CHAPTER 5
VARIATIONAL EVOLUTION
Variation played no role in transmutationism nor in either of the two kinds of transformationism. All three theories were strictly based on essentialism. “Evolution” takes place in transmutationism through the origin of a new essence and in the transformationist theories through a gradual change of the essence.
VARIATION AND POPULATION THINKING
Darwin showed that one simply could not understand evolution as long as one accepted essentialism. Species and populations are not types, they are not essentialistically defined classes, but rather are biopopulations composed of genetically unique individuals. This revolutionary insight required an equally revolutionary explanatory theory of evolution: Darwin’s theory of variation and selection. Two sources of evidence led Darwin to this new concept. One was the empirical study of variable natural populations (particularly during his study of the barnacles), and the other was the observation by animal and plant breeders that no two individuals of their herds or breeding stocks were identical. These individuals were not members of essentialistic classes, and, as we now know, all individuals in a sexual population are genetically unique.
Apparently, most people find it difficult to grasp the significance of this uniqueness. Let them remember that no two individuals among the 6 billion humans are identical, not even so-called identical (monozygous) twins. An understanding of the fundamental difference between a class of essentially identical objects and a biopopulation of unique individuals is the foundation of so-called “population thinking,” one of the most important concepts of modern biology.
The assumptions of population thinking are diametrically opposed to those of the typologist. The populationist stresses the uniqueness of everything in the organic world. What is true for the human species—that no two individuals are alike—is equally true for all other species of animals and plants. Indeed, even the same individual changes continuously throughout its lifetime and when placed into different environments. All organisms and organic phenomena are composed of unique features and can be described collectively only in statistical terms. Individuals, or any kind of organic entities, form populations of which we can determine the arithmetic mean and the statistics of variation. Averages are merely statistical abstractions, only the individuals of which the populations are composed have reality. The ultimate conclusions of the population thinker and of the typologist are precisely the opposite. For the typologist, the type (eidos) is real and the variation an illusion, while for the populationist the type (average) is an abstraction and only the variation is real. No two ways of looking at nature could be more different. (Mayr 1959)
Darwin’s Variational Evolution
It was Darwin who introduced this new way of thinking into science. His basic insight was that the living world consists not of invariable essences (Platonian classes), but of highly variable populations. And it is the change of populations of organisms that is designated as evolution. Thus, evolution is the turnover of the individuals of every population from generation to generation.
When Darwin, in 1837, became an evolutionist (see Chapter 2), he asked himself, How can the process of evolution be explained? Could he adopt one of the already proposed explanations? He realized eventually that neither transmutationism nor transformationism nor any other theory based on essentialism would do. And he was right. All essentialistic theories of organic evolution are badly flawed, as was convincingly established during the post-Darwinian controversies.
Darwin had to develop an entirely new kind of explanation that accounted for the abundance of variation in nature. This led him to his theory of natural selection, which was based on population thinking (see Chapter 6). The same theory was found independently by Alfred Russel Wallace.
Although Darwin published On the Origin of Species in 1859 (actually Wallace and Darwin published a first statement in 1858), the explanatory theory of variational evolution was not universally adopted until ca. 80 years later. It is a theory based on the variability of populations. There were two sets of practitioners who had already appreciated this variability for a long time, the taxonomists and the animal and plant breeders, and Darwin had close connections to both of them.
When sorting out the collections he had made on the voyage of the Beagle, Darwin encountered the same question again and again: Are some slightly different specimens merely variants within a population or are they different species? Indeed, in the 1840s when he wrote his monographs on the classification of the barnacles, Darwin came to the conclusion that no two specimens in a collection from a single population were exactly identical. They all were as uniquely different from each other as are human individuals. And the animal and plant breeders, with whom Darwin was associated since his Cambridge student days, told him the same. They always knew which individuals in their herds they should select as the breeding stock for the next generation. Individuality made this possible.
Since the terms “transmutationism” and “transformationism” are not suitable for this new theory, Darwin’s theory of evolution through natural selection is best referred to as the theory of variational evolution. According to this theory, an enormous amount of genetic variation is produced in every generation, but only a few individuals of the vast number of offspring will survive to produce the next generation. The theory postulates that those individuals with the highest probability of surviving and reproducing successfully are the ones best adapted, owing to their possession of a particular combination of attributes. Since these attributes are largely determined by genes, the genotypes of these individuals will be favored during the process of selection. As a consequence of the continuous survival of individuals (phenotypes) with genotypes best able to cope with the changes of the environment, there will be a continuing change in the genetic composition of every population. This unequal survival of individuals is due in part to competition among the new recombinant genotypes within the population, and in part to chance processes affecting the frequency of genes. The resulting change of a population is called evolution. Since all changes take place in populations of genetically unique individuals, evolution is by necessity a gradual and continuous process.
