“There is no beginning, no middle, no end, no suspense, no moral, no causes, no effects. What we love in our books are the depths of many marvelous moments seen all at one time.”
Kurt Vonnegut, Slaughterhouse-Five
Vonnegut was half right. There is definitely no beginning, and if there is an end, it’s not in sight. We are always in the middle, and we are all missing links. Just like there was no absolute point when your life began, there was no moment of creation when our species began, no spark of life, no breath of God into the nostrils of an Adam molded in the red earth, no cracking of a cosmic egg. So it goes. Nothing living is fixed, and all creatures are four dimensional, existing in space, and also through time.
Life is transition: The only things that are truly static are already dead. Your parents had parents, and theirs had parents, and so on, two by two, back through the whole of history, and prehistory. If you keep going back and back, your ancestors will slowly and inevitably become unrecognizable to you, via apes and monkeys, two-legged then quadrupedal, and ratty mammals and brutish beasts on land, and before them in wading sea creatures and fishy swimmers, and worms and weedy sea plants, and around two billion years ago, you don’t even need two parents, but just the binary fission of a single cell, one becomes two. Eventually, at the beginning of life on Earth around 4 billion years ago, you’re locked in a rock at the bottom of the oceans, inside the hot bubbling tumult of a hydrothermal vent. This geologically slow, incremental change is like a color chart, where pixel-by-pixel white becomes black, whether it’s the gap from reptile to mammal, or from four-legged to upright. On occasion there will be a splash of color thrown into the mix, but for the most part, the pathway to your ancestors creeps rather than jerks,* and all of it gray in its depths.
Life on Earth has been continuous in that time, and we are a dot on that gray continuum. Conjure up that image of a hairy monkey-like ape on all fours, to the left of a crouching ape, to the left of a hunched stooping ape, to the left of an upright, modern bearded man-ape like us wielding a flint-tipped spear with his right leg cocked coyly forward to protect us from seeing his immodest instruments of biological transition. This iconic image implies something that we now know is untrue. We just don’t know the pathway of the apes that led to us. We know many of the creatures en route, but the map is full of gaps and smears. The second untruth is that there is a direction to our evolution, to our bipedal gait, and our big beefy brains, and our tools and culture. With that arrow we are to infer progress, from simplicity to an inescapable advance into the erect future, an inevitable cognitive revolution of the mind.
Alas, we are no more or less evolved than any creature. Uniqueness is terribly overrated. We’re only as unique as every other species, each uniquely evolved to extract the best possible hope for our genes to be passed on into infinity given the present unique circumstances. With all the bones of evolution, and a modern understanding of evolution and genetics, it’s impossible to conceive of a twenty-step progress of apes from the left to the right, let alone those neat discrete jumps in five moves. There is no measure of the progress of evolution, and the language we once used, where species were “higher” or “lower,” no longer carries any meaning for science.
Charles Darwin used those words,* as was the style of his time, when he outlined the mechanism for the origin of species in 1859. We had scant evidence for other upright apes then, with or without their spears. He had no mechanism for how that modification was passed from generation to generation. Since the end of the nineteenth century we’ve known the patterns by which characteristics are passed from parent to child. In the 1940s we discovered that DNA was the molecule that transmitted that information down the generations. Since 1953, we’ve known that the double helix is how DNA is built, giving it the impressive ability to copy itself and allow those copies to build cells just like the ones they came from. And since the 1960s we’ve known how DNA encodes proteins, and that all life is built of, or by, proteins. Those titans of science, Gregor Mendel, Francis Crick, James Watson, Rosalind Franklin, and Maurice Wilkins, stood on their predecessors’ and colleagues’ shoulders, and would in turn be the giants from whose shoulders all biologists would see into the future. The unraveling of these mysteries was the great science story of the twentieth century, and by the beginning of the twenty-first the principles of biology were set in place. In cracking the universal genetic code, and unwinding the double helix, we had unveiled a set of simple rules of life. Yet they turned out to be profoundly complex, as we will soon see.
But Darwin didn’t know any of that. When he published his second great work, The Descent of Man, in 1871, his primary concern was the question of
whether man, like every other species, is descended from some pre-existing form . . .
Then, just a handful of Neanderthal remains were known: a skull from Belgium, another from Gibraltar, and a bag of bones from central Germany. As early as 1837, Darwin had sketched out a visionary version of an evolutionary tree in a notebook, showing how one branch of life became two and more, selected by nature in response to the changing environment. How these ancient apes fitted onto the human tree was entirely unknown.
“I think,” he scrawled at the top of the page in that notebook, in his inimitably dreadful handwriting, but never finished that thought. What was set in motion in the nineteenth century was the idea that, alongside all animals, we were part of a continuum—a species begotten not created. Nowadays, only the willfully ignorant dismiss the truth that we evolved from earlier ancestors. The images of gigglemug skulls of our long-dead forebears are commonplace, and they become front-page news when a new species is claimed. Dozens of lines of evidence bellow incontrovertibly that we are an ape, with an ape ancestor common to chimpanzees, bonobos, gorillas, and orangutans.
Sometimes people say, as a way of revealing the paucity of the fossil record, that all the specimens of ancient human evolution could be placed on a large table or in a single coffin. That’s not true either. We have literally thousands of ancient, hardened bones, found all around the world; many in the nursery of the human story in eastern Africa, many in Europe, and the more we look, the more we find. For Darwin, though, we were effectively alone at the end of a mysterious branch on our family tree.
But for all the sheer grit of the diggers who devote their lives to sitting in dugout caves or dusty ancient riverbeds armed with toothbrushes and tiny picks, there are not nearly enough physical specimens to reveal anything resembling a complete picture when it comes to human evolution; there are individual fossils arranged into groups according to shared characteristics such as the shapes of their brows, the arch of an instep, the cusps on their molars. These were dated according to where they were found, in which layer in the ground, and what other things are found nearby—tools, the shadows of cooking, or traces of hunting.
Or if they’re young enough, by the ratio of radioactive carbon atoms that, instead of being replenished via living metabolism, in death are slowly ticking down at a regular rate. It’s all good, robust science, contentious as research often is, and frequently fractious, but the analysis of old bones is precise, complex, and highly sophisticated. In the 200 years since the first other human species was discovered, our understanding of how we came to be has undoubtedly increased immeasurably, but our confidence in that path has changed, and continues to evolve. For decades that image of the progress of monkey-ape to ape-man to man-ape has been on display in museums around the world, and in textbooks, a nice line of clear evolution that says “this is how we got here.” In Down House, in the English county of Kent, where Darwin beetled away, meticulously drawing up the best idea anyone ever had, you can still buy coffee mugs with that image on it.
When I was young and falling in love with science in the 1980s, the evolutionary trees looked just like that. My father would collect articles for me from New Scientist or Scientific American showing neat branching diagrams suggesting that one species morphed into another, or one becoming two, with the other gruff ape-men perishing along the way. The picture seemed clearer, the fewer specimens we had. By the end of the twentieth century, more and more human species and specimens had risen from their graves, different enough to blur those nice clean lines, and the branches got fatter, less distinct and more pollarded.
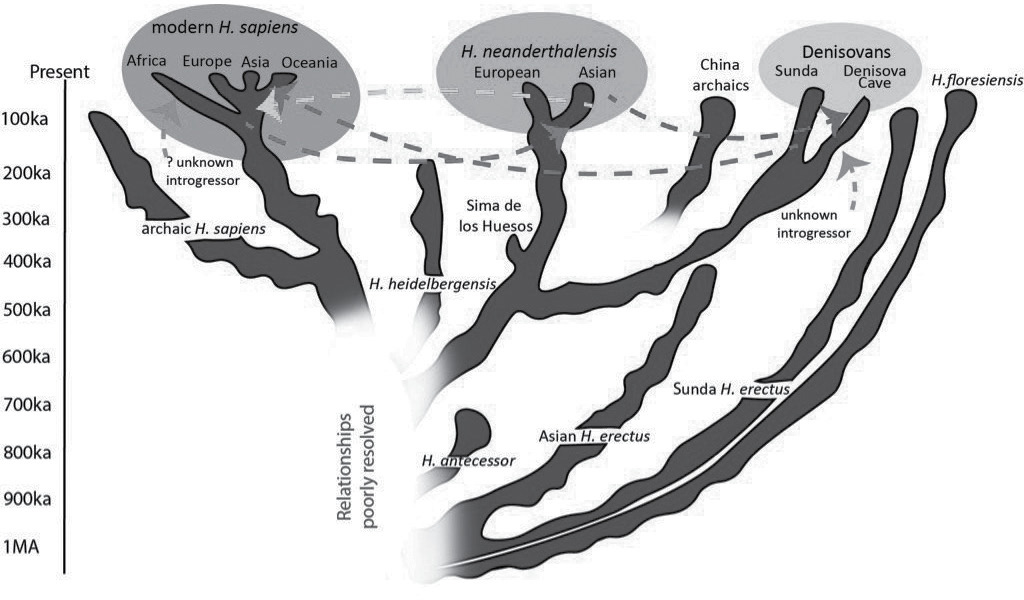
Maybe it’s time for us to retire the long-serving metaphor of the evolutionary tree of life, and certainly the picture of apes to ape-men to man-apes. Today, you’d be hard pushed to call it a bush, shrub, or anything arboreal at all. Instead, it is represented in graphical form as more of a set of upside-down dribbly blobs running upward into the pool that is us, streams, rivers, and rivulets, some running into the ocean, others petering out en route (see the evolutionary shrub of mankind). An alternative version is to place the specimens in their species clusters on a chart, oldest at the bottom, us the sole survivors at the top, the width showing the geography of where all the bones were found, and you must accept that the lines between them are dotted, meaning hypothetical. If this was a detective story, we have the bodies, but the clues are scant and disconnected. The case is far from settled.
