WE SAW IN CHAPTER 4 that the genetic code is universal; that is, codons are the same in all living organisms. This means that genes from one type of organism, animal cells for example, should be able to guide the synthesis of the corresponding protein in bacterial cells. In order to accomplish this, genes from one organism must be transferred to bacteria. We already saw in chapter 1 that DNA from one strain of bacteria can be transferred to another. This process is called bacterial transformation. Since DNA from different organisms has the same basic structure, only differing in the sequence of bases, we should be able to transfer DNA from any organism into bacteria. Given this ability, we should be able to manufacture animal, or even human, proteins in bacteria. This feat has indeed been achieved and is described in this chapter. Today, many important human proteins are made in bacteria, and these proteins are actually used in human medicine.
Many proteins are medically useful. How would you go about making a lot of a medically important human protein? Isolating such proteins from humans can be problematic. For example, many proteins can only be found in substantial amounts in vital organs. Thus quantities are limited to those that can be obtained from the cadavers of people willing to donate their organs. Some proteins can be isolated from blood. However, blood can be contaminated with viruses such as HIV, hepatitis, and, more recently, West Nile virus. Expensive screening is required to ensure safety. You can see that protein availability is limited from these sources. Thus making the desirable proteins in bacteria is an attractive option. This involves a technique known as recombinant DNA technology, or cloning, and makes possible the production of human proteins in bacteria. The DNA technology that is used to do this relies on two critically important elements: restriction enzymes and plasmid vectors. Let us start with restriction enzymes.
These enzymes, also called restriction endonucleases, exist in all bacterial cells and have one specific function: they cut the sugar-phosphate backbones of “foreign” DNA, such as the DNA from viruses that infect and kill bacteria. Thus, restriction enzymes can be seen as defense mechanisms evolved by bacteria to defend themselves against viral attacks. Indeed, when restriction enzymes cut DNA, they break up the genes located on this piece of DNA. However, restriction enzymes do not cut DNA randomly, they only cut it at a specific sequence. For example, one restriction enzyme from the bacterium Escherichia coli (E. coli) cuts DNA when it encounters the sequence shown in in figure 5.1.A.
Another restriction enzyme from the bacterium Haemophilus influenza cuts it at a difference sequence, as shown in figure 5.1.B.
Note that the two sites where the restriction enzymes cut have an interesting structure. If one reads the nucleotides on one strand, the opposite strand read backward is identical. This is called a palindrome. A great majority of restriction enzymes cut at palindromic sequences. Restriction enzymes cut at these palindromic sequences of DNA double helix in a very special manner: they produce DNA fragments with complementary single stranded sequences at their ends. One can think of them as “sticky” ends of the DNA. That is, if another piece of DNA is cut with the same restriction enzyme, the two cut ends will have short complementary pieces that can base pair together. You will see below how scientists have taken advantage of this property. Over a hundred bacterial restriction enzymes in pure form are currently available commercially. This means that when scientists isolate DNA from any source, they can cut it with any of the available restriction enzymes. The DNA molecules obtained after cutting are much shorter and therefore much more manageable than the original very long stretches of DNA found in cells. Also, since a given restriction enzyme always cuts DNA at the same sequence, these DNA fragments are a collection of pieces that begin and end with the same base sequence. In addition, these short DNA fragments each contain a limited number of genes. Let us leave restriction enzymes for the time being and focus now on the second ingredient of cloning experiments: plasmid vectors.

Figure 5.1 Examples of Restriction Enzyme Sites. The sites are shown whole on the left, and on the right they are shown after being cut by the enzyme. A. Restriction enzyme site for a restriction enzyme from Escherichia coli. B. Restriction enzyme site for a restriction enzyme from Haemophilus influenza.
