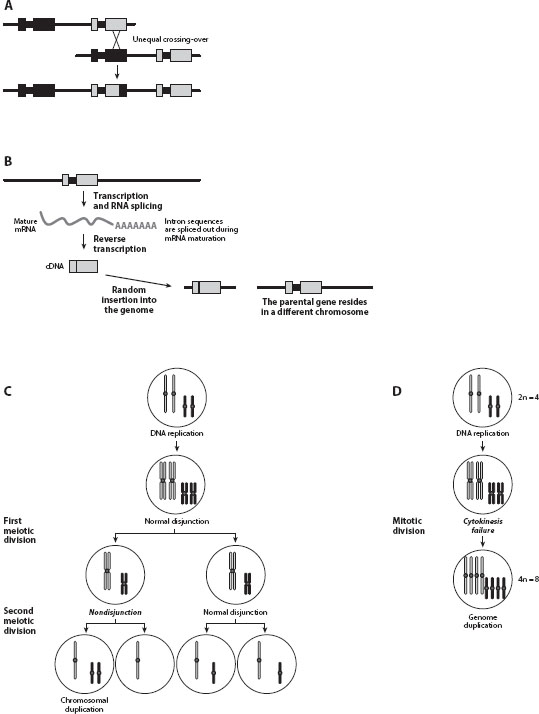
Gene duplication occurs by one of the three general mutational mechanisms: unequal crossing-over, retroposition, and chromosomal (or genome) duplication. Unequal crossing-over refers to crossing-over between homologous chromosomes that are not precisely paired, resulting in nonreciprocal exchange of material and chromosomes of unequal length (figure 1A). That is, one of the resultant chromosomes contains an extra copy of a chromosomal segment, while the other loses this segment. This mechanism typically generates tandem gene duplicates that are arrayed next to each other along the chromosome. Depending on the position of crossing-over, the duplicated region may contain part of a gene, an entire gene, or several genes.
Figure 1. Mechanisms of gene duplication. (A) Unequal crossing-over, which results in a recombination event in which the two recombining sites lie at nonidentical locations in the two parental DNA molecules. (B) Retroposition, a process by which a messenger RNA (mRNA) is retrotranscribed to complementary DNA (cDNA) and then inserted into the genome. In (A) and (B), squares represent exons and bold lines represent introns. (C) Chromosomal duplication via nondisjunction during meiosis. (D) Genome duplication via cytokinesis failure in mitosis.
Retroposition occurs through a completely different mechanism: a messenger RNA (mRNA) of a gene is retrotranscribed to complementary DNA (cDNA) and is then inserted back into the genome, resulting in an extra gene copy (figure 1B). Genes duplicated by this mechanism are also known as retroduplicates. Because they arise from mRNAs, retroduplicates lack introns and regulatory sequences such as the promoter, and often contain poly A tracts at the end. Furthermore, in contrast to genes duplicated by unequal crossing-over, a retroduplicate is unlinked to its mother gene, because the insertion of cDNA into the genome is more or less random. It is also impossible to have blocks of genes duplicated together by retroposition. Because retroposition must occur in the germ line to be heritable, only genes expressed in the germ line are subject to heritable retroposition.
Chromosomal duplication refers to the phenomenon whereby one to several (but not all) chromosomes in a genome are duplicated. It occurs by nondisjunction of homologous chromosomes during meiosis and leads to aneuploidy (figure 1C). By contrast, genome duplication refers to the duplication of the entire genome, and is also known as polyploidization. Autopolyploids are polyploids with multiple chromosome sets derived from a single species. They arise from a spontaneous, naturally occurring genome doubling (figure 1D), or the fusion of unreduced gametes. In comparison, allopolyploids are those with chromosomes originated from different species, as a result of doubling of chromosome number in an F1 hybrid of the two species.
2. FIXATION OF DUPLICATE GENES
The probability that a duplicate gene gets fixed (i.e., is found in every individual) in a population is determined by the fitness effect of duplication, as for other types of mutations. Depending on the type of gene duplication and the genes involved, gene duplication may be deleterious, beneficial, or neutral to the organism in which the duplication occurs (see chapter V.1).