We’re utterly fascinated with ourselves, and with some justification. We are just another animal, but we’re the only one evolved to have scrutinized our own existence, to look in the mirror and really squint at it. Many, many books have been written about the origin of our species, but this story deals specifically with just those from which we can reconstruct the past, and past relationships, using the newest tool in the paleoanthropologist’s shed, and that is DNA. That molecule has revolutionized much of our understanding of human history in unprecedented ways, all in the space of the first few years of this century. It’s a field changing so rapidly that researchers have told me of their reluctance to publish new findings for fear of them being superseded not within years or months, but within weeks or even days. Keeping track is not an easy task, as the study of human evolution is mutating into an unremitting revolution. The picture of how we humans came to be what we are is more detailed than ever before, and we still have a long way to go. Before we get to all that, here is a brief, scant overview of the story so far. Let us begin not at the beginning, because there was no beginning, but, somewhat arbitrarily, with two feet.

The murky evolutionary shrub of humankind.
Old bones, combined with new analysis of old DNA, has meant that what was once a confident branching tree has been pollarded and pruned and replanted as an unrooted bush. The broad blobs represent individual human species, and the dotted lines are the flow of genes via sex between them. The more we learn, the messier the picture becomes.
Bipedal apes walked the earth at least 4 million years ago. In fact, all apes are capable of two-footed movement, but what concerns us is habitual bipedalism—walking as a primary means of travel. Standing upright was a quintessential step in our own evolution, as it prompted and coincided with a number of anatomical changes, such as the position and shape of the spine, how it connects to the skull, and so on. Why this happened is not agreed upon, and there are plenty of theories: Some focus on an increased efficiency of movement in being upright; others on its being an adaptation to suit a life on the savannah rather than swinging in the trees, or the changing climate of the Rift Valley. The most famous of these early walkers is Lucy, born around 3.2 million years ago. Forty percent of her fossilized skeleton (which is a lot to be preserved for remains that old) was discovered in 1974 by Donald Johanson, and named after the Beatles song “Lucy in the Sky with Diamonds” that was playing back at the researcher’s base camp in the Awash Valley in Ethiopia that heady night. Lucy was one of the first members of the species Australopithecus afarensis discovered. We cannot say whether her species was a direct ancestor of us. What we can say is that there were many other primates living at this time, and she looks more closely related to us than any other.
The classification of animals is also a frequently unsatisfactory business, but to tell the story of our species we need to dive in and hope for the best. The system that we exclusively use was invented in the eighteenth century by the Swedish naturalist Carl Linnaeus, and gives creatures two Latin names: a genus, and a species.* An English oak tree is a Quercus robur. There’s a wasp called Lalapa lusa, and a Fijian snail called Ba humbugi. Enema pan is the rhinoceros beetle. The common toad is Bufo bufo, which might seem a bit lazy as it’s Latin for “toad toad,” but lots of common animals have this form, including the mollusc Extra extra, and our fellow great apes, Gorilla gorilla.* You may well have a Felis catus at home, which at least comprises two different words for “cat.”
Lucy’s type, Australopithecus afarensis roughly translates as “southern apelike thing from Afar.” And there are other species of southern apelike things—sediba, anamensis, and africanus. Earlier apes are slotted into genus categories with names like Sivapithecus (Shiva’s ape, having been found in India), and Ardipithecus (ground ape), and Gigantopithecus (really big ape).
We are genus: Homo; species: sapiens—Homo sapiens: the wise man. That’s the short version. There’s an equivalent in biology to when children write their addresses from street to town to country to continent, hemisphere, solar system, and galaxy. Several classification ranks sit above genus and species to place us precisely in the living universe:
Domain: Eukaryota (complex life)
Phylum: Chordata (animals with a central column, akin to a backbone)
Class: Mammalia (milk producing)
Order: Primates (monkeys, apes, tarsiers, and a few more)
Suborder: Haplorhini (dry-nosed apes)
Family: Hominidae (great apes)
Everything in this book from here on in will be Homo. Neanderthals are classified as Homo neanderthalensis—the humans from the Neander valley, in Germany; Homo habilis—the handy man.
You belong to a strangely exclusive club. Membership of a genus doesn’t necessarily show relatedness between the members, but instead shows that members are more similar to each other than they are to organisms not in that genus. This is the best system we have. Species is a definition also riven with problems, but the most accepted form is that two species are defined as distinct when they are incapable of producing fertile offspring together. Zebroids, ligers, mules, hinnies, grolar bears* are all relatively rare, relatively healthy hybrids. But none of them produce fertile offspring of their own. Soon, we shall see why this species definition for humans is not at all adequate.
The current convention is to list around seven species that fit into the genus Homo, and I will be referring to them as human. This is not uncontroversial, but one of the key problems of taxonomy is that in trying to name things, we are attempting to describe how things are, and this doesn’t necessarily acknowledge the essential temporal nature of life, that evolution is universal, and change over time is the norm. Remember that the subject of evolutionary change is DNA, but classification does not depend upon that.
For the time being, though, let us think of species as distinct groups of animals, who are different enough to be incapable of producing fertile offspring together, and in the genus Homo, there have been at least seven.* Ones for whom we have fossil remains from the period starting a million years ago can be called archaic humans, and there are a few. Homos ergaster, heidelbergensis, antecessor, and a few more are all present in different places, and with subtly different anatomical details during this period, and they are all thought to have evolved from earlier Homo erectus, the upright human. They did an excellent job of populating the world, but we don’t believe so far that they left any DNA for us to recover, and here we are tracking the past with DNA. Most of the others have also not yielded any DNA samples (yet), probably because they are too old, or died in places too hot for it to endure, and so our understanding of our relationship to them is limited to fossils and paleoarchaeology.
The ground of human evolution trembled with the discovery of a tiny woman on the Indonesian island of Flores in 2003. The skeletal remains of a meter-high female, and parts of at least eight other people were unearthed in a cave called Liang Bua. Immediately these miniature humans, classified as Homo floresiensis, were referred to as Hobbits, and though their feet are large, there was no evidence of them being hairy. They appear to have lived in this humid cave as recently as 13,000 years ago, which is only a few centuries before the beginning of farming. These tiny people cooked with fire, and probably butchered meat of giant rats and stegodons (a species of tiny elephant) that lived alongside them.
Who were these small humans? The initial reports in the journal Nature were clear that their bodies were similar enough to be included in the genus Homo, but different enough to warrant being a separate species from any other known Homo. A vocal minority of scientists criticized this position, and asserted that they were actually like us, but diseased and shrunken through some speculative pathology. Down syndrome, microcephaly, Laron syndrome, and endemic cretinism have all been suggested, but the evidence for these is slight and dicey. Populations on islands often evolve to be very small or big, as the forces of selection might be limited and specific to insular isolation, and indeed the Hobbits shared their island with rodents of unusual size, tiny hippos, and dwarf elephants. All are now extinct, but it seems most likely that Homo floresiensis was a separate species of human, probably sharing a common ancestor with us at some point within the last 2 million years, but had shrunk in size due to the pressures of a tropical island life.
But we couldn’t get DNA out of the remains of these midgets. The bones were not fossilized, and were soft, like wet cardboard. An attempt to extract DNA was made in 2009, using a tooth, which is hard on the outside, and so offers some protection from the ages. It failed, and their DNA is lost in time, like tears in rain. Maybe the heat and humidity of the tropics over a few millennia were enough to annihilate all of the DNA that might have been skulking in those teeth and decaying bones. It is a shame that this is the case, as there have been heated arguments about the provenance of these people, and DNA would’ve solved them in a heartbeat. Their island status, the position and limitations of their range, and their physical characteristics suggest that the Hobbits of Flores were not ancestors of ours but distant cousins. Nevertheless, the number of human species who lived into the past 50,000 years had suddenly gone from two to three, and deservedly the Hobbit and its freakish giant and dwarf island cohabitants were famous. Overnight, our planet started to look a bit more like Middle Earth.*,†
And it was going to get more so as DNA reading technology matured. For more than a hundred years, the study of human evolution had been dominated by bones, and a few tools—anatomy and culture. Reading DNA is really a form of anatomy on a molecular scale; it contains clues to how bones are shaped, and how evolution has shaped them. The invention of the technology to read DNA was primarily born of a desire to understand disease, but it was clear that decoding genomes would illuminate human history too.