Back in the 1960s and 1970s, scientists discovered that bacterial cells often contained more than one piece of DNA. The majority of the DNA was associated with the bacterial chromosome, a single piece of DNA containing several million base pairs and carrying several thousand genes. However, many bacterial species also harbored a circular “minichromosome” that only contained a few thousand base pairs and carried just a few genes. These “minichromosomes” are called “plasmids.” An important property of plasmids is that a bacterial cell can often carry multiple copies, sometimes hundreds, of these “minichromosomes.” Another important feature is that, being small, plasmids often contain just a single cutting site for a number of restriction enzymes. Thus, cutting a circular plasmid with a restriction enzyme generates a linear molecule with complementary single-stranded ends (figure 5.2).
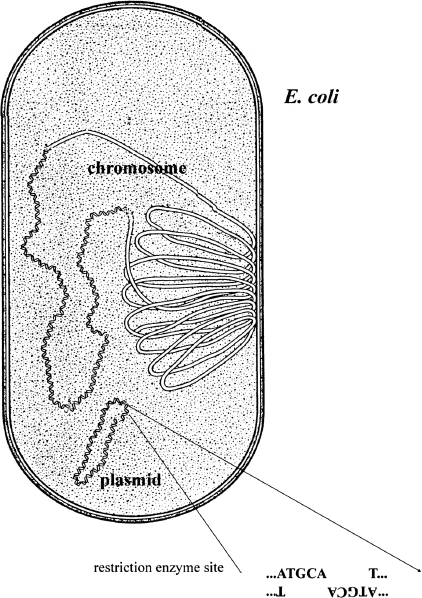
Figure 5.2 E. Coli Chromosomes. A diagram of an E. coli cell showing its main chromosome and a minichromosome or plasmid. Because plasmids are much smaller, a restriction enzyme often cuts it at only a single site.
We will use the example of the insulin gene to explain how gene cloning works (figure 5.3). Insulin was the first protein to have its amino acid sequence deciphered. This is because it is relatively small (see table 4.1). It is also one that is very important medically. Figure 5.3.A shows a part of the amino acid sequence of human insulin. Because we know the genetic code if we know the sequence of amino acids of a protein, we can make a sequence of DNA, a gene, that corresponds to the sequence of amino acids in that protein. Figure 5.3.B shows the sequence of DNA corresponding to the amino acid sequence shown in figure 5.3.A. Because many amino acids can be coded for by a number of different codons, the DNA sequence shown is one of many that corresponds to the amino acid sequence. The gene coding for insulin was synthesized in this way in the laboratory. In order to have this gene transcribed at a high level in bacteria, a promoter and terminator are added to the beginning and end of the gene (figure 5.3.C). Finally, in order to clone the piece of DNA with the insulin gene on it, single-stranded ends complementary to the ends of plasmid DNA cut with a restriction enzyme are attached (figure 5.3.D). Next, scientists grow a culture of bacterial cells that contain plasmids. This plasmid DNA is purified and cut with the restriction enzyme that will produce the same complementary or “sticky” ends that were put on the piece of the insulin gene (figure 5.3.E). The engineered insulin gene is then mixed with the cut plasmid DNA. Because the cut plasmid molecules and the engineered insulin gene possess the same complementary sequences at their ends, these two DNA pieces will stick together (figure 5.3.F). Finally, the sugar-phosphate backbones of the two DNA molecules are joined together with an enzyme known as DNA ligase. The resultant piece of DNA is referred to as a “recombinant DNA molecule” because two pieces of DNA from different sources are combined.
A process similar to making a gene from a known amino acid sequence, the product of which we shall refer to as “synthetic genes,” is used for other relatively small proteins of medical interest. It is also used to make the Bt toxin gene used in genetically modified plants, which will be discussed in chapter 6. However, other proteins of medical interest are far too large to use this procedure. For those proteins, a more arduous and complex cloning procedure is necessary.