Gene duplication could be deleterious for several reasons. First, because gene transcription and translation costs energy, gene duplication may impose a fitness cost. Second, gene duplication may break the often-sensitive balance in dosage (i.e., the precise amount of RNA or protein relative to other genes) that is required for certain genes. This is why trisomy, the presence of three instead of two copies of a chromosome in a diploid individual, is usually deleterious. In humans, all autosomal trisomies are lethal, except for trisomy 21, which causes Down syndrome and, less often, trisomy 13 (Patau syndrome) and 18 (Edwards syndrome). Third, a retroduplicate may be inserted into a gene or a functional element, causing a deleterious effect.
Gene duplication, however, may be beneficial when extra gene product is useful to the organism. For example, cells need a large number of ribosomes for rapid protein synthesis; thus duplication of ribosomal protein and RNA (rRNA) genes is typically beneficial. A recent study found that duplication of the human salivary amylase gene is advantageous in certain human populations with high starch diets, apparently because the increased amount of amylase helps digest starch.
Gene duplication can also be neutral or nearly neutral. For instance, duplication of a gene expressed at low levels imposes very little energy cost and hence virtually no fitness cost unless it has other effects. Most retroduplicates are not expressed because of the lack of promoters; thus they have effectively no fitness effect if they do not interfere with the expression or function of other genes.
As is true for other types of mutations, most duplicate genes do not get fixed in a population; however, among those duplicates that did get fixed and are observed today, a major question remains: were they fixed mostly by positive selection or by random genetic drift? Some authors proposed that positive selection for enhanced gene dosage is the primary mechanism for duplicate gene fixation, but available genomic data do not seem to support this view, although positive selection is clearly involved in a few cases.
A large number of duplicate genes are segregating within populations in their paths to fixation or loss. These polymorphisms constitute a form of copy number variation (CNV). Recent studies have shown a surprisingly large number of CNVs in humans. While most CNVs seem to have no visible phenotypic effect, others are associated with human diseases. It is likely that CNVs cause disease by disturbing the dosage of the gene involved or the dosage of the involved gene relative to those of other genes in the genome (i.e., dosage balance).
3. PSEUDOGENIZATION AFTER DUPLICATION
Even after a duplicate gene gets fixed in a population, it may not remain functional and thus be stably retained in the genome for a long time. This is because the daughter gene is usually identical in function to its mother gene; their functional redundancy implies that the loss of one of them has no fitness consequence. In other words, mutations that knock out the expression or function of one of the duplicates can accumulate and the gene gradually becomes a pseudogene, defined as a dysfunctional relative of known genes that has lost its function (i.e., its protein-coding ability or its expression). Given enough time, pseudogenes are no longer recognizable because they either diverge too much from their functional relatives or get deleted from the genome. Analysis of the age distribution of duplicate genes in model eukaryotes demonstrated convincingly that pseudogenization is the most common fate of duplicate genes, as the number of (nonpseudogenized) duplicate genes declines sharply with age during the first few million years after gene duplication.
While pseudogenization after duplication seems uninteresting because of its lack of impact on phenotype or fitness, it is important to note that this process may contribute to the formation of reproductive isolation between populations and hence speciation (see chapter VI.8). Let us imagine a duplication event that results in a pair of chromosomally unlinked genes A and B in one species. Shortly after the duplication, the species is split into two populations that are geographically separated. Assume that gene A is pseudogenized in one population, while B is pseudogenized in the other. If the geographic barrier is removed and the two populations merge, the hybrid from a cross between the two populations will have one functional allele and one null allele at locus A as well as at locus B. Thus, a quarter of the gametes produced by the hybrid contain null alleles at both loci (figure 2). If the functions of the genes involved are important, these gametes may malfunction. For example, if the functions of A and B are required for gamete survival, one-quarter of gametes will die. Consequently, the hybrid has a fecundity of f = 0.75, relative to an individual from either population. It is easy to see that if n pairs of duplicate genes are reciprocally pseudogenized in the two populations, f = 0.75n, which drops quickly with n. Thus, this model, known as divergent resolution, provides an explanation for a rapid rise in genetic reproductive isolation between geographically isolated populations simply by random degeneration of redundant duplicate genes. This model may be particularly important in lineages that experience whole-genome duplication (WGD), because of the abundance of unlinked duplicate pairs that are subject to divergent resolution.
