Evolution of New Genes
Manyuan Long
OUTLINE
1. Mutational mechanisms to generate new genes
2. Rates of new gene origination
3. Patterns of new gene evolution
4. Evolutionary forces acting on new genes
5. Functions and phenotypic effects of new genes
Every gene has its first moment: this is its origination, when a new gene appears in a genome and evolves a distinct or new function(s) that did not previously exist. The genes in extant organisms are of different ages, from ancient to very young. To understand the origination of a gene is to understand the earliest stage of its evolution; however, the origination process cannot be directly observed for most genes because they are ancient. For these ancient genes, there were likely multiple evolutionary events, which may have obliterated the early signature of the gene’s origination process. An alternative is to examine the genes that have formed recently, which are called new genes or young genes; in these cases, a reconstruction of the origination process is feasible and provides an exciting glimpse into the evolution of new genes.
Several questions are relevant to understanding the origination of new genes. First, what mutational processes generate new genes in a genome? Second, how often do new genes reach 100 percent frequency (i.e., fixation) in a species population? Third, if the new genes frequently appear in genomes during evolution, are there any patterns or rules underlying their origination? Fourth, what evolutionary forces are responsible for the fixation of a new gene within a genome? And how did the new gene accumulate mutations to optimize its function? Finally, what are the roles of new genes in phenotypic evolution? Understanding the functions and phenotypic effects of new genes is critical to determine their role in evolution.
Since the first truly young gene, Jingwei, was found in Drosophila by Manyuan Long and Charles Langley two decades ago, new techniques for molecular and genomic analyses have been invented, and sequence databases have expanded at a previously unimaginable rate. This large amount of data has shed new light on the origination of new genes, such that a general picture is emerging of the general process of genetic and phenotypic evolution.
GLOSSARY
Chimeric Gene. A type of new gene whose domains or encoded exons originated from a combination of different genes.
Copy Number Variation. Newly formed gene duplicates or gene deletions that have not been fixed within a population.
Gene Trafficking. The transfer of gene copies between sex chromosomes and autosomes during evolution.
Neofunctionalization. An evolutionary process in which a new function is acquired by a gene.
New Gene. A gene that appears in a genomic location where it had not previously existed.
Orthologue. A gene found in different species that originated from a common ancestral gene and diverged after speciation.
Paralogue. A gene formed by duplication within a genome.
Retrogene. A gene that originated via retroposition, in which a parental gene is transcribed, the subsequent RNA is reverse transcribed, and the new copy is inserted into a new location in the genome. When retrogenes have no function because of lack of regulatory systems for expression, they are called retropseudogenes or processed pseudogenes.
Transposable Element. A segment of DNA that is capable of moving within and between genomes.
1. MUTATIONAL MECHANISMS TO GENERATE NEW GENES
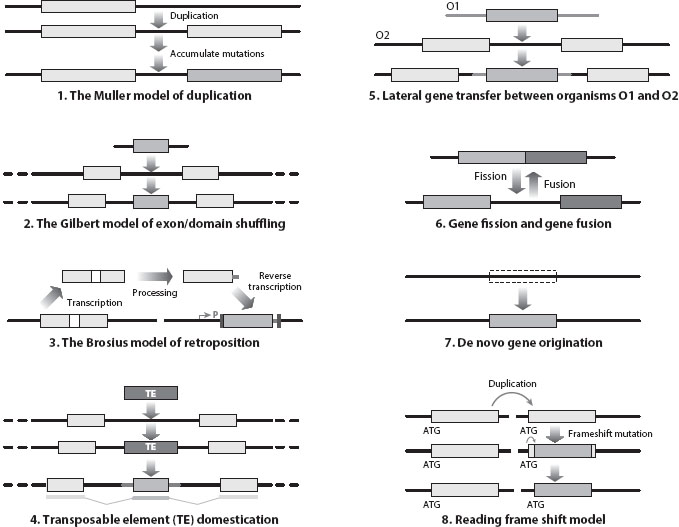
What genetic mechanisms underlie the formation of a new gene? Until the early 1990s, three general models, termed the Muller model of duplication (DNA-based duplication), the Gilbert model of exon/domain shuffling, and the Brosius model of retroposition (RNA-based duplication), had been proposed for the origination of new genes (figure 1). Importantly, these three mechanisms are not mutually exclusive and can be used simultaneously to create a new gene. For example, in the ancestor of African fruit flies (Drosophila yakuba and D. teissieri), a retrogene from the alcohol dehydrogenase (Adh) gene was inserted into a previously existing duplicate of the yellow emperor (Ymp) gene, which led to the creation of a chimeric gene (Jingwei) that functions in the pheromone metabolism recruitment pathway. Additional mechanisms of new gene formation, via transposable element insertion, lateral gene transfer, and frameshift mutations (see below), have since been discovered, as well as gene fission and fusion mechanisms (figure 1).