Here is a short interlude about how we learned to read DNA. In 1997, in the world of living human genetics, the largest scientific project in history was going full steam ahead. Hundreds of scientists—some former competitors—had effectively teamed up with a common goal: to provide the world with a complete readout of every single letter of DNA in a human being, all 3 billion of them. The tale of the Human Genome Project is told in Chapter 6, but for this story the most important thing is that this was a technological grand scheme designed to make reading the letters of DNA easy and cheap. In doing so, medicine, evolution, and the mysteries of being human would be revolutionized. The ability to read DNA was pioneered by the unassuming English genius Fred Sanger in the late 1970s, using a process that copied the original sequence millions of times. To do that, your ingredients need to include the alphabet you’re writing in; DNA consists of only four letters, more formally called nucleotide bases—A, T, C, and G. You also need an enzyme whose job it is simply to copy and link up the bases of DNA, called a polymerase. Throw all these ingredients into a tube and set the temperature right, and the double helix will separate into single strands, which serve as templates to replace the letters that would form the missing strand. You end up with millions of copies of the original template. Each one of the letters of DNA physically links to the one that precedes it and the one that follows, whereas English periods halt any sentence. The polymerase molecule trundles along adding the next letter one at a time like a typewriter copying a line of text. In DNA sequencing, you add not just the correct letter molecules, but also a few that act as periods.
Because so many copies are made during this process, and because the periods are added randomly, what you end up with is a muddle of DNA molecules that stop at
e
ev
eve
ever
every
every s
every si
every sin
every sing
every singl
every single
every single l
every single le
every single let
every single lett
every single lette
every single letter
every single letter.
Sequencing DNA is reconstruction. You make millions of copies that are fragmented at every letter. You then order them by size. DNA is a molecule that carries a negative electrical charge, and this means that if you stick it in some salty water, and put a voltage across that water, the DNA will head toward the positive electrode. The speed at which it migrates is determined by its mass, which is determined by how long that fragment is—a large piece will move more slowly than a small one. So, if instead of putting it in water, you put the DNA in a jellylike gel to slow it down, and run an electric charge across the gel, then the DNA will separate very precisely according to size, just like sieving dirt.
There’s one more trick to this technique. There are only four letters of DNA, unlike the English alphabet’s cumbersome twenty-six. So, take your original gene, and separate it into four tubes. In each of the tubes you add all the ingredients, but in the first you also add some A bases that halt the chain, ones that add the period, but only when there is an A on the template. In the second, you include everything as well as chain-terminating C bases, and so on in tubes three and four, Ts and Gs. After the reactions are complete you have one tube that contains every fragment of DNA that ends with an A, in the second that ends with a C, the third with a T, and the fourth with a G. If you stick these four solutions in four columns on a gel and apply the current, they will be drawn out and separated, and every position of every letter revealed:
Your A column will look like this (though the letters would merely be smudges on the gel):
*****AA**A*****A******A*A***A*
And the T column:
**T*T**T**T*T*T*T*T*T****T*T**
The C column:
C**C****C**C*********C*C**C**C
And G:
*G***********G***G*G**********
If you overlay these four together the asterisks become letters, and you get a complete read:
CGTCTAATCATCTGTATGTGTCACATCTAC
When you see TV scientists holding up X-ray sheets covered in dotted black lines in neat columns, that is what they’re looking at. It’s a sequence of the letters of DNA that sit inside a cell in your body, unreadable for 4 billion years, but now rendered so commonplace that it can be done in minutes for a few dollars. It’s an incredibly clever way of reading DNA, and Fred Sanger quite rightly picked up his second Nobel Prize for Chemistry* for inventing it.
During the 1990s, with the Human Genome Project’s aim being to sequence 3 billion letters, Sanger’s technique was evolved, improved, and automated. There is an account in Chapter 6 as to why this was still a gargantuan task that took years and billions of dollars to complete. When I was a student in the 1990s, I would send off purified samples of short lengths of DNA to a specialist sequencing department and await the results (not on nicely photogenic X-rays, but in computer files) for a few days. Now, most genetics labs have their own sequencers and they churn out megabytes of data in hours. New techniques have been invented that have not completely replaced Sanger sequencing, but are even quicker and cheaper, and were you to begin a career as a geneticist today you would probably never employ this technique. We even have sequencers smaller than a deck of cards that will plug directly into your laptop via a USB port, so they can be taken out into the field to sequence the genomes of animals and plants in the wild. All these technologies are fueling the revolution in genetics for everyone alive. Since the turn of the century, we have been able to do the same for people who have been deceased for quite some time.
In a hole in the ground, a man lay extremely dead. He was either left in this tomb by his family or perished right there, with no idea that he was one of the more important people in millions of years. Posthumously—very posthumously—this man did two things: The first is that his emergence out of that cave kick-started the study of ancient humans. It had been his home, we presume, in what we now call Germany, around 40,000 years ago. Kleine Feldhofer Grotte is no longer there; it was discovered but destroyed by quarry miners in the nineteenth century. The entrance stood a few meters above the valley floor, a man-sized squeeze into a rocky room around three by five meters, with a high ceiling. Of the things pulled out by amateur sleuths in the 1850s, and subsequently in excavations of the buried site this century, thousands of artifacts have been found, and the remains of at least three people. Fossilized bones were found in 1856 by the quarry miners—a drink-coaster sized chunk of skull, two femurs, more arm bones than one person requires, and fragments of a shoulder blade and ribs—who handed them over to a local anthropologist.
The remains of this man were not the first (he was probably the third non–Homo sapiens human skeleton to have been discovered), but he became what is known as the “type specimen”—the one that defines the species, against which all are subsequently compared. The name of the species is formally attached to the type specimen, so whatever his name was in life, as far as we are concerned, he became known as “Neanderthal 1.” With the formal identification of this man, the field of paleoanthropology—the study of ancient humans—truly began.
But they wouldn’t let him lie. The second revolutionary thing Neanderthal 1 did occurred another 150 years later. He volunteered his DNA. Up in that cold cave his remains were protected, to a degree, from the elements, from hungry animals, and most significantly from voracious bacteria, all of which would happily eventually destroy all the evidence that he once had lived. Instead, due to his unusual domicile, his bones were left as untouched as someone lying dead for 40,000 years can be, and that meant that he became the first non–Homo sapiens human to enter what was, in 1997, a very exclusive club. Tucked inside the slowly decaying cells of what was probably his throwing arm, were the molecules that faithfully carry ancestry from the past into the future.
We modern humans weren’t the only ones in the Human Genome Project. Somewhat counterintuitively, there were six species included in the project’s primary aims. A genome is much more useful if it can be compared to another, and that includes genomes from other species. So, the original mission of the first creatures to join the genome club aside from us included the most commonly used model organisms—the fly Drosophila melanogaster; the rat and the mouse; our closest ape relative, the chimp; and an oddity, the honey bee, for it is a social beast, and almost all members don’t get to reproduce at all, but serve their queen with whom they share exactly half their DNA. All of these were due to have their entire genomes read, deciphered, and interrogated over the final years of the twentieth century.
In 1997, using precisely the same techniques being developed for living humans, a Swedish researcher working in Leipzig quietly laid the foundation stones for a new, utterly revolutionary field—paleogenetics. Svante Pääbo had borrowed the right humerus, the bone between the elbow and the shoulder, of Neanderthal 1 from Rheinisches Landesmuseum Bonn. With a precision saw, he sliced an inch-long segment out of the middle, exposing what was once soft vibrant marrow where blood and immune cells would have once been thriving. Bone marrow is the site of a panoply of new cell generation, and that means they are dividing energetically, which means replicating their genetic material boisterously. There lay the first treasure trove of Neanderthal DNA.
DNA is universal in all living species. It’s packaged up in different ways, a language organized into books, chapters, origami, and pamphlets. And passed around the generations in different ways too. In animals, DNA is bundled up into chromosomes, huge chunks of double helix, wrapped around itself, and wrapped around little lumpy proteins, which again spiral tight again until they look like those iconic X shapes that we see in textbooks. In most cells, you carry two complete sets of chromosomes, one set inherited from your mother, one from your father, twenty-three pairs in total all neatly stored in the nucleus, the little nut in the center of cells.
Biology is a science of exceptions and endless qualifications, and twenty-two of those pairs are the same as each other (called autosomes), and one of those pairs is not a pair. The pair that is not a pair are the sex chromosomes. I have a Y and an X, whereas women have two Xs. Women get one X from each parent, but men only get their Y chromosomes from their fathers. But, though the Y is important for determining maleness, it’s a weedy piece of DNA compared to the others, and makes up very little of the total amount of DNA. The X is the second biggest of all the human chromosomes.