Darwin’s Theories of Evolution
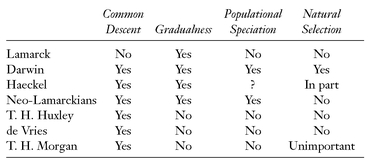
Darwin’s views on evolution are often referred to as The Darwinian Theory. Actually they consist of a number of different theories that are best understood when clearly distinguished from each other. The most important of Darwin’s theories of evolution are discussed below (see Box 5.1). That they are indeed five independent theories is documented by the fact that the leading “Darwinians” among Darwin’s contemporaries accepted some and rejected others (see Box 5.2).
Two of these five theories, evolution as such and the theory of common descent, were widely accepted by biologists within a few years of the publication of the Origin. This represented the first Darwinian revolution. The acceptance of man as a primate in the animal kingdom was a particularly revolutionary step. Three other theories, gradualism, speciation, and natural selection, were strongly resisted and were not generally accepted until the evolutionary synthesis. This was the second Darwinian revolution. The Darwinism proposed by Weismann and Wallace, in which an inheritance of acquired characters is rejected, was named Neodarwinism by George John Romanes. The Darwinism accepted since the evolutionary synthesis is best simply called Darwinism, because in most crucial aspects it agrees with the original Darwinism of 1859, while the belief in an inheritance of acquired characters is by now totally obsolete.
Box 5.1 Darwin’s Five Major Theories of Evolution
1. The nonconstancy of species (the basic theory of evolution)
2. The descent of all organisms from common ancestors (branching evolution)
3. The gradualness of evolution (no saltations, no discontinuities)
4. The multiplication of species (the origin of diversity)
5. Natural selection
Box 5.2 Rejection of Some of Darwin’s Theories by Early Evolutionists
The following table shows the composition of the evolutionary theories of various evolutionists. All of these authors accepted a fifth theory, that of evolution as opposed to a constant, unchanging world. They differed in accepting or rejecting some of Darwin’s four other evolutionary theories.
Darwin’s theory of gradualism fitted well into the thinking of the transformationists, but the resistance of the saltationists was sufficiently great that the universal acceptance of the gradualness of evolution had to await the evolutionary synthesis. Darwin’s concept of gradualness, however, was of an entirely different nature from that of the transformationists. Their gradualness was due to the gradual change of an essential type, whereas Darwinian gradualism is due to the gradual restructuring of populations. This makes it quite clear why Darwinian evolution, being a populational phenomenon, must always be gradual (see Chapter 4). A Darwinian must be able to show that every seeming case of saltation or discontinuous evolution can be explained as being caused by a gradual restructuring of populations.
VARIATION
The availability of variation is the indispensable prerequisite of evolution, and the study of the nature of variation is therefore a most important part of the study of evolution. Variation, the uniqueness of every individual, is, as we said, characteristic of every sexually reproducing species. To be sure, at first sight all the individuals of a species of snail or butterfly or fish might seem identical, but a closer study of these individuals will reveal all sorts of differences in size, proportions, color pattern, scaling, bristles, and whatever characteristic one studies. Further studies have shown that variability affects not only visible characters, but also physiological traits, patterns of behavior, aspects of ecology (e.g., adaptation to climatic conditions), and molecular patterns, all of this reinforcing the conclusion that in one way or another every individual is unique. And it is this always available variability that makes the process of natural selection possible.
Although the variability of the phenotype was appreciated by naturalists as far back as Darwin’s day, the early geneticists treated the genotype as rather uniform. When the studies by the population geneticists from the 1920s to the 1960s revealed the presence of a great deal of cryptic variation, this was questioned by some of the classical authors. Yet not even the most enthusiastic Darwinians suspected the amount of genetic variation in populations that was eventually revealed by the methods of molecular genetics. Not only was it discovered that much of the DNA consists of noncoding DNA (“junk”), but it was also found that many, perhaps the majority of alleles are “neutral,” that is, their mutation does not affect the fitness of the phenotype (see below). As a result, it is now realized that seemingly identical phenotypes may conceal considerable variation at the level of the gene.