Figure 1. Molecular mechanisms of new gene evolution. The first three are general models: the Muller model, Gilbert model, and Brosius model, followed by five mechanisms that are also frequently used in various organisms. In the Brosius retroposition model, the poly(A) tail and short flanking sequences are also labeled as two dark gray bars with the poly(A) tail before the second bar, with fortuitously recruited regulatory system R. Although lateral gene transfer from one species (O1) to the other (O2) is more often observed in prokaryotes, it does contribute to new gene formation in eukaryotes. Frameshift mutations can be caused by insertion or deletion of nonintegers of 3 (e.g., one or two nucleotide insertion or deletion in a codon).
In the mid-1990s, it was observed that transposable elements (TEs) could be “domesticated” to create a new coding portion of a nuclear gene. For example, in the human genome, an Alu TE was inserted into the coding portion of the decay-accelerating factor (DAF) and created a new hydrophilic carboxy-terminal region in DAF that later evolved a new function to inhibit DAF from moving into the membrane. As many as 400 human gene families have since been found to be hybrids between a nuclear gene and a TE; these examples represent many different types of TEs. Furthermore, TEs are also found to facilitate recombination, leading to the formation of chimeric genes. For example, in two dozen cases in Drosophila, DNAX TEs are associated with new duplicate copies and parental copies in the melanogaster subgroup, and Pack-Mule TEs in rice have been involved in the formation of approximately 2000 chimeric genes.
Lateral gene transfer (LGT) was known to happen frequently among prokaryotic organisms, but it was previously thought not to occur in eukaryotes; however, several examples have recently been documented in eukaryotes. For example, genes and genome fragments of the parasitic bacteria Wolbachia have been observed in the genomes of Drosophila, mosquitoes, and bees; several mitochondrial genes that encode ribosomal and respiratory proteins were subject to horizontal transfer between distantly related species in flowering plants; and the pea aphid has recruited genes encoding multiple enzymes for carotenoid biosynthesis from the genome of an ancestral Phycomyces fungus. Thus, the role of LGT in eukaryotic genome evolution was previously underappreciated.
Finally, frameshift mutations, which are usually thought to be deleterious, have been found to contribute to the formation of new genes. In a survey of human genomes, there were about 470 novel protein families that could have been created by a reading frameshift mutation in duplicate copies. Although many of these observations might derive from sequencing and assembling errors, this model is interesting in that it proposed the rapid creation of novel proteins.
In all the aforementioned mechanisms, new genes are derived from previously existing genes; thus, it was surprising to discover that several dozens of new protein-coding genes in D. melanogaster appear to have no orthologues in closely related species (even those that diverged only a few million years ago), suggesting that new genes can arise de novo (figure 1). Besides, de novo genes have also been reported in plants, mice, humans, fish, and viruses. One simple interpretation is that previously noncoding or intergenic regions can accumulate enough mutations to create functional open reading frames.