There’s another exception to the way DNA is passed from parent to child. All the autosomes and the sex chromosomes never leave the nucleus, a bound space in the center of most cells. But there’s a minuscule but terribly important bit of DNA that is not in the nucleus, but instead sits inside the mitochondria—the tiny but powerful energy generation units that all complex life relies on. They were almost certainly acquired around 2 billion years ago when two single-celled organisms fused for mutual benefit. What that meant was that these new cells formed a new branch of life—the eukaryotes—different from everything that had come before, which were small single-celled beings, either bacteria or archaea. These three groups are called domains, and are at the very top of the hierarchy of living things, above the five kingdoms. The three domains are Bacteria, Archaea, and Eukaryota, which is basically everything that is not in the first two categories. The eukaryotes carry inside them a tiny amount of very important DNA that is not stored in the nucleus but bound inside these subcellular power stations. In contrast to the scraggy Y chromosome, mitochondrial DNA (mtDNA) is only passed from mother to child. The sperm swims along with only half the genetic information to make a new person—twenty-two chromosomes and an X (if that child is to be a woman) or a Y—and wheedles its way into the egg, which also carries twenty-two chromosomes and an X, and also the mtDNA of the mother.
Almost all—more than 97 percent—of your DNA is carried in the twenty-two pairs of autosomes and the X, and all this genetic information is inherited from both parents in a roughly equal manner. Each of the autosomes is a unique combination of the pair of chromosomes that your mother or father inherited from their mother and father. When a sperm or egg is being forged in your father’s testes or in the ovaries of your mother* the two matching chromosomes line up and shuffle. Imagine lining up two suits of cards, hearts and clubs, in a row, and then swapping some of the same numbers with each other. The result would be two complete suits, all the right numbers in the right order, but a mix of hearts and clubs. This is what the chromosomes do in making sex cells. But instead of ace to king each chromosome has millions of possible swaps. So the result is, for each of the twenty-two autosomes, a new combination. It is this process, called “recombination,” that guarantees that your precise genetic makeup is unique to you, for all time.
Mitochondria and the Y don’t do this though. One comes from your mother, which came from her mother, and from her mother, and all the way back only on your maternal side, and the Y exactly the same on your paternal side. For those in pursuit of ancestry, these make for interesting tools, and ones that have been the focus of many studies, not least because they have been historically the easiest, smallest, and therefore the first chunks of DNA available for ancestral scrutiny. Mitochondria exist in their millions in the busy milieu inside cells, and so the chances of their surviving the onslaught of time is higher. The autosomes and sex chromosomes exist as one complete set kept exclusively inside the nucleus—the cell’s central office. So compared to the nuclear DNA, there’s stacks of it, millions of identical copies all much more readily available for analysis. Both mtDNA and Y will make frequent appearances in these pages; not just because they are informative, but also because their value is sometimes overstated in the hunt for ancestry.
The Neanderthals were a people who lived all over western Europe from the easternmost tip of Spain, to the caves of north Wales, into the mountains of central Asia, and as far south as Israel. The oldest true Neanderthal bones we’ve found are 300,000 years old, and we haven’t discovered any younger than 30,000. That is a reasonable longevity for a human species. Homo erectus, an earlier upright ape, spread all over the world from an exodus out of Africa that began 1.9 million years ago. But the Neanderthals still clocked up a longer run than we have so far. We anatomically modern humans are generally thought to have evolved primarily in eastern Africa around 200,000 years ago, and emerged out of Africa in our own exodus sometime in the last 100,000 years. This number inches up every few years, as more specimens are found. A discovery in October 2015, from the Fuyan cave in the Daoxian region in southern China, dug up forty-seven modern teeth at least 80,000 years old, and it’s not unreasonable to presume that the owners of those teeth took some tens of thousands of years to get that far east from the motherland.
According to traditional paleoanthropology based on bones, by the time Homo sapiens reached Europe, probably around 60,000 years ago, the Neanderthals were already there and well established, albeit in small communities. But with DNA as evidence, these dates are due for serious revision, as we shall see later in this chapter.
Nevertheless, Neanderthal anatomy clearly shows that they were visibly different from the interlopers. Brain capacity is one of the key measures in paleoanthropology, and the Neanderthals had bigger ones than us; a modern man’s averages about 1.4 liters by volume, women’s being a little smaller. The Neanderthals range between 1.2 and 1.7 liters. That cranial capacity is not something we can specifically correlate with intellectual capabilities, though in general, in apes, bigger brains mean more sophistication.
They were shorter and stockier than us, thicker set, barrel chested, with broader noses and clunkier brows. It’s surely for these simple physical reasons that they have a pretty bad rep. In common parlance they are synonymous with and stigmatized as brutish cavemen, grunting oafs, and neanderthal acts as a byword for lowbrow dimwit thuggery. When the classification of the early specimens was being wrangled in the nineteenth century, the great German biologist Ernst Haeckel suggested one specimen be known as Homo stupidus.
Nothing indicates that the Neanderthals were anything of the sort, nor significantly different from their Homo sapiens contemporaries. They hunted, butchered, and cooked large prey. In the last 100,000 years there is some evidence that they sewed, made clothes and jewelry, and these artifacts predate the arrival of anatomically modern humans, meaning that the Neanderthals developed these skills themselves, rather than learning them from the new kids on the block. Recently researchers have claimed that hand paintings in a cave in Nerja on the pebbly Mediterranean coast of Spain were theirs and not ours. Some have argued that the presence of pollen in graves in Shanidar in Iraq and in southern France* were traces from flowers left there as part of ritual burials, though this remains controversial.
Because of the paucity of remains, the evolutionary relationship between Homo neanderthalensis and Homo sapiens has long been disputed. The full gamut of suggestions has been made over the years, from their being the direct ancestors of modern Europeans, to their existence on a completely different bough of the evolutionary tree, who left no extant descendants. The last common ancestor of us and them is thought to have existed around 600,000 years before today.
Svante Pääbo’s digging within Neanderthal 1’s arm bone was the first step in answering this. They extracted 0.4g of matter—the weight of a decent pinch of salt—from the section of precision-butchered bone, and from it pulled fragments of mtDNA. This was, in 1997, the most ancient DNA yet recovered. Much of that first study was devoted to showing that it was possible, and that the DNA extracted was not contamination.
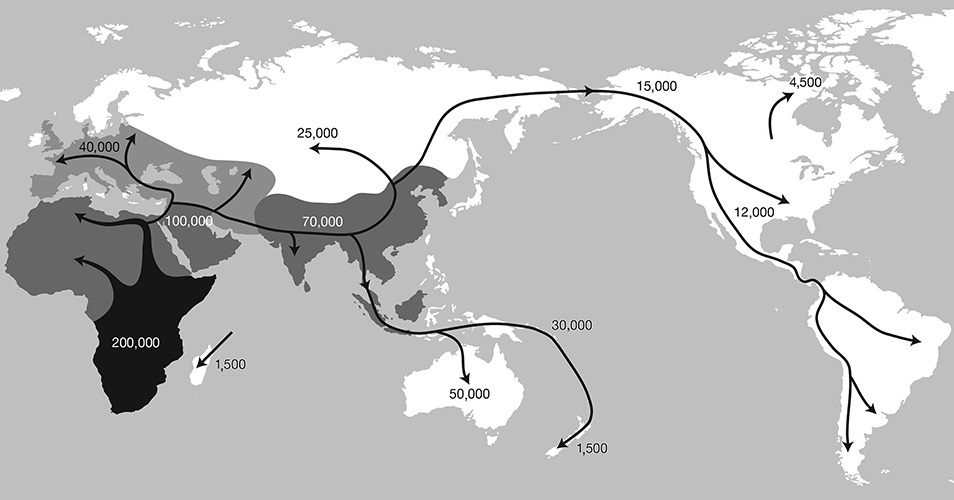
 The migration of Homo sapiens out of Africa.
The migration of Homo sapiens out of Africa.
Anatomically modern humans began their tenure on Earth primarily in eastern Africa around 200,000 years ago, though more archaic Homo sapiens have been found as far away in time and space as 300,000 years ago in Morocco. Our ancestors had begun to trickle out of Africa at least 100,000 years ago. They met Neanderthals in Europe, and other human species en route, and according to our DNA, bred with many of them.
As with all ancient DNA, much of it is not human in origin. Some contamination might be the DNA of organisms that have inveigled their way into the rotting corpse looking for a feed. Some might have been the organisms—mostly harmless or even beneficial bacteria—that live in and on us and outnumber our own human cells. And some might be malicious pathogens, maybe even the thing that killed them.
Jurassic Park and its lesser sequels had become record-breaking hits in the 1990s, and the idea of DNA recovered from long-dead species was very clearly in our cultural consciousness. The reality, as ever, was rather short of the stories in the movies. The DNA was chunked up into short fragments, and all profoundly damaged, as if recovering frayed tatters from a decrepit book. These bones were only 40,000 years old, in contrast to the more than 65-million-year-old resurrected dinosaurs* of Jurassic Park, and already not in a terribly good state. The fact that Pääbo and his team got anything out is testament to the newfound skills of geneticists that were emerging in the shadow of the Human Genome Project. This was a baby step into deciphering and reconstructing the past in an entirely unprecedented way.