Polymorphism
Sometimes variation falls into definite classes, a phenomenon referred to as polymorphism. In the human species we have polymorphisms for eye color, hair color, straightness or curliness of the hair, different blood groups, and many other of the genetic variants of our species. The study of polymorphisms has greatly contributed to our understanding of the strength and direction of natural selection, as well as the causal factors underlying variation. Two outstanding studies are those on the color polymorphism of banded snails (Cepaea) by Cain and Sheppard and on chromosome arrangements in Drosophila by Dobzhansky. In most cases it is unknown what is responsible for the maintenance of polymorphism in a population over long periods. A balance of selection pressures is usually assumed, but it may be reinforced by some superiority of the heterozygous carriers that favors the retention of the rarer gene in the population. In a highly diverse environment, phenotypic diversity may be selected, as in the case of the banded snails.
THE SOURCE OF VARIABILITY
What is the source of this variability? Where does it come from? How is it maintained from generation to generation? This is what puzzled Darwin all of his life, but in spite of all his efforts he never found the answer. An understanding of the nature of this variability was finally made possible, after 1900, by advancements in genetics and molecular biology. One can never fully understand the process of evolution unless one has an understanding of the basic facts of inheritance, which explain variation. Therefore the study of genetics is an integral part of the study of evolution. But only the heritable part of variation plays a role in evolution.
Genotype and Phenotype
As early as the 1880s it was recognized by perceptive biologists that the genetic material (germ plasm) was something different from the body of an organism (soma), and this distinction was satisfied when the early Mendelians introduced the terms
genotype and
phenotype. But the prevailing opinion at that time was that the genetic material consisted of proteins like those that make up the body. It came as a real shock when Avery demonstrated in 1944 that the genetic material consisted of nucleic acids. The terminological distinction between an organism and its genes now acquired a new meaning. The genetic material itself is the genome (haploid) or the genotype (diploid), which controls the production of the body of an organism and all of its attributes, the phenotype. This phenotype is the result of the interaction of the genotype with the environment during development. The amplitude of variation of the phenotype produced by a given genotype under different environmental conditions is called its
norm of reaction. For instance, a given plant may grow to be larger and more luxuriant under favorable conditions of fertilizing and watering than without these environmental factors. Leaves of the Water Buttercup
(Ranunculus flabellaris) produced under water are feathery and very different from the broadened leaves on the branches above water (see
Fig. 6.3). As we shall see, it is the phenotype that is exposed to natural selection, and not individual genes directly.
It has been heatedly argued in the past whether a particular property of an organism was due to “nature” (its genes) or “nurture” (its environment). All research in the last 100 years indicates that most characteristics of an organism are affected by both factors. This is particularly true for characters that are controlled by multiple genes. There are two sources of variation in a sexually reproducing population, superimposed on each other: the variation of the genotype (because in a sexual species no two individuals are genetically identical) and the variation of the phenotype (because each genotype has its own norm of reaction). Different norms of reaction may react rather differently to the same environmental conditions.
THE GENETICS OF VARIATION
We owe our understanding of variation to the branch of biology called genetics, which is devoted to the study of the nature of inheritance. This science has grown, since its founding in 1900, into one of the largest biological disciplines and is extremely rich in fact and theory. Even textbooks restricted to evolutionary genetics may run to more than 300 pages. I am forced in this work on evolution to limit my treatment to an analysis of the basic principles of genetics, leaving a more detailed treatment to the special texts of the field. Maynard Smith (1989) and Hartl and Jones (1999) are recommended for fuller detail. For a beginner, the genetics chapter of any biology text, such as that of Campbell (1999), will be helpful, or the more extensive genetics chapters in the evolution books of Futuyma (1998), Ridley (1996), and Strickberger (1996). Fortunately, an understanding of the basic principles of genetics necessary for an understanding of evolution does not require all the detail offered in these books. I feel that it is sufficient to understand a limited number of basic principles, but these must be understood thoroughly. The seventeen principles listed here would seem to be the most important ones.
Seventeen Principles of Inheritance
1. The genetic material is constant (“hard”); it cannot be changed by the environment or by use and disuse of the phenotype. The inheritance of constant genetic material is called hard inheritance. Genes cannot be modified by the environment. Properties acquired by the proteins of the phenotype cannot be transmitted to the nucleic acids of the germ cells. There is no inheritance of acquired characters.