2. RATES OF NEW GENE ORIGINATION
At present, the genomes of thousands of species have been sequenced from almost all major types of organisms, including bacteria, archaea, protozoa, fungi, plants, and animals. Comparative analyses from eukaryotic genomes, especially from the various model organisms, have identified thousands of young genes (see chapter V.3). These observations suggest that the origination of new genes is a common process and that genomes have been modified frequently by adding new genes with new functions. These genomic sequences also provide data to estimate the rate of new gene origination.
The origination rates of new genes created from a few mechanisms have been analyzed. The first rate was estimated for gene duplication (see chapter V.5). Michael Lynch and colleagues extensively analyzed the duplication rates in major model organisms, including humans, mice, Arabidopsis thaliana, and Saccharomyces cerevisiae, and observed an average duplication rate of 0.01 per gene per million years, although a majority of new duplicates became silenced in a few million years. This duplication rate implied that the fixation of duplicate genes occurs at a high rate: 100 new duplicates per million years per 10,000 genes. This would suggest that one to three new duplicates are fixed in the genomes of Drosophila and humans every 10,000 years. These estimates may be impacted by gene conversion and other factors (e.g., insufficient annotation that tends to ignore the duplicates that have recently appeared); however, recent estimations based on young duplications in mammalian and Drosophila genomes give a rate with the same order of magnitude.
It is still unknown how many of these new duplicates will evolve novel functions, rather than maintain redundant functions. A gene created by a structural change (e.g., a chimeric gene) is likely to have functions that diverge from its parental copy. A change in the temporal or spatial expression of a gene can also allow it to acquire a new function. Extensive surveys of the origination rates of new genes have been conducted in Drosophila because of the availability of genomic sequence data in multiple species in the genus. In Drosophila, the origination rate of new genes including DNA-based duplication, retrogenes, and de novo origination was approximately 23 per genome per million years, and the majority of these genes evolved via a chimeric structure by recruiting new exons and new untranslated regions (UTRs). These data suggest that new gene functions, rather than redundant functions, evolve frequently, with one functional chimeric gene originating every 50,000 years in Drosophila.
In the human genome, a majority of retrogenes are chimeric genes that have recruited new exon regions from the surrounding insertion sites. In primates, it has been estimated that the rates of retroposition, and the formation of chimeric genes by retroposition, are 1 and 0.01 per million years per genome, respectively. In the grass family, the rate of chimeric genes created by retroposition is estimated to be as high as 7 per genome per million years. These observations suggest a surprisingly rapid rate of new gene evolution.
3. PATTERNS OF NEW GENE EVOLUTION
Are there any rules governing the origination of new genes? The large numbers of new genes detected in various organisms have provided an excellent set of data to detect possible patterns or rules underlying the processes of new gene origination. It is important to understand the evolutionary patterns of new genes because these patterns may provide clues for understanding the mechanisms underlying the formation of new genes and, in turn, for formulating theories from which to make predictions. So far, three evolutionary patterns (discussed below) associated with the origination of new genes have been detected; these discoveries have stimulated further interest in the mechanisms responsible for these patterns.
Soon after the genomic sequence of D. melanogaster became available, it was realized that a computational identification of retrogenes and their parental copies across the genome would be feasible. It was possible to discriminate the derived retrogene copies from the parental genes because the retrogenes are copied from processed messenger RNA transcripts; newly created retrocopies do not have introns but do have a poly(A) tail and a pair of short duplicate sequences flanking the retrogenes (figure 1). Although the molecular signatures of the poly(A) tail and flanking duplicate sequences might be eroded over substantial evolutionary time (e.g., longer than 10 million years), the loss of introns becomes a permanent feature of retrogenes. Thus, by looking at the paralogous copies from a single species, the ancestral relationship between parental copies and the derived copies can be explicitly characterized.