Once the above operations are finished, it is necessary to make many copies of the pieces of recombinant DNA. Copying uses the natural copying mechanism of live bacteria by introducing the recombinant DNA molecule that was made in the test tube into bacterial cells, generally Escherichia coli (E. coli). When treated with appropriate chemicals, E. coli cells can take up DNA molecules in their growth medium. Once inside the bacterial cells, the recombinant DNA molecules can be copied, just like plasmids naturally found in E. coli. In fact, the transformed E. coli cells do not “know” that the plasmids now contain human DNA, and they replicate it along with the plasmid sequences to which they are joined. This is why plasmids used in cloning are called vectors; they are used to ferry pieces of foreign DNA into bacterial cells.
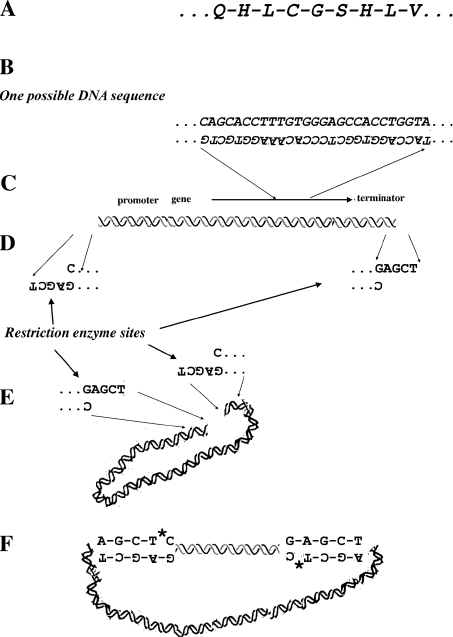
Figure 5.3 Making Human Insulin Using Recombinant DNA. A. Partial amino acid sequence of human insulin. B. The DNA sequence corresponding to the amino acid sequence shown in A. C. Promoter and terminator suitable for high-level transcription in E. coli are added. D. The insulin gene with the promoter and terminator is flanked by restriction enzyme sites that correspond to the restriction enzyme used to cut the plasmid. E. Plasmid DNA is cut with the restriction enzyme. F. Pieces of DNA with the insulin gene and the cut plasmid DNA join spontaneously at their complementary ends. Finally, an enzyme is used to glue the sugar-phosphate backbones of the two DNA pieces. The * indicates where the ligase enzyme glues the sugar phosphate backbone on the recombinant DNA. This process results in a recombinant DNA molecule containing the human-insulin gene in a bacterial-plasmid DNA. The engineered human-insulin gene is shown as the straight, short double helix, and plasmid DNA is shown as a curved double helix.
As we just saw, E. coli cells can make hundreds of copies of recombinant plasmids containing a human gene. This means that E. coli in the laboratory contains hundreds of copies of the human gene you want to express, or, in other words, the dosage of human genes in these bacterial cells is very high. With the appropriate promoter and terminator, these human-gene copies are transcribed at high level (many mRNA molecules are made), and consequently, translation of these mRNAs will generate large amounts of proteins corresponding to that gene. This is possible thanks to the fact that the genetic code is universal. In fact, not only does E. coli not “know” that it harbors and actively replicates a piece of human DNA, it is also “unaware” that it is making large amounts of human mRNA and large amounts of the corresponding human protein. Today, large vats, called fermenters, are used to grow engineered E. coli on an industrial scale. The protein of interest is purified from these cells and sold by pharmaceutical companies.
Human proteins manufactured in E. coli cells include, among others, insulin, erythropoietin, blood-clotting factors, somatotropin, osteogenic protein, and tPA, or tissue plasminogen activator. Insulin is used to assist individuals who cannot make their own insulin, people suffering from type I diabetes. Erythropoietin is a protein factor that plays a key role in the production of red blood cells. It is used to treat anemia, in particular anemia that may follow cancer chemotherapy. It is also a drug abused by some athletes to increase their amount of red blood cells. Blood-clotting factors are administered to patients suffering from hemophilia, a disease that prevents blood from clotting properly. The osteogenic protein can be used to assist in bone healing. Somatotropin, also called growth hormone, is administered to young people of short stature who do not make enough of this hormone. Finally, tissue plasminogen activator is frequently used in the emergency rooms of hospitals to treat patients who have just suffered a severe heart attack. At least a dozen pharmaceutical companies manufacture these and other protein factors, representing a market of several billion dollars (table 5.1).