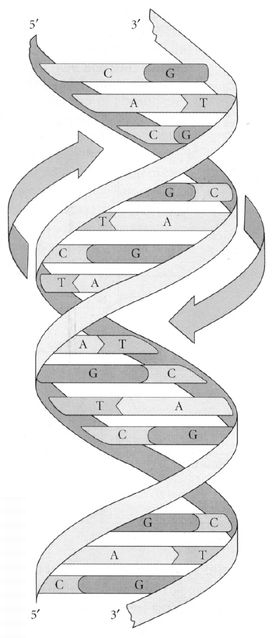
2. The genetic material, as was discovered by Avery in 1944, consists of DNA (deoxyribose nucleic acid) molecules (in some viruses also RNA). The DNA molecule has a double-helix structure, as discovered by Watson and Crick in 1953 (
Fig. 5.1).
3. The DNA contains the information that permits the production of the proteins that (together with lipids and other molecules) make up the phenotype of every organism. It controls the assemblage of amino acids that are converted into proteins with the help of cellular structures and mechanisms.
4. In the eukaryotes most DNA is located in the nucleus of every cell and is organized into a number of longitudinal bodies called
chromosomes (
Fig. 5.2). (Small amounts of DNA and RNA occur also in cellular organelles, such as mitochondria and chloroplasts.)
5. Sexually reproducing organisms are normally diploid, that is, they have two homologous sets of chromosomes, one inherited from the male parent and the other from the female parent.
6. Both male and female gametes have only one chromosome set, and so are haploid. When the egg is fertilized, diploidy is restored to the newly formed organism (zygote), because the chromosomes of the two parents do not fuse but remain discrete (see principle 7). This is why Mendelian inheritance is called particulate.
7. During the fertilization of an egg by a spermatozoon, the chromosomes of the male parent (containing the paternal genes) do not fuse or blend with the chromosomes of the female parent (containing the maternal genes) but rather coexist in the fertilized egg (zygote). The genetic material is thus handed unchanged from generation to generation, except for an occasional mutation (see principle 11).
8. Characteristics of organisms are controlled by genes, which are located on the chromosomes.
The well-known double helix of DNA. The base pairs, always one purine and one pyrimidine, are the horizontal “steps” of the helical staircase.
Source: Futuyma, Douglas J. (1998). Evolutionary Biology 3rd ed. Sinauer: Sunderland, MA.
9. A gene is a sequence of nucleic acid base pairs that encodes a program with a specific function.
10. On the whole, the nuclei of all cells of the body contain the same genes.
11. Although a gene is normally constant from generation to generation, it has the capacity to “mutate” occasionally into a different form. Such a newly mutated gene (mutant) will again be constant, unless another new mutation occurs.
12. The totality of the genes of an individual constitute its genotype.
13. Each gene has a number of different forms, called
alleles, which are responsible for most of the differences among the different individuals of a population (
Fig. 5.3).
14. A diploid organism has a pair of each gene, one from the male parent and one from the female parent. If these two genes are the same allele, the organism is called homozygous for this gene; if they belong to different alleles, the organism is called heterozygous.
15. When, in a heterozygote, only one of the two alleles is expressed in the phenotype, it is called the dominant allele; the other allele is called recessive.
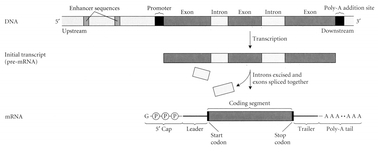
16. A gene has a complex structure, consisting of exons, introns, and flanking sequences (
Fig. 5.4).
17. There are several different kinds of genes, some of which control the actions of other genes (see below).
Age of Genes. Perhaps the most unexpected result of modern molecular studies of the genome was the discovery of the great age of many genes. The sequence of base pairs is often so conservative that one can determine that a certain mammalian gene is also part of the genome of the fruit fly Drosophila or the nematode Caenorhabditis. Indeed it seems possible to trace some genes all the way from animals or plants to bacteria. This fact is particularly important in the study of disease genes. For instance, one can treat a mouse with an inserted human disease gene with all sorts of drugs to test their curative capacity. It is also of great potential for the application of genetic engineering. Even where such practical applications are not possible, a comparison of the same gene in different kinds of organisms usually makes an important contribution to our understanding of gene functions.