The fixation of a retrogene and the genomic location of its parental copy are influenced both by mutational events and subsequent evolutionary forces. To determine the relative roles of these processes, the observed genomic distribution of retrogenes can be compared to a hypothetical distribution based on the mutation rate, often the null hypothesis based on the neutral theory of molecular evolution (see chapter V.1). Because retropseudogenes are likely to be evolving neutrally, their distribution can be tested against neutral expectations and used to estimate a distribution of neutral mutations. If they are neutral and the incidence of mutations is not biased among chromosomes, two simple predictions can be derived: (1) the number of parental genes on a chromosome should be proportional to the number of genes on that chromosome, and (2) the number of retrogenes on a chromosome should be proportional to the length of the chromosome. In these analyses, it is important to take the relative population sizes of the X chromosomes and autosomes into account, because they are impacted by additional population genetic factors (e.g., sex ratio) (see chapter V.4). Using data from human genomes, these predictions were tested and confirmed in the functionless retrogenes (processed pseudogenes), suggesting that retroposition is a neutral process.
Trafficking of New Genes between Sex Chromosomes and Autosomes
Computational analysis of D. melanogaster genomic sequences was used to characterize the distribution of retrogenes and parental genes from a database containing all possible retrogenes identified in genome sequences. Compared with the neutral expectation, the retrogene data revealed unique patterns: (1) the observed distribution is significantly different from the expectation; (2) a significant excess of X-linked genes are the parental genes of retrogenes; (3) an excess of the retrogenes are found on autosomes; and (4) retroposition events between the two autosomes or from the autosomes to the X were significantly lower than expected. Thus, the D. melanogaster genome showed evidence for directional trafficking of retroposed genes from the X to the autosomes. The genome sequences of 11 additional Drosophila species revealed the same trend—X-linked genes copied and then pasted in autosomes—suggesting that X-to-autosome gene trafficking is a general process of gene evolution in the Drosophila genus. By contrast, similar analyses applied to the human and mouse genomes revealed bidirectional gene trafficking in mammalian genomes: there was a high excess of retroposition both from the X to the autosomes and from the autosomes to the X chromosome.
Association of Sex-Specific Expression with Trafficking of New Genes
Analyzing the expression pattern of retrogenes and parental genes revealed another pattern: the vast majority of the X-derived autosomal retrogenes have evolved a testis-specific expression pattern in both Drosophila and mammals. Conversely, very few of the X-linked retrogenes copied from autosomal parental genes are expressed in the testes, but instead often evolved female-specific expression. In addition, the parental genes had significantly lower expression in testis. In D. pseudoobscura, gene movement out of the neo-X chromosomes was also observed, with unidirectional gene movement from the X to autosomes and the subsequent evolution of expression in the testes. Similar patterns have been observed in other organisms, including mosquitoes (Anopheles gambiae), stalked-eye flies (Teleopsis), and mammals.
New Genes Are Preferentially Located in Specific Chromosomal Environments
Gene trafficking between the X and autosomes reflects a preference of new genes for a genomic environment that distinguishes sex chromosomes and autosomes. Is there preference for specific genomic environments within chromosomes? The answer is yes, based on evidence from the particular genomic regions flanking or adjacent to new genes. For example, in the human genome, examination of about 50 functional retrogenes revealed a significant connection between the presence of a functional retrogene and the transcriptional potential of the flanking regions: on average, more expressed genes were identified from the regions surrounding the functional retrogenes in comparison to the regions flanking the retrogenes that are transcriptionally silenced. This observation indicated that retrogenes might take advantage of the regulatory environment formed by nearby genes for their expression.
4. EVOLUTIONARY FORCES ACTING ON NEW GENES
What are the underlying evolutionary forces responsible for the evolution of new genes? Two significant events occur as stages in the evolution of new genes. The first stage involves the fate of a new gene within a species in which the new gene can either be lost from the population or spread to fixation in the population. The second stage involves the further accumulation of mutations in the new gene sequence to further improve its function. After this stage, the new gene is subject to the same processes of molecular evolution as any other gene in the genomes (see chapter V.1); however, the roles of evolutionary forces, particularly natural selection and genetic drift (see chapter IV.1), are interesting and peculiar in these first two stages of the evolution of new genes.