The first thing it said was that the DNA plucked from the Neanderthal man was different from all modern human mtDNA. The sequence of the fragments of DNA analyzed is different enough to say with some certainty that this part of their genome had separated from the lineages that led to all modern humans well before a common ancestor of all modern humans. DNA changes over time in a relatively predictable manner, like a slowly ticking clock, and so by taking two sequences that are similar but different, we can estimate how long ago they diverged. This technique is not perfect, but it has value in broad terms. In the case of this first study of Neanderthal DNA, the age of the divergence between us and them was put at between 550,000 and 690,000 years ago. These were both reassuring for the traditional forms of human evolution: Neanderthals were not us, and have not been us for an age pretty much in alignment with what the paleontology and archaeology said. The status quo had not been upended by this technological feat. In unlocking this door to the past though, over the next decade, everything would change.
The revolution accelerated alarmingly, but always the brake was the profound skill required in the process. Extracting ancient DNA is not an easy thing to do, and the volume of ancient DNA studies is testament to truly expert skill. Gene sequencing from living cells these days is easy as pie, and anyone can do it with a couple of day’s training and the right equipment: It’s the analysis, the number crunching that takes real expertise. Compared to its living equivalent, ancient DNA is a fragile wisp, and because of the delicacies involved, reading the genes of the long dead will never be normalized in such a way that anyone can do it.
But, as with the Human Genome Project, part of the deal is that when you sequence these chunks of DNA, they become public. These ancient genomes are published as databases, free for all to plunder. Geneticists don’t have to go near a fossilized bone or a dank cave nowadays to quiz the genetics of our millennia-dead ancestors. You just need the Internet. The first few extractions did require trailblazing development of new techniques in preserving and analyzing ancient DNA, because they were limited in what they could actually extract. In 2006, another team successfully pulled out DNA from a 38,000-year-old Croatian Neanderthal, and used it to answer some old questions. Two papers, with some shared authors, were published in the two top research journals, Science and Nature, in the same week, and the results were robustly similar, and subtly different. The key finding was that the sequences of DNA generated implied that the human that led to us diverged from those who led to the Neanderthals around 500,000 years ago. One hinted at what was to come: that there might have been a touch of interbreeding at a later stage, a tease of surprising sexual dalliances. The other said there was none.
Then in 2010 came the complete Neanderthal genome. Svante Pääbo’s team had radically improved their techniques for extracting DNA from ancient bones. Using dust, chips, and flakes drilled out of fossils, they assembled a full draft—rough though it was—of the complete DNA from a Neanderthal.
Let’s take a moment to revel in this. The speed of progress has been truly breathtaking. Our contemporary genomes had only been deciphered in any near-complete form in 2001 (and, as described in Chapter 6, really properly in 2003), and yet within a few years we had completed a draft genome of an extinct human species, from bones untouched for tens of thousands of years. Pääbo and his colleagues had invented a time machine.
What were they like? What genes are and what they tell us about people are very closely related, but not, in almost all cases, definitive. This is a seam that will run throughout this book, confronting and dispelling the culturally ubiquitous idea that genes are fate, and a certain type of any one gene will determine exactly what an individual is like. That this is a fallacy is universally known among geneticists, yet it is still an idea that carries a lot of cultural significance, fueled frequently by the media and an ultra-simplistic understanding of the absurd complexities of human biology. Knowing the gene sequence in an individual provides some limited information, unless they have one of a relatively small set of genes that have a very significant effect. This is discussed in much greater detail later on, but for here it is, for better or worse, the main way we can analyze the genes of the long dead.
A popular exam question for undergraduates studying paleoanthropology is “Did Neanderthals speak?” The correct answer, to be spread over 3,000 words of supporting anatomical evidence please, is that Neanderthals were capable of speech, very probably. The structure of their throats is not dissimilar to our own and, in particular, the discovery of a hyoid bone in the Kebara Cave in Israel in 1989 indicated that their capability of speech must have been similar to our own. The hyoid is a horseshoe-shaped bone where your neck meets the underside of your chin; feel it with your thumb and forefinger (where you might throttle someone) and then swallow. Its structure in humans is uniquely ridged and supported by delicately tuned muscles that connect up in all directions, to the tongue, to the mouth floor under the tongue, to the larynx, the pharynx, and the epiglottis, twelve in total. That’s a lot of very fine muscle for a bone so small, and indicates that it’s quite specially adapted for something that we do uniquely. The Kebara hyoid has been the subject of many detailed microanatomical studies because of its potential role in speech. And the answer is still the same: Neanderthals probably had the capacity for speech, like us.
From a neurological point of view, we can also make not much progress. The capacity for speech occupies a large chunk of our brains, as you would expect. Although something as complicated as speech engages many parts of the brain, the key zone is known as Broca’s area, named after the nineteenth-century French neuroanatomist who treated two patients who had lost the ability to speak after injury to this region. This scale of neuroanatomy is not that useful to us in answering the question of Neanderthal communications, as it’s a largish piece of brain, a hunk of meat that you might find in a stew. It’s there in the other great apes too, though they cannot speak. So, given that Neanderthals had larger brains on average than ourselves, we can reasonably assume that their Broca’s area was present and correct as with all great apes.
You might think the answer would spring up out of the past by using the new genetics. There is a gene, much studied and much lauded, that is inextricably bound to speech. It’s called FOXP2, and although we don’t really have a great understanding of what it does in the body, it is clear that it is essential for the type of verbal communication and dexterity that we find so trivially easy, compared to our nearest ape relatives, and indeed all other life on Earth. One of the key ways we know how genes work is to look at what happens when they go wrong. We do this deliberately in experimental animals, precisely or randomly disrupting genes to see what happens. For obvious reasons, we don’t do that in people, but the equivalent is to study the genetics of disease and disorders. Several mutations in FOXP2 have been identified since the gene was formally described in 2001 by Simon Fisher and his team at Oxford University. The identification relied on a single family (known only by the initials KE) that had been referred to Great Ormond Street Hospital for Children and its research wing (and my alma mater), the Institute of Child Health at University College London. The KE family had a heritable form of developmental verbal dyspraxia, where children present a phalanx of vowel and consonant problems. The passage of inheritance in this family, sixteen members profoundly affected, led to the identification of a region on the seventh chromosome that was different from normal, and subsequent studies showed that they had a spelling mistake in this particular gene.* Since then much has been made of FOXP2, including popular press fanfares about its being “the grammar gene” or “the language gene.” It is neither, because language and speech are complex behaviors controlled by many genetic factors, and not merely a single gene. But clearly it has a major role in how we communicate.† Furthermore, when we look at the equivalent versions of FOXP2 in other animals, we see that it plays a big part in vocalizations across the animal kingdom. Male zebra finches cannot direct their songs to females with a suppressed FoxP2, and mice pups that have had their Foxp2* deleted cannot make the ultrasonic peeps that are an essential part of communicating with their mothers.
FOXP2 is certainly essential for speech. The 2006 analysis of the Neanderthal genome revealed that they had exactly the same type of FOXP2 gene as us, and different from chimpanzees. The differences are subtle but clearly important. There’s only two changes between the protein sequence of FOXP2 in chimps and us, and we can talk and they cannot.
Can we say the same about Neanderthals? The basic answer to that undergraduate exam question “Did Neanderthals speak?” remains the same as it did when I answered it in 1995, though, if you are an undergraduate looking for an easy pass, the content of your essay today should be entirely different. Plus ça change, plus c’est la même chose. Neanderthals very probably had the capacity for speech. Until we genuinely do invent time travel, it is going to be impossible to prove.
If that story doesn’t offer a satisfying conclusion (and the study of old bones frequently does not), here’s another one of the senses for which we can offer a solid, perfect answer: smell. Before we get to a singular sweet conclusion though, here is a customary scientific disclaimer: We don’t really understand how smell works. In our noses there are cells that are triggered when particular molecules float onto them. We capture airborne whiffs with proteins called olfactory receptors, which sit astride the membrane of the neurons in our noses that wire directly to the brain. They’re very similar to the proteins in our rods and cones in our retina that trigger vision, but instead of being stimulated by photons of light, they fire their signal after trapping odorous chemicals, and thus begins the perception of smell. Unlike with vision, there are many types of olfactory receptors and each one seems to capture a range of smelly molecules. To complicate matters even more, each smelly molecule seems to stimulate multiple receptors.
As with all proteins, olfactory receptors are encoded in genes, written in DNA and set in our genomes. We have around 400 olfactory genes, ones that relate to smells, and these combine in myriad ways to create the rich smellscape that we enjoy. How these combine to create complex perception remains something of a mystery. But there are some exceptions, and a neat study in 2015 focused on one of these, presumably so we didn’t all get too despondent that human genetics is all a bit inscrutable. OR7D4 is an olfactory gene, but mercifully has a very direct relationship with how we smell, as it only detects one molecule. Different people have different variants of this gene, and unusually, they correlate pretty precisely with what we think of the smell of a chemical called androstenone. We don’t really know what this steroid does in or for people, if anything at all, though it is found in sweat. But if you’re a pig, then it’s the main way you get laid.