Figure 2. Divergent resolution of duplicate genes can lead to reproductive isolation and speciation. Horizontal boxes represent chromosomes and black bars represent genes. A and B are a pair of functionally redundant duplicate genes. A cross on a gene name indicates pseudogenization. The bold circled gamete has neither functional A nor functional B, and thus is less fit than the other three gametes.
In addition to those pseudogenes that arise gradually from functional duplicate genes, there are also so-called dead-on-arrival pseudogenes that have never been functional. For instance, retroduplicates lack their own promoters and hence do not have the machinery to have ever been expressed. These pseudogenes, also known as processed pseudogenes for their lack of introns, are highly abundant in many eukaryotic genomes. Because the higher the expression of a gene in the germ line, the greater the probability of retroduplication, one can infer the germ line expression of a gene in ancient times from the numbers of its processed pseudogenes that belong to certain age groups. Furthermore, because retroduplicates arise from mRNAs, processed pseudogenes of different ages provide information on ancient mRNA processing such as alternative splicing that may be absent today. Thus, processed pseudogenes in a genome can be viewed as fossilized ancient transcriptomes that permit an otherwise-impossible glimpse into ancient gene expressions.
4. STABLE RETENTION OF DUPLICATE GENES
For a duplicate gene to be stably retained in a genome, it must be useful to the organism, such that loss of the gene would cause an immediate decrease in fitness too large to spread through the population, because of natural selection. Several mechanisms have been proposed to explain the ways in which duplicates make fitness contributions. First, when gene duplication is immediately beneficial as a result of increased gene dosage, both gene copies can be stably retained because the loss of either gene decreases the dosage and thus fitness (figure 3A). For example, the retention of multiple rRNA genes is easily explained by this mechanism. Duplicate rRNA genes within a genome tend to be highly similar in sequence despite the fact that the duplication may be quite ancient. The high sequence similarity is the result of gene conversion, which is a mutational process homogenizing DNA sequences within a genome. Presumably, highly homogenized rRNA genes are beneficial over heterogeneous rRNA genes, so the product of gene conversion is selectively maintained. This mode of duplicate gene evolution is known as concerted evolution.
Figure 3. Mechanisms allowing the stable retention of duplicate genes. (A) Duplicate genes maintain sequence and functional similarity, typically by concerted evolution, when having a high concentration of the gene product is beneficial. (B) Neofunctionalization, in which one daughter gene acquires a new function while the other performs the old function. (C) Subfunctionalization, in which each daughter gene inherits one of the ancestral functions. (D) Escape from adaptive conflict, through which each of the two daughter genes inherits one ancestral function and improves it. The improvement is impossible in the progenitor gene because of the adaptive conflict between the two functions, represented by a lightning bolt. Rectangles represent genes, and circles and stars represent different functions. Larger symbols indicate enhanced activities or improved functions.
Second, gene duplication allows one gene to perform the ancestral (and presumably important) function and the other to adopt a new function that may be prohibited prior to the duplication because of the impossibility of one gene performing both functions (figure 3B). This route, known as neofunctionalization, is generally thought to be the most important contribution of gene duplication to evolution. For example, the primate eosinophil cationic protein (ECP) gene was duplicated from the eosinophil-derived neurotoxin (EDN) gene; in a relatively short evolutionary time after the duplication, ECP acquired an antibacterial activity that is not found in EDN. Rapid sequence evolution driven by positive Darwinian selection (see chapter V.1) was detected in the neofunctionalization of ECP.