Evolutionary Forces Acting on the Fixation of New Genes
Recent genomic technologies have enabled the investigation of the trajectory of a newly arisen gene toward its final fixation or loss from the population. For example, a population-genetic study was conducted on copy number variation (CNV) in D. melanogaster. It was observed that a majority of polymorphic duplicates are found in intergenic regions. This result suggests a role for purifying selection against gene duplication, especially complete gene duplication, consistent with the conjecture that the initial gene duplication is slightly deleterious; however, five recent gene duplicate events, involving genes responding to toxins, were found at high frequency (>70%), suggesting that positive selection is favoring these new gene duplicates.
Daniel Schrider and colleagues recently presented the first study of retrogene polymorphisms, using next-generation sequencing in 37 inbred lines derived from a North Carolina D. melanogaster population. By comparing between-species divergence and within-species polymorphism, they found an excess of fixed retrogenes that were copied from X-linked parental genes on autosomes. This recent result reveals a significant role of positive selection in fixation of new retrogenes within species. They also conducted a similar study in humans and detected a positive selection in fixation of retrogenes.
Evolutionary Forces Acting on New Genes Subsequent to Their Fixation
It is conceivable that when a new gene is fixed, further evolutionary modification may be necessary to optimize its function. Such a verbal model predicts a period of rapid sequence evolution in a new gene, which will eventually slow down in later stages. A very young gene, Jingwei, that originated 3 million years ago in the common ancestor of the three African Drosophila species (D. yakuba, D. teissieri, D. santomea) provided data in support of this model (plate 3). While there were no fixed synonymous changes in this new gene, nine amino acid substitutions occurred in the ancestral stage before the first speciation event that led to D. yakuba and D. teissieri; moreover, after the divergence of D. yakuba and D. teissieri, there was an excess of amino acid substitutions over synonymous changes, in comparison to within-species polymorphism, suggesting a role of Darwinian positive selection in the evolution of Jingwei (see chapter V.14).
The Adh gene has been involved in the formation of two additional chimeric genes in two different Drosophila species groups: Adh-Finnegan, a DNA-based duplication in the repleta group, and Adh-Twain, an RNA-based duplication (retroposition) in D. subobscura, D. guanche, and D. madeirensis. Comparison of all three Adh-derived new genes (including Jingwei) reveals two interesting patterns. First, there is evidence for convergent evolution (see chapter V.12), because the same amino acid substitutions are fixed in these different genes in different organisms! Second, a recent analysis of a large set of new genes in 12 Drosophila species revealed early and rapid substitutions driven by positive selection, with later and slower evolution shaped by purifying selection. These data provide clear evidence that the new genes continue to be under positive selection subsequent to their fixation.
The Targets of Selection
The analysis of duplicate genes described above clearly reveals a role for natural selection and mutational mechanisms in determining the genomic positions of new genes. For example, the testis-expression patterns shown by the new X-derived autosomal retrogenes (described above) have been interpreted as resulting from natural selection. Several explanations for this pattern have been put forward, including sexual antagonism, sexual genomic conflict, degree of dominance, sexual selection, dosage compensation, and male sex-chromosome inactivation.
The classical sexual antagonism model proposes that a mutation with sexually antagonistic effect (i.e., those that are advantageous for males and disadvantageous for females) would be spread from very low frequency to high frequency in a population if its genetic effect is recessive. If a new modifier inhibits its expression in females, or if the gene appears in a small population (in which the effect of genetic drift effect is large), such a sexually antagonistic new gene can be preferably fixed on the X. By contrast, if the new gene is genetically dominant, the fixation of a new antagonistic gene is favored in an autosomal location. It is also likely that dosage compensation can restrict the development of male-biased expression of the X-linked genes, thus favoring the genes that moved to autosomes.