Male pigs and boar produce it in their saliva, and it’s the principal component in “boar taint,” the smell of uncastrated males. When a female gets a sniff, she may well adopt a mating stance, if the mood takes her. This, as far as I am aware, does not happen in humans, but some of us can smell androstenone and others can’t. To many, it honks like stale urine;* to some it is sweet; and to me, well, I can’t smell it at all. These perceptions are determined by what allele of the OR7D4 gene you carry on your chromosome 19.
OR7D4 is a perfectly ordinary gene. It’s one of roughly 20,000 that we carry around in our cells, just under a thousand letters—nucleotides—long, which is a very typical length for many genes. Variations in a few of these letters determine what you think when some boar taint wafts up your nose, and these have a very direct correlation with a very real and visceral sense, whereas most other smell receptor genes are more complex and nuanced in how the sequence in the gene relates to what we actually perceive.
I asked Matthew Cobb, one of the scientists behind this ancient whiff, why they had asked the Neanderthals this question, and the answer was simple: “Because we can.” I like that answer. The real purpose of the paper was not an insight into the smellscape of the long dead, but to see how the various versions of the gene are distributed across the world today. In doing so, Cobb and his colleagues speculate that the variations might share a distribution pattern with pig farmers over recent human evolution; later we’ll see how milk farming had a similar effect in the same way. But this slight sidestep into the deep past typifies how easy contemporary genetics renders quizzing the ancients. This simple, not particularly important gene, says something almost magical. Our desire to reconstruct the past has taken a new turn into the realms of the senses. We now know that if a Neanderthal came across a frisky boar, he or she would be revolted, unlike me, who remains utterly unmoved.
One of the analyses of the Neanderthal genome looked in on a gene called MC1R. This is one that encodes a pigment in skin and hair. There’s a rare version of this gene that some people carry, and if they have the same rare version on their sixteenth chromosome inherited from both parents, they will have red hair. There are a few alleles of MC1R that result in being pale-skinned and ginger,* but one of the most common ones is a single letter change, from G to C, three quarters of the way along the gene. Looking at the Neanderthal MC1R reveals some of them had a different mutation, one that we haven’t seen in living humans, but which may have meant that some of them had reddish hair and pale skin too. There’s no hair left from those people, and pigmentation doesn’t survive well over time. So we test it by taking the version of the gene from the Neanderthals and inserting it into a bacteria or other small organism that we can genetically manipulate, and see what it makes. This system doesn’t grow a hair, but simply determines the melanin type that will fill the melanosomes, that will color the skin and hair. The results were equivocal. So it goes. Were they ginger? Maybe, but if they were, they weren’t ginger like us.
A continuing theme of this book is the limits to what genes can tell us about us. I could list a few more of the genes that have been identified in Neanderthals as being the same as, similar to, or noticeably different from our own, but the relationship between how genes are spelled in our genomes (the genotype) and how they manifest themselves as proteins and ultimately showable characteristics (phenotype) is frequently not very clear. Genes with clunky abbreviations are there in the Neanderthal genome as they are in our own, and we can speculate about precisely what they do. One of the grand endeavors of twenty-first-century genetics has been to test the function of genes by manipulating their equivalents in mice and seeing what happens, or by identifying human diseases in which those genes are mangled. We could speculate about SRGAP2, which has been linked with intelligence in some studies. We (and Neanderthals) have more copies of this gene than chimps, and probably more connections in our neurons as a result. Or we could talk about HACNS1, which is involved in hand development before birth, and is very different from the version that chimps have, whose manual dexterity is not as sophisticated and articulated as our own. But we don’t really understand what HACNS1 does during our own development, and therefore its role in the evolution of our handy skills. With all of the genes of Neanderthals potentially known, the truth remains that in most cases, knowing precisely how a gene is spelled is not enough to say precisely what it does.
This is not to undermine a century of genetics. I’ve picked out a few things about Neanderthals that can be determined by looking at individual genes, which are of interest—mostly trivial interest—hair color and smell. What turns me on is the broader arc of prehistory. Genetics turned into genomics when we began looking at all the DNA in an organism rather than just a handful of genes. Sweeping the whole Neanderthal genome is much more interesting than inspecting individual genes because it tells us about us. The question of our relationship with Neanderthals has been refined with genetics, in terms of our shared ancestors; our lineage moved away from theirs around half a million years ago. But what DNA analysis revealed more categorically than anything else was that we had sex with them, repeatedly, probably as soon as these two peoples met, and every time afterward.
So what happened? Humans are both horny and mobile. The language we use feels deceptive in these terms, at least in the time-scales we’re referring to. When we say humans migrated out of Africa, as our ancestors surely did, it sounds a little like they packed their bags and headed north to the Promised Land. The whole basis of current thinking about the origin of us is referred to as the Out of Africa hypothesis, defined by our migration away from the first site of anatomically modern humans. The timescales are not really known precisely, other than to say that they were over thousands of years. Our Homo sapiens ancestors inched into Europe around 60,000 years ago, and that story is told in Chapter 2. They didn’t turn up overnight with suitcases though. The spread of small groups or tribes expanded in all directions, including fundamentally away from where their ancestors were. That’s the best we can say. The first studies showed that there were at least five of those euphemistic gene flow events, but again, that doesn’t mean that five individuals had sex and produced offspring that lived their lives, and on into the distant future. It means that populations, tribes, interbred and shared their DNA across those populations.
All the Neanderthal DNA sequences are available as online databases, and nowadays the latest sequencing technologies mean that everyone can have their genome scanned (though not fully sequenced) and analyzed for many different things. 23andMe is one such company, and I had my genome parsed with them, the results of which are discussed in more detail later on. But one of the things that emerges out of these personal genomics is what Neanderthal DNA you carry. For me, a solid 2.7 percent of my total DNA is drawn from these people (which rather uninterestingly, according to their data, is exactly average for most Europeans; academic results suggest that this is an overestimate, and the proportion is lower in Europeans). Three billion letters of DNA make up my genome, and based on the 23andMe data, around 81 million of those come from Neanderthals, spread in chunks of varying size across my twenty-three pairs of chromosomes. Six whole human chromosomes have less DNA than that, including the Y, which makes me manly. Admittedly, this contribution is not all in one lump, nor is its influence felt in a singular way. But it is there in me.
For a century the Neanderthal people have worn a stigma of hunched brutish grunting cavemen. Facial reconstructions of their skulls show them to look not exactly like us, and not exactly pretty. But beauty is a very subjective matter, and just because you don’t fancy them doesn’t mean that your ancestors didn’t. They definitely had sex with them.
I carry Neanderthal DNA. Therefore, Neanderthals were my ancestors. If you carry their DNA, then they were your ancestors. If you are broadly of European descent, then it is almost certain that you also carry around Neanderthal DNA. This idea is called introgression—the introduction of DNA from a separate group over repeated familial backcrosses. It is a form of fusing some bits of DNA from distinct populations. Since their discovery, Neanderthals were referred to as cousins, or close relatives. It seems clear to me, using genetics as my ally, that Neanderthals categorically were also our ancestors.* This is one of the key concepts of DNA ancestry, the one that confuses all the nice clean hypothetical branches of a family tree. It’s called admixture—the mixing of genes from populations who were previously separated.
We carry their DNA. It’s not present in all human populations: Most Africans have very little; some eastern Asians have more than Europeans. In the longer term, “gene flow events” is how admixture occurs. But let’s be functionally crude, we’re talking about sex. Anatomically modern humans had sex with anatomically Neanderthal humans on many occasions in our history.
We learned from Croatian Neanderthal bones that we had interbred around 60,000 years ago, a time when it was presumed Homo sapiens had first reached Eurasia. As soon as we met, we mated. Romanian bones showed that it happened again around 40,000 years ago. It was beginning to look as though whenever our ancestors encountered Homo neanderthalensis, they got it on.
A female Neanderthal who died 50,000 years ago in the Altai Mountains in Siberia had joined the genome club in 2014, her toe bone being the source of the most detailed DNA yet recovered for her kind of human. But two years later, in February 2016, more meticulous analysis of her genome yet again upended what we had thought was true. It showed that she carried some modern human DNA, and by comparing it to others, we can put a date on when that introgression happened. It had occurred in one of her ancestors around 50,000 years before she was born. We don’t know who it came from, but whoever they were, they were like us, and they were a long way in time and space from when and where we thought the African diaspora of Homo sapiens had reached. It may be that these people represented the first wave of African emigrants, perhaps a kind of scout party pioneering east, tens of thousands of years before we had thought any of us had left the motherland. The Out of Africa hypothesis remains completely intact in principle, but the dates and the overall flow have profoundly changed with evidence provided by ancient DNA.