Third, an ancestral gene may already possess dual functions; after duplication, each copy may adopt one of the ancestral functions such that they together possess both functions of the ancestral gene (figure 3C). This molecular division of labor, known as subfunctionalization, does not increase organismal fitness, but permits each duplicate to make a fitness contribution. For instance, the zebra fish engrailed-1 gene is expressed in the pectoral appendage bud, while its paralogue eng1b is expressed in a specific set of neurons in the hindbrain/spinal cord. This pair of duplicates was generated in teleosts after they diverged from tetrapods. In tetrapods such as mice and chickens, the single-copy En1 gene is expressed in both expression domains: the developing pectoral appendage bud and specific neurons of the hindbrain and spinal cord.
Fourth, a model that is becoming increasingly popular is called the escape from adaptive conflict (EAC), or the specialization model, which can be viewed as a hybrid of the neofunctionalization and subfunctionalization models (figure 3D). In EAC, the ancestral gene already possesses dual functions, but neither function can be optimized because optimizing one function compromises the other. After duplication, the ancestral functions can be subdivided into the duplicate copies, and the removal of the conflict allows each function to be optimized. Different from the pure neofunctionalization model, EAC asserts that both duplicates will acquire enhanced functions yet no entirely novel function is gained in either copy. EAC is also distinct from the pure subfunctionalization model in that it requires an improvement of ancestral functions. EAC also implies that the improvement of one function in a gene is realized by the degeneration of the other function in the same gene, requiring that these two processes are coupled both in time and in molecular mechanism. The EAC model is best illustrated by the evolution of the duplicated GAL1 and GAL3 genes in the yeast galactose use pathway, where the dual functions of their progenitor gene have been divided into the duplicates and further improved. The replacement of GAL1 with the progenitor gene reduces the fitness, so does the replacement of GAL3 with the progenitor gene. It should be noted, however, that while the evolutionary patterns of the GAL genes fit the EAC model, the initial force that permitted the evolutionary retention of these genes could be pure subfunctionalization.
This last point also emphasizes that the apparent mechanism responsible for the retention of a pair of duplicate genes today may be different from the mechanism underlying the initial retention of the duplicates. Most empirical studies, including the examples provided above, have revealed the mechanisms for current rather than initial retention. Mechanisms for initial retentions are necessarily inferred from comparisons of present-day properties of duplicate genes, and thus should be taken with a grain of salt.
Which of the four models above best explains the initial retention of duplicate genes? To address this question, it is more productive to analyze gene function/expression data than their proxies such as the evolution rate of gene sequences, because the four models are all about gene function or expression. At the genomic scale, yeast duplicate genes exhibit large differences in patterns of expression and protein-protein interaction (PPI). This observation suggests that the model of dosage advantage or concerted evolution cannot be the primary mechanism for duplicate gene retention, because divergences in function and expression are prohibited under this model. Examination of accurately measured genome-wide expression levels of duplicate genes in two yeast species and two mammal species showed that duplicated genes have significantly reduced expression levels compared to their unduplicated progenitor genes. This result further rejects the dosage advantage model and suggests that at least with respect to the amount of gene expression, subfunctionalization has occurred; however, this finding per se is insufficient to establish the role of subfunctionalization in the initial retention of duplicates. In an analysis of tissue expressions of human genes and PPIs of yeast genes at the genomic scale, it has been shown that duplicate genes experience substantial subfunctionalization as well as substantial neofunctionalization, but the former happens quickly after gene duplication, while the latter is a much slower process. This analysis was based on the comparison among groups of duplicate genes that were generated at different time points in the past and found high degrees of subfunctionalization among all age groups of duplicates but high levels of neofunctionalization only among old duplicates. These findings suggest that subfunctionalization is more likely than neofunctionalization to underlie the initial retention of duplicate genes. Because of the lag of neofunctionalization compared to subfunctionalization, this result appears to be inconsistent with the EAC model; nonetheless, in EAC, neofunctionalization is a quantitative improvement rather than a qualitative change in function. EAC could still occur right after duplication, because such quantitative functional improvements are underdetected in the above study of gene expression and PPI, and it is possible that the observed slow neofunctionalization is of a fundamentally different type that is unrelated to EAC.