Much interest in recent years has been focused on another aspect of the mechanisms involved in new gene evolution: male sex-chromosome inactivation. In mammals (e.g., humans and mice), it has been observed that when the male germ line cells enter the meiotic stage, the X and Y sex chromosomes are condensed into an X/Y body, and genes on these chromosomes are “silenced” (i.e., not expressed). Thus, there should be strong selection for any genes necessary at these stages of spermatogenesis to be located on autosomes rather than on the X chromosome. This prediction of gene trafficking from the X to the autosomes in mammals, also likely in Drosophila and Anopheles, is confirmed by large-scale analyses of gene expression; however, the mechanisms responsible for the biased genomic distributions of new genes remain to be further elucidated.
5. FUNCTIONS AND PHENOTYPIC EFFECTS OF NEW GENES
The aforementioned observations and analyses, made primarily with data from Drosophila and mammals, have revealed that new genes have originated and fixed frequently in the genomes of various organisms. Expression analyses of these new genes suggest that many have reproductive functions (e.g., expressed specifically in testis); however, this tissue-specific expression pattern is just a first step toward understanding the function and phenotypic effects of new genes. Additional information is critical to understanding how new genes have evolved and how their evolution has contributed to organismal evolution. Analysis of sequence evolution of gene duplicates has provided ample information about their functional evolution. The new functions and phenotypic effects of genes that have arisen in recent evolutionary time are particularly informative.
The previously mentioned new gene, Jingwei, was extensively investigated for the evolution of its biochemical function. Because of the high sequence similarity in its Adh-derived domain, it was initially expected that the new gene might have maintained the functions of its Adh parental gene and that new functions might have been added from its N-terminal domains (plate 3). However, the enzymatic activities of the Jingwei protein were assayed by testing its activity on more than 30 different alcohol substrates. It was observed that the gene evolved new metabolic activities: two chemicals, farnesol (involved in the biosynthesis of juvenile hormone) and geraniol (the pheromone for communication among the individuals), became the specific substrates of Jingwei. These evolutionary and experimental analyses revealed that the new enzymatic function involving new substrates had evolved in a short evolutionary time.
Additional evidence for the rapid evolution of new functions for new genes comes from a study knocking down the expression of more than 195 new genes that originated within the past 35–3 million years in Drosophila. The conventional expectation was that only ancient and conserved genes would be functionally important and that the recently evolved genes would be associated with interesting but dispensable minor functions. Surprisingly, 30 percent of new genes were observed to be lethal when knocked down, which was the same percent of old genes found to be essential. Further assays of the phenotypic effects of 59 of these essential new genes revealed that all affected the development of D. melanogaster. These observations together suggest that the genetic program of development contains species-specific or lineage-specific components. An important conceptual connection from this study can be made: the developmental effects of new genes appear to be adaptive, as there was significant evidence for positive selection on these genes. To this end, the relationship between adaptive evolution and evolution of development can be explicitly linked, suggesting that microevolution and evolution of development are not mutually exclusive but combined under the same mechanism found by Darwin: natural selection.
FURTHER READING
Betrán, E., K. Thornton, and M. Long. 2002. Retroposed new genes out of the X in Drosophila. Genome Research 12: 1854–1859. This is the first paper reporting the directional movement of retrogenes from the sex chromosome to autosomes and shows that the vast majority of autosomal retrogenes have evolved male-biased functions. This finding of the dynamic process of gene evolution provides a mechanistic interpretation for the preferential distribution of male-biased genes on autosomes.
Brosius, J. 1991. Retroposons: Seeds of evolution. Science 251: 753. This paper presents a model of gene evolution and points out the important role of retroposition (RNA-based duplication) in the origin of new genes.
Chen, L. B., A. L. Devries, and C.H.C. Cheng. 1997. Evolution of antifreeze glycoprotein gene from a trypsinogen gene in Antarctic notothenioid fish. Proceedings of the National Academy of Sciences, USA 94: 3811–3816. This report presents evidence of de novo origination of antifreeze protein in Antarctic fish, showing a clear role for environmental factors in driving the evolution of a novel antifreeze protein.