Whenever they encountered each other, the genes flowed. When we talk of gene flow events, the word events is potentially misleading, just as the word migration is. Given the widespread distribution of Neanderthal DNA in populations across Europe today, it is unlikely that this arrived in our gene pool after one of your ancestors mated with another probably shorter, stockier one. These events refer to interbreeding on a population scale, and the question of why this admixture introgression occurred is an interesting one.
When eggs and sperm are made, the gene shuffle of recombination is random, so the genes that an individual will acquire as the result of an interspecies hookup will be just the luck of the draw. As they spread through the population as a result of that individual breeding, natural selection can cast its hand over the usefulness of those genes. In autumn 2015, a sweep of studies analyzed how well evolution received the various genomic acquisitions from Neanderthals. Because DNA tends to be inherited in chunks, we can learn how useful bits of genome are by the size of chunks that are shared. A version of a gene that is definitely useful and therefore likely to be selected by nature over time may carry other bits of flanking DNA with it as it passes through the generations, like a peloton in a bike race being driven forward by the presence of a top rider, bringing all the pack with them. Or the genome may slowly ditch them if they are harmful. By comparing sections of Neanderthal DNA with sections of DNA in modern humans that we think have come from Neanderthals, we can build up a very finely tuned model of the success, from an evolutionary point of view, of these hybridizations. When Graham Coop and his colleagues did this, they found that our genomes are slowly purging themselves of Neanderthal DNA, which suggests that these matings were not to our advantage, but not massively disadvantageous. Our DNA around Neanderthal chunks is undergoing weak negative selection, the peloton slowly decelerating as a result of a weak rider. It might have to do with population size. It’s likely that their numbers were always low; mtDNA taken from various Neanderthal bones is all pretty similar, which indicates low genetic diversity, which suggests a small breeding population, maybe just a few thousand. We think that when these meetings were taking place, we were much more populous than them. Once their DNA entered ours, even if it were slightly not to our advantage, then it could be swamped out by a much bigger gene pool. While it was in them, it could be perpetuated as there was no notable better DNA with which to compare. A second paper, by Kelley Harris and Rasmus Nielsen, found the same thing, and also that the selection against introgression was not strong enough to suggest an inherent barrier to reproduction, as you might see in distantly related species.
Another quirky detail that helps us understand these dalliances a tiny bit more emerges from this number crunching. The amount of introgression from Neanderthals is proportionally lower on the modern X than on the rest of the chromosomes. X chromosomes are only passed on by males half of the time because we also have a Y, but all of the time by women, who have two Xs. The observation that there is less Neanderthal DNA on our Xs implies that the first encounters we had with them that resulted in procreation were male Neanderthals with female Homo sapiens.
What did the Neanderthals ever do for us? Not that much. We can’t tell why we have been slowly rejecting their DNA for thousands of generations. One of the important lessons here is that it demonstrates the speed of evolution, or rather, its breathtakingly slow burn. Any seriously deleterious effects would have been wiped out immediately, and maybe lost to time. However, the fact that their DNA mixed with ours, and is only being selected out of thousands of generations, indicates that some form of hybrid incompatibility was not apparent. These analyses are extreme fine-tuning, only visible through the microscope of statistical inspection of DNA sequences. And they most certainly do not suggest that Neanderthals were a separate species.
We can’t say how this interbreeding happened. Was it forced? Or mutually consensual? We don’t know. We first met in Siberia 100,000 years ago. We coexisted in the main body of Europe for more than 5,000 years, which is by comparison almost as long as written human history. If you consider our understanding of the last 5,000 years of history, during which time much of it has been explicitly documented, and then consider what has been recorded by proxy in the prehistory of Europe, then you see the scale of the problems of reconstructing the deep past. Our relationship with the Neanderthals has been scrutinized for decades and we know that we lived and bred with them. But some archaeological research suggests we may also have hunted and eaten others. The distribution of the Neanderthal people was widespread, and we probably encountered them all over Eurasia, but it may be that their low numbers resulted in a bottleneck, a lack of genetic diversity that renders a population less healthy overall. It may be that we brought with us diseases that they had not evolved to counter. Our existence ultimately subsumed theirs. The Neanderthals were a proto-species, an embryonic light that flickered in evolutionary time, but was not strong enough to stand across epochs. Whatever the reason for their dwindling from not many to none, we carry their genes, and their immortality will be as enduring as our own.
The Altai Mountains loom out of the ground near the Russian borders with China and Mongolia, and they are icy cold. This land is harsh. There’s a cave in this hinterland of Siberia, called Denisova, named after an eighteenth-century eremite called Denis who lived there. Due to the brutal weather it’s inaccessible for much of the year. Modern human and Neanderthal remains have been recovered from Denis’ cave over the forty years it has been explored, as well as dozens of species of animals, from lions and hyenas to woolly rhinos and, as befits the Russian motherland, a lot of bears. Soviet researchers had pulled out over 50,000 artifacts from this cave right up to the Middle Ages, indicating that it had been occupied in some form for more than 230,000 years. Siberia is one of the least populous areas on Earth; today it averages three people per square kilometer. For Denis’ cave to be such a regular haunt for hundreds of thousands of years is not usual. You can see why though. Despite the gritty weather, it’s a highly desirable residence; a waterfront property overlooking a picturesque river, the rustic estate boasts a wide rectangular south-facing entrance, into a nine-by-eleven-meter main chamber served by a working vertical chimney for a fireplace or kitchen stove, with three smaller secluded side galleries for bedrooms or even a study. Total floor plan: roughly 270 square meters.
In 2008, the remains of one of the former residents surfaced in the cave. Calling them remains is generous, as it was only a tooth and single distal manual phalanx of the fifth digit of a juvenile, or in more normal language, the last joint of a little finger of a child. The layer in which it was found puts its date as being somewhere between 30,000 and 50,000 years old. A single finger bone is enough for paleoanthropologists to classify the former owner as a hominin, a taxonomic division that includes all the Homos, as well as the gorillas and chimpanzees. But for more precision than that, a fingertip is not nearly enough.
Except that it was. The tooth was big, bigger than expected for both Homo sapiens and Homo neanderthalensis.
But what was in the finger was enough in fact to overturn human evolution yet again. Russian diggers who found the bone passed it on to Svante Pääbo for DNA analysis, and here they were extremely lucky.
Pääbo’s team managed to extract the mitochondrial genome from the single tiny bone—in doing so they destroyed it—and the results were published in Nature just before Easter in 2010. That sequence, patched up from the inevitable decay of several hundred centuries, was enough to be revolutionary. It wasn’t us, and it wasn’t Neanderthal. No other species in the genus Homo was known to exist at that time in Europe or Asia, and it was not a sequence akin to our primate cousins, chimps, or bonobos. It—she, we would soon discover—was a new type of human.
Of the limited information that was extracted from this little loop of DNA, the number of key differences in the DNA pointed to a human whose ancestors moved out of Africa in a migration that was different from the ancestors of the Neanderthals (around half a million years ago) and our own exodus from the motherland that began at least 100,000 years ago. The number of differences in her DNA was twice as many as between us and the Neanderthals, and that number can be used to calculate the last common ancestor of these three people. Around a million years ago, somewhere in Africa, a group of humans lived who were to be separated into us, the Neanderthals, and the Denisovans.
That separation was temporary, lasting just a few hundreds of thousands of years. By Christmas 2010, the rest of her genome was complete. An “unknown hominin from Southern Siberia” is how she was described in that first study. Fossils are rare by their very nature, and to get fossilized requires a lot of luck. With this finger, one bone out of more than 200 that made up her body, they got extremely lucky. It seems that these conditions preserved the DNA locked inside this tiny bone better than any one of the Neanderthal remains found so far.
The bone size, when compared to Neanderthal or modern human finger bones, indicates that its owner was a juvenile, a child. It says nothing about sex. The DNA says it all. Sex is determined by DNA, but a chromosome count is not an option in ancient samples. Chromosomes only make those nice identifiable shapes at a particular time in the cell’s natural cycle, and can only be achieved in cells taken from the living. So a visible Y or a pair of Xs was never in the cards for the Denisovan finger. Instead, Pääbo’s team extracted the DNA in fragments and inserted them into a kind of database made of other bits of DNA designed to host the unknown sections. Once this library is established you can use it to generate the sequence of the letters of code on a computer, and make all the comparisons you like without touching the bone ever again. One of the first steps was to look to see if there were any fragments that looked like a modern Y chromosome. The answer was no, so it was very probably a girl. This is an absence of evidence, which we scientists like to remind people is not the same as evidence of absence. But in this case, the nonappearance of Y is enough for her to be believably female. Other characteristics that emerged from her genome suggest she had brown eyes, brown hair, and dark skin, all traits common to a species of Homo before the modern advent of light skin and blue eyes (all discussed in Chapter 2).