These empirical findings are generally consistent with theoretical predictions. Specifically, Lynch and colleagues showed that the probability of duplicate gene retention by subfunctionalization is much greater than that by neofunctionalization, especially when the population size is not very large. When the population size gets larger, the chance that a beneficial neofunctionalizing mutation occurs and gets fixed before the occurrence of subfunctionalization increases, and the relative role of neofunctionalization in duplicate gene retention expands.
5. RATE OF GENE DUPLICATION
Examining the numbers of duplicate genes of different ages in a genome, Lynch and Conery (2000) estimated the first genome-wide rate of gene duplication. They estimated from a number of eukaryotic model organisms that fixed duplicate genes arise at a rate of about 0.01 per gene per million years, but the vast majority become pseudogenes within a few million years. Their estimate, however, was only approximate, because of the limited genomic data available at that time and some simplifying assumptions. For example, it was later pointed out that gene conversions between duplicate genes make them look younger than they are, resulting in an overestimation of the duplication rate; also, whole genome duplication (WGD) was not separated from individual gene duplications in the above estimation. WGD is much more frequent in plants than in animals, although a dozen or so WGD events are known in animals.
Another way to estimate the rate of gene duplication is through the examination of mutation accumulation (MA) lines, which are very small populations of organisms maintained in a constant environment for hundreds to thousands of generations. The lack of virtually any natural selection in small populations allows the estimation of the duplication rate per generation at the mutational level. Recently, the genomes of several MA lines of the yeast Saccharomyces cerevisiae and nematode Caenorhabditis elegans were sequenced. Surprisingly, the rate of the appearance of new duplicates was found to be on the order of 10-6 per gene per generation in yeast, about 105 times that measured for duplicate genes that are eventually fixed in a population. Similarly, the rate was on the order of 10-7 per gene per generation in C. elegans, about two orders of magnitude greater than that measured for fixed duplicate genes. This discrepancy—between the rate of appearance of new duplicates and the rate of fixation of duplicates—strongly suggests that the vast majority of gene duplication events are deleterious and thus do not reach fixation. This conclusion is also supported by recent surveys of CNVs in fruit flies.
6. DETERMINANTS OF GENE DUPLICABILITY
What factors determine gene duplicability, the probability that a gene is duplicated at the mutational level, fixed, and stably retained in the genome? Although three processes (mutation, fixation, and retention) are involved here, it is often difficult to differentiate them in the study of gene duplicability because typically only retained duplicates are observed; nevertheless, under the assumption that duplication rate at the mutational level is not widely different among genes, gene duplicability studies help identify important factors influencing the fixation and retention of duplicate genes.
An important observation of gene duplicability is that compared to other genes, those encoding members of protein complexes have reduced rates of individual gene duplication. Because components of a protein complex need to be balanced in concentration, dosage imbalance brought about by doubling the concentration of one but not other members of the protein complex is likely deleterious. WGD, however, creates an opposite situation. The individual carrying WGD is usually reproductively isolated from other individuals of the same species. In other words, WGD is immediately fixed if the lineage with WGD will survive in evolution; thus, duplicate genes that are present long after WGD tell us what genes tend to be retained after fixation. Interestingly, genes encoding protein complex members tend to be maintained after WGD, because individual gene losses would cause dosage imbalance just as individual gene duplication does.
It has also been reported that the more complex a gene is, the higher its duplicability, where gene complexity is measured by protein length, number of protein domains, and number of cis-regulatory motifs in the promoter of the gene. This phenomenon appears to be attributable to higher retention probabilities for more complex genes, presumably because more complex genes are subject to faster subfunctionalization and hence greater likelihood of stable retention. Hence, gene duplication increases both gene number and gene complexity, two factors in the origin of genomic and organismal complexity.