Chen, S., Y. Zhang, and M. Long. 2010. New genes in Drosophila quickly become essential. Science 330: 1682–1685. This report shows that many young genes have evolved essential functions in development in Drosophila. Thus, the developmental program of a species can evolve rapidly with the birth of species-specific and lineage-specific components of the genetic systems underlying development.
Emerson, J. J., H. Kaessmann, E. Betrán, and M. Long. 2004. Extensive gene traffic on the mammalian X chromosome. Science 303: 537–540. This report reveals bidirectional gene trafficking between the X and autosomes in the human genome. In one direction, X-linked parental genes were copied onto autosomes and evolved testis-biased expression; in the other direction, an excess of female genes and nonsex-related genes were moved to the X.
Gilbert, W. 1978. Why genes in pieces? Nature 271: 501. This essay presents a model of gene evolution by exon shuffling. The evolutionary products derived from exon shuffling, chimeric genes, have since been widely observed.
Kaessmann, H., N. Vinckenbosch, and M. Long. 2009. RNA-based gene duplication: Mechanistic and evolutionary insights. Nature Reviews Genetics 10: 19–31. This article provides a comprehensive review of new gene origination by retroposition (RNA-based duplication) from the underlying mechanisms to the generation of new functions.
Levine, M. T., C. D. Jones, A. D. Kern, H. A. Lindfors, and D. J. Begun. 2006. Novel genes derived from noncoding DNA in Drosophila melanogaster are frequently X-linked and exhibit testis-biased expression. Proceedings of the National Academy of Sciences USA 103: 9935–9939. This research article reports five de novo genes that evolved from noncoding regions, mostly on the X chromosome and all predominantly expressed in the testis in Drosophila.
Long, M. Forthcoming. Evolution and function of new genes from flies to humans. Annual Review of Genetics 47.
Long, M., E. Betrán, K. Thornton, and W. Wang. 2003. The origin of new genes: Glimpses from the young and old. Nature Reviews Genetics 4: 865–875. This review presents the first general picture of new gene evolution known at the time. It emphasizes the advantage of using young genes to investigate gene evolution.
Long, M., and C. H. Langley. 1993. Natural selection and the origin of Jingwei, a chimeric processed functional gene in Drosophila. Science 260: 91–95. This report presents the first discovery of the evolution of a new gene. Population genetic and molecular analyses revealed that Darwinian positive selection acted on this newly emerged chimeric gene, which consists of an Adh-derived retrogene and a duplicate of an unrelated gene.
Makalowski, W., G. A. Mitchell, and D. Labuda. 1994. Alu sequences in the coding regions of mRNA: A source of protein variability. Trends in Genetics 10: 188–193. Transposable elements were conventionally viewed as selfish DNAs. This paper presented the first evidence that Alu transposable elements can contribute to protein diversity.
Muller, H. J. 1936. Bar duplication. Science 83: 528–530. In this report of Bar duplication, the model of new gene evolution by DNA-based duplication was explicitly described for the first time.
Schrider, D. R., K. Stevens, C. M. Cardeño, C. H. Langley, and M. W. Hahn. 2011. Genome-wide analysis of retrogene polymorphisms in Drosophila melanogaster. Genome Research 21: 2087–2095. Using molecular population genetics, this analysis detects a significant role for positive selection in the fixation of retrogenes.
Vibranovski, M. D., Y. Zhang, and M. Long. 2009. General gene movement off the X chromosome in the Drosophila genus. Genome Research 19: 897–903. This paper shows that retrogene trafficking, first proposed by Betrán et al. (2002), is generally true in the genus Drosophila. It further demonstrates that DNA-based duplication has contributed to gene trafficking from the X chromosome to autosomes; thus gene trafficking is not a property of biased mutation patterns but a consequence of natural selection.
Wang, W., H. Yu, and M. Long 2004. Duplication-degeneration as a mechanism of gene fission and the origin of new genes in Drosophila species. Nature Genetics 36: 523–527. This research report presented the first mechanistic analysis of gene fission, a process whereby one gene splits into two genes, revealing that duplication is an intermediate step.