Table 5.1 Examples of Recombinant Products on the Market
| |
| Product |
Purpose |
| |
| DNase |
Treating cystic fibrosis |
| Erythropoietin (EPO) |
Promotion of red-blood-cell growth, including for patients undergoing chemotherapy |
| Blood-clotting factors |
|
| Factor VII, VIII, IX |
Clotting factors for people with hemophilia |
| G-CSF |
Reduction of infection in cancer patients |
| |
receiving anticancer drugs |
| Glucagon |
Treating severe hypoglycemia |
| Glucocerebrosidase |
Long term enzyme-replacement therapy for type I |
| |
Gaucher disease |
| IL-2 (interleukin-2) |
Treating metastatic cancers |
| Insulin |
Managing diabetes |
| Interferons |
Treating chronic hepatitis C and multiple sclerosis |
| Interleukin |
Treating deficiency in blood-clotting cells after chemotherapy |
| Natriuretic peptide |
Assisting heart function in patients with congestive heart failure |
| Osteogenic protein |
Assisting in bone healing |
| Somatotropin |
Promotion of growth for those who normally lack this hormone |
| tPA |
Treating acute myocardial infarctions |
| |
What is the advantage of making human protein products in E. coli? Let us take the example of insulin. Before the advent of recombinant DNA technology, diabetics had to purchase insulin made from pig pancreases collected from the slaughterhouse. Pig insulin is not very antigenic; that is, this protein does not trigger a quick immunological response in humans. However, after years of daily use, a percentage of patients started developing immune reactions toward pig insulin. They then had to switch to sheep insulin, for example. These inconveniences are now a thing of the past; human insulin made in E. coli does not trigger an immunological response in diabetics because it is genuine human insulin. In addition, human insulin made in bacteria can never be contaminated with animal viruses because these viruses cannot be propagated in bacteria.
A number of blood-clotting factors provide further examples of human products made by using recombinant DNA technology. The clotting process of your blood involves a cascade of events involving different factors. Each factor is encoded by a separate gene. The hemophilia in Queen Victoria’s family is due to a dysfunctional gene located on the X chromosome. There is a second gene on the X chromosome that encodes another clotting factor, and there is a third gene, not on the X chromosome, that encodes yet another clotting factor. These factors are now available for hemophiliac patients via recombinant DNA technology as injectable solutions. Before the advent of recombinant technology, hemophilia patients relied upon plasma from donated blood. Factors from donated blood have the potential of containing dangerous viruses such as HIV or hepatitis. In fact until clotting factors were available via the recombinant DNA technology, 90 percent of adult hemophilia patients were infected with either HIV or hepatitis C.
Finally, other types of cells in addition to bacteria can be used to produce recombinant human proteins of medical importance. Yeast cells, normally used to make bread, beer, or wine, do express human proteins when properly engineered with recombinant human genes. Mammalian cells, including human cells, grown in a culture medium are also used to make recombinant human protein.
The scientists who discovered key tools for recombinant DNA technology, such as restriction enzymes and plasmids, did not know that their work would lead to such important practical applications. This is a beautiful example of the power of science: most of the time, basic discoveries are made without any particular practical goal in mind. However, some basic discoveries can be put to practical use in a very short period of time with important results, in particular for the field of human medicine.
We have seen that the recombinant DNA technology now makes available significant amounts of many human proteins that were not available before the development of gene cloning. Gene cloning requires the use of restriction enzymes and plasmid vectors. Restriction enzymes cut DNA pieces at specific palindromic sequences leaving short, single-stranded, complementary or “sticky” ends. Plasmid vectors are small circular pieces of DNA found in bacteria that can be used to shuttle DNA pieces into bacteria. Genes to be put into bacteria must have appropriate promoters and terminators so that the bacterial cells that are transformed by the recombinant DNA can make many copies of the gene and the protein that it encodes.