Interestingly, in terms of the duplicability bias among genes of different importance to the fitness of the organism, it has been shown in yeast that less important genes have a greater duplicability than more important ones. There are two apparent reasons for this phenomenon. First, yeast genes encoding members of protein complexes tend to be more important than other genes, and the balance hypothesis explains why the former should have a lower rate of individual gene duplication than the latter. Second, in yeast, gene importance is negatively correlated with the number of cis-regulatory motifs in the promoter of the gene, and the gene complexity hypothesis explains why less important genes, which have more cis-regulatory motifs, should duplicate more than important genes.
There are several other gene functional properties that have been observed to correlate positively with gene duplicability, although the underlying mechanisms are often unclear. These include functioning as metabolic enzymes, interacting with the external environment, interacting with fewer protein partners, controlling physiological traits (rather than morphological traits), having more phosphorylation sites, and functioning as intermediary proteins (rather than receptors) in signaling pathways.
7. FUNCTIONAL REDUNDANCY AMONG DUPLICATE GENES
Functional redundancy refers to the functional similarity between genes, and is typically demonstrated by measuring the fitness effect of gene deletion. For example, it has been shown at the genomic scale in S. cerevisiae and C. elegans that deleting a duplicate gene has a significantly smaller fitness effect than deleting a singleton gene. Furthermore, deleting a pair of duplicate genes has a much greater fitness effect than deleting either gene alone. Evolutionary theory predicts that the degree of functional redundancy between a pair of duplicates gradually declines with the time since duplication, as a result of subfunctionalization and/or neofunctionalization; however, it has been reported that some duplicate genes are highly redundant even hundreds of millions of years after duplication. Of course, in the case of ribosomal proteins, rRNAs, histones, tRNAs, and other molecules in high demand in the cell, functional similarity among duplicates is selectively favored and thus requires no other explanation.
For other duplicate genes, several hypotheses have been proposed to explain the unexpectedly long retention of functional redundancy. First, some believe that redundancy is beneficial in itself because it protects the organism from the potential harm of deleterious mutations, much like the backup role of a spare tire for a car. This backup hypothesis, however, cannot on its own be correct, because the benefit of backup cannot be detected by natural selection unless the product of population size and mutation rate is orders of magnitude greater than that observed in cellular organisms. Second, the piggyback hypothesis asserts that paralogous genes have some nonoverlapping functions as well as some overlapping functions, and the existence of the latter is a by-product of the former owing to strong protein structural constraints. Third, reduction of gene expression after duplication, a special form of subfunctionalization, is commonly observed. Suppose 100 protein molecules is the optimal expression level for a gene. On duplication of the gene and subsequent evolution, one daughter gene may produce 60 protein molecules, and the other, 40. In this scenario, both gene copies can be stably retained, yet no functional divergence between them is expected even long after duplication. With respect to fitness, there is no redundancy between the two genes because both copies are required to reach the highest fitness; however, because of the nonlinear relationship between dosage and fitness, deleting both genes would almost always have a much greater fitness effect than deleting either one of them, creating the apparent phenomenon of redundancy. The relative importance of the second and third hypotheses explaining the functional redundancy is unclear and will likely remain a topic of intensive study.
8. FUNCTIONAL DIVERSIFICATION OF DUPLICATE GENES
While the maintenance of functional redundancy among duplicate genes is not unusual, the most common observation among stably retained duplicates is their functional divergence. The degree of functional divergence varies greatly. In many cases, duplicate genes perform similar types of function, but with different activities or specificities, or at different times or locations. For example, isozymes catalyze the same biochemical reaction but usually have different catalytic parameters; they are encoded by duplicate genes that are often expressed in different tissues or at different developmental stages. Duplication also expands the scope of a basic function. For example, odorant receptor (OR) genes form the largest gene family in the vertebrate genome. Each OR is able to recognize only a limited number of odorants. Vertebrates are believed to be able to detect 10,000 or more odorants because of the possession of hundreds of functional OR genes that recognize different ligands.