The second key comparison was to try to establish what the ancestral distance was between us, them, and Neanderthals. The way to do this is to look at a stretch of DNA and compare the precise sequence in several species. Very simply, the more similar they are, the more closely related the owners are. This applies at every level of living thing, from twins to bacteria. The comparison in the new genomes showed that Denisovans and Neanderthals were more closely related to each other than either was to any living human. But the real kicker came with the revelation that Denisovan DNA was alive and well in contemporary Melanesians—the indigenous people of Fiji, Papua New Guinea, and a scattering of islands off the northeast coast of Australia. Just as the Neanderthals left their permanent mark in me and you if you are of Eurasian descent, these other people, known only from this single bone, imprinted their genetic mark through the ages in the ancestors of these island people, up to 5 percent of their genomes.
The people of Tibet carry adaptations to living at altitude, as they do in the deeply inhospitable plains around Everest with its lower levels of oxygen. The people of China to the north and India to the south do not. Mostly, that adaptation is crystallized in a gene called EPAS1, which sits in a region of DNA whose sequence is notably different from the Tibetans’ neighbors. By comparing this highly unusual bit of DNA to other local and global known sequences, it seems that this adaptation was plucked from the Denisovans. Admixture introgression had provided an adaptation that allowed Tibetans to thrive in an environment in which probably you and I would struggle.
We haven’t been able to determine how many chromosomes either the Neanderthals or the Denisovans had. It seems likely that they had the same number as us, and interbreeding as we did so successfully reinforces this assumption. There is something significant that we do now know about the large-scale arrangement of the genomes of these peoples. There are four living genera of hominidae, aka the great apes: Pan (chimps and bonobos), Pongo (orangutans), Gorilla (gorillas), and Homo (us). The first three have twenty-four pairs of chromosomes, whereas we have twenty-three. But all great apes, including us, share effectively all the same genes in our genomes. The discrepancy is found in our chromosome 2—the second largest single chunk of DNA we carry. It’s such a whopper because it is an end-to-end fusion of two chromosomes found in chimps, orangs, and gorillas. We know this because the genes on the two hairy ape chromosomes are effectively the same and in the same order as those found on our chromosome 2. And we can also see the remnants of chromosome architecture, no longer needed after this massive joint. Chromosomes have a pinched waist, like a knot in a long balloon, called the centromere. The two chimp chromosomes have a centromere each, but our fused chromosome 2 has one obvious centromere, and the shadowy remnants of another, betraying our common ancestry with our hairy quadrupedal cousins. We can spot these in the DNA sequence, and there they are in both the Neanderthal and Denisovan genomes. This says that this uniquely Homo characteristic occurred before the three of us diverged.
Pope John Paul II pontificated, as popes do, in 1996 that evolution was more “than just a theory,” which while being a generous bridging gesture between the magisteria of science and religion, misunderstands that theories in science, unlike in the vernacular, are the top of the intellectual pile, the zenith of descriptions of the true nature of nature. Theories are the best we’ve got. Anyway, he tried to reconcile the idea of our divine special creation with the irrefutable evidence of our evolution from earlier apes by suggesting that there was an “ontological discontinuity”—if we were looking for a moment where the metaphorical breath of God metaphorically entered us, then it could be when those two chromosomes fused. If that were the case, then the Denisovans and the Neanderthals are in our special club too. Otherwise, there is no discontinuity, just a bumpy slide into our present.
What happened to these peoples? We don’t really know. The Denisovans are almost entirely mysterious, and while there are rumors of other bones being found in that Siberian cave, we currently are relying on a fingertip and a big molar. A second tooth was discovered in late 2015, and I am told that more are on their way. We see their genomes, mostly of just that young girl in fact (the first tooth is from a different young adult), and we see that they are neither Homo sapiens nor Homo neanderthalensis (though the most recent studies show that just like with Homo sapiens, the Neanderthals mated with the Denisovans too). They have not been us for a million years, nor Neanderthal for 800,000. Yet, at some point, a group of them hooked up with a tribe of Homo sapiens whose descendants ended up populating large chunks of the east, and in particular the Melanesian Islands.
There is one more almost surreal twist in this tale, so far (for I am sure there will be many more in the months and years to come). In 2013, David Reich, a Harvard geneticist essential to many of the stories in this book, further looked at the Denisovan genome with a very close squint (in the form of some sophisticated and sensitive statistical analysis) and saw something difficult to explain. The Neanderthals and the Denisovans split off from the lineage that ended in us some 400,000 years ago. But if you look hard enough, the Denisovan genome looks slightly more different from ours than it should. What this implies, according to Reich and others, is that they also had admixed and interbred with another species, one for whom we have no DNA to compare. A ghost population, a group of humans who are only known by the spectral presence they left in DNA as an outcome of sex.
Speculation is rife about who they were. Some, such as the UK’s doyen of ancient human bones, Chris Stringer, from the Natural History Museum in London, thinks they might be a species known from bones from which we have not fished out any DNA yet—Homo heidelbergensis. There’s another possibility, featuring another set of bones that will only further muddy the once-clear picture of human evolution. In the Longlin Cave, in Guangxi Zhuang in southwest China, remains of some odd people have been found. They died somewhere between 14,000 and 11,000 years ago, right on the turn of the Pleistocene to the Holocene. By this stage we see only anatomically modern humans. But these guys were not that. They shared many characteristics with us but also many primitive features. In the cave were the cultural remnants of lots of cooked venison, and they have become known as the Red Deer Cave people. Colin Groves, one of the Australian researchers who catalogued the find, said that they were “close to us but not quite ‘us,’” and may be classed as yet another human species. DNA extraction has so far failed, but some have suggested that these people might be the fruit of Denisovan and modern human matings. Until we get the DNA, the answer is another enigma in this increasingly populous field.
So there it is, for now. If you want to stick to the old definitions of a species, then it is impossible to call Neanderthals a separate species. The Denisovans have not yet been classified with the official taxonomy classifications that we adhere to. DNA is not how it is done, and a finger bone and a tooth are just not meaty enough. They were not us, and they were not Neanderthal, but we happily bred with them. This is a strange conundrum that betrays our difficulties with classifying life, and our adherence to a system that was designed to show the perfection of divine creation, organisms static in time and set in stone as they stand before us. Darwin’s great idea ruined that ideal, because he recognized that life passes through time, and changes continually. The only life forms that don’t change are dead ones.
Obviously, a worm is not the same as a monkey, nor conceivably capable of impregnating one. These are creatures separated by hundreds of millions of years of evolution. We know that chimpanzees and us, only 6 million years apart, cannot interbreed, though we can conceivably have sex, however distasteful that sounds. But what we now know as a result of DNA is that the physical distinctions brought on by tribes of physically distinct human species a million years ago, separating from each other for many thousands of generations and years, was not enough to prevent us from having successful reproductive sex.
The seven billion of us alive today are, according to all the evidence available to us, the last remaining group of human great apes from a set of at least four that existed 50,000 years ago. One of them, the little people of Flores, was remote and unusual, as decreed by the evolutionary weirdness of living an insular life. But the others were not that different from us. In the years to come, we will scrutinize the bones of dozens, or hundreds of ancient humans, remains that have been difficult to place, or controversial, or who have unknowingly caused furious arguments. Some of these battles will be settled with DNA, and others will be stoked. One thing seems inevitable: The ancient genomes left to discover will reveal that the world was a whole lot more cosmopolitan in the millennia before we came to be the last representative of the genus Homo. The landscape of the last million years of our past has been flattened by DNA, and the rebuilding has just begun. The family tree is cut down, and we’re mapping out the tributaries and rivers and brooks that led to the pool that we all swim in today.
This description is just skimming the surface of our new genetic history. As the techniques become more robust, and bones continue to be unearthed, we will expose more and more sexual dalliances from our ancient past, between all sorts of peoples. The rate at which new studies about the DNA of Neanderthals and Denisovans are being generated is such that on an almost weekly basis, dates, locations, and impact of the relations between those early people are being revised. It’s now clear, more than ever before, that the old, simplistic view of how we came to be who we are is just wrong. Gone are the days of neat branching trees, or the hunched ape step by step standing tall. The American beat poet Edward Sanders invented a word that I think is brilliantly apposite here. It was coined to describe a chaotic situation, and is often deployed by the military when maneuvers go badly wrong. For our purposes, if we are to look at the evolution that led to where we are now, instead of the nice neat tree, I think it could reasonably be described as one big, million-year clusterfuck. Whenever humans met—sapiens, Neanderthal, Denisovan—they had sex. What a time to be alive!
We’ll discover in the next few chapters that family trees are never trees in the way that we draw them in historical pedigrees, or in science papers when looking at a few closely related species, or even as Darwin scribbled so perceptively in his notebook in 1837. As long as we have been erect, mobile, and frisky, the branches have been entangled within our species, and now we know with cousins that have been apart from us for so long that they looked deceptively different. The Neanderthals, the Denisovans, and our other as yet undiscovered phantom cousins were different from us, and soon we will know them better.* But they also became us, and we will find them in the old bones and inside our own cells. We carry the past with us. There was no beginning, and there are no missing links, just the ebb and flow and ebb again of living through epochs. Those ancient people never went extinct—we just merged.