As mentioned, retroduplicates are usually dead on arrival because of the lack of a promoter; occasionally, however, they may be expressed when they are fortuitously inserted into a genomic region that harbors a promoter, for example, into the intron of another gene. There is now accumulating evidence that retroduplication is also an important source of new genes (see chapter V.6). Probably because of completely different expression patterns and the involvement of gene fusion, functions of retroduplicates can sometimes differ dramatically from those of their mother genes.
9. FUTURE DIRECTIONS IN THE STUDY OF GENE DUPLICATION
Two theoretical questions about the functional divergence of duplicate genes are yet to be resolved. First, is the functional difference between duplicates attributable mainly to subfunctionalization or to neofunctionalization? While some authors stress the former, others emphasize the latter. Still others believe that both happen in each duplicate pair and that their relative contributions depend on the time since duplication. Second, what is the role of natural selection in the functional divergence of duplicate genes? This question is related to the first, because the role of selection is different in different types of functional changes. For instance, subfunctionalization by degenerate mutations does not require positive selection. By contrast, EAC must involve positive selection. It is commonly thought and has been demonstrated in case studies that positive selection is involved in neofunctionalization, but in theory, neofuctionalization can also occur by random fixation of neutral mutations; the utility of the new function may be realized only after its fixation, on an alteration of the genetic background or environment. The role of purifying selection in neofunctionalization is also understudied. Clearly, neofunctionalization in a daughter gene requires at least a partial relaxation of the selective constraint associated with the functions of the progenitor gene. But whether neofunctionalization has a greater chance of occurring in the presence of some functional constraints or in the presence of no constraint is not entirely clear, because in the presence of no constraint, the gene may become a pseudogene before acquiring new function, as has been recently demonstrated experimentally. These uncertainties notwithstanding, it is apparent that gene duplication is the primary source of new genes in evolution and that it has contributed to biodiversity at the genomic, functional, and organismal levels.
FURTHER READING
Conant, G. C., and K. H. Wolfe. 2008. Turning a hobby into a job: How duplicated genes find new functions. Nature Reviews Genetics 9: 938–950. A recent review on the mechanisms of functional changes after gene duplication.
Force, A., M. Lynch, F. B. Pickett, A. Amores, Y. L. Yan, and J. Postlethwait. 1999. Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151: 1531–1545. A classic paper that proposed the subfunctionalization model of duplicate gene evolution.
Gu, Z., L. M. Steinmetz, X. Gu, C. Scharfe, R. W. Davis, and W. H. Li. 2003. Role of duplicate genes in genetic robustness against null mutations. Nature 421: 63–66. A genome-wide study that revealed substantial functional redundancy among duplicate genes.
He, X., and J. Zhang. 2005. Rapid subfunctionalization accompanied by prolonged and substantial neofunctionalization in duplicate gene evolution. Genetics 169: 1157–1164. Temporal patterns of subfunctionalization and neofunctionalization in duplicate genes were revealed by genome-wide data on protein-protein interaction and gene expression.
Kaessmann, H., N. Vinckenbosch, and M. Long. 2009. RNA-based gene duplication: Mechanistic and evolutionary insights. Nature Reviews Genetics 10: 19–31. A recent review on retroduplication.
Lynch, M., and J. S. Conery. 2000. The evolutionary fate and consequences of duplicate genes. Science 290: 1151–1155. Most fixed duplicate genes become pseudogenes in a few million years.
Ohno, S. 1970. Evolution by Gene Duplication. New York: Springer-Verlag. A classic book that greatly stimulated the study of gene duplication.
Papp, B., C. Pal, and L. D. Hurst. 2003. Dosage sensitivity and the evolution of gene families in yeast. Nature 424: 194–197. Gene duplication is deleterious when breaking dosage balance.
Qian, W., B. Y. Liao, A. Y. Chang, and J. Zhang. 2010. Maintenance of duplicate genes and their functional redundancy by reduced expression. Trends in Genetics 26: 425–430. Expression reduction after gene duplication allows the long-term retention of functionally redundant duplicate genes in the genome.
Zhang, J. 2003. Evolution by gene duplication: An update. Trends in Ecology & Evolution 18: 292–298. A review on the role of gene duplication in genomic and organismal evolution.