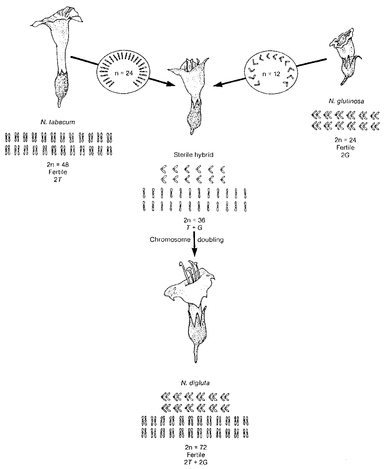
Origin of polyploidy. A cross between two species of plants often produces a sterile hybrid. A doubling of the chromosome number may, in certain crosses, produce a fertile allopolyploid species with the double chromosome number.
Source: Strickberger, Monroe, W.,
Evolution, 1990, Jones and Bartlett, Publishers, Sudbury, MA.
www.jbpub.com. Reprinted with permission.
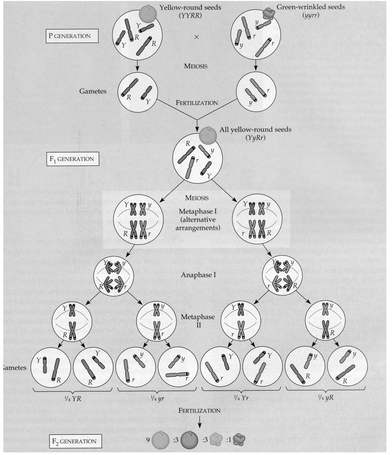
A gene may have different versions called alleles. In one of Mendel’s crosses he used gene Y with two alleles Y (dominant; yellow seeds) and y (recessive; green seeds), and gene R with two alleles R (dominant; round seeds) and r (recessive; wrinkled seeds). Crosses with these two sets of alleles gave the results shown in this figure.
Source: Figure 15.1, p. 262 from Biology 5th edition, by Neil A. Campbell, Jane B. Reece, and Lawrence G. Mitchell. Copyright © 1999 by Benjamin/Cummings, an imprint of Addison Wesley Longman, Inc. Reprinted by permission of Pearson Education, Inc.
Structure of a eukaryotic gene, with its exons, introns, and flanking sequences.
Source: Futuyma, Douglas J. (1998). Evolutionary Biology 3rd ed. Sinauer: Sunderland, MA.
GENETIC TURNOVER IN A POPULATION
According to the Hardy-Weinberg equation, the gene contents of a population would remain the same from generation to generation if it were not for a number of processes that may lead to the loss of existing genes or to the acquisition of new genes. These processes are responsible for evolution (see Box 5.3).
Seven such processes are of particular evolutionary importance: selection, mutation, gene flow, genetic drift, biased variation, movable elements, and nonrandom mating. Selection will be treated in Chapter 6; the other six will now be discussed.
Mutation
The use of the term mutation in biology has had a checkered history (Mayr 1963: 168–178). Prior to 1910 it was used for any drastic change of the type, particularly when such change instantaneously produced a new species. Morgan (1910) restricted mutation to a spontaneous change of the genotype, more precisely to a sudden change of a gene. Gene mutations are due to errors of replication during cell division. Even though the replication of the DNA molecules during cell division and gamete formation is remarkably accurate, occasional errors do occur. The replacement of a base pair by a different one is called a gene mutation. However, there are also larger changes of the genotype, such as polyploidy or changes of the gene arrangement, as occurs in chromosomal inversion. These are referred to as chromosomal mutations. Any changes on the pathway from the DNA of the gene (messenger RNA, ribosomes) to the amino acids or polypeptides of the phenotype are also classified as mutations. Mutations may also be caused by the insertion of a transposable element in the chromosome. Any mutation that induces changes in the phenotype will either be favored or discriminated against by natural selection.
Box 5.3 The Hardy-Weinberg Principle
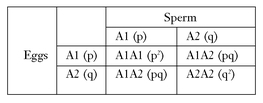
In the early years of genetics there was much confusion about what determined allele frequencies in a population. However, in 1908 G. H. Hardy in England and W. R.Weinberg in Germany showed mathematically that allele frequencies in populations would remain constant, generation after generation, if certain processes did not occur that would lead to the loss of existing genes or the acquisition of new genes. They expressed this as a mathematical formula, which is a reapplication of a mathematical law, the binomial expansion. Being a strictly mathematical solution, it is not a biological law.
Let us illustrate this by an example. Let us assume a gene is represented by two alleles in a population, A1 and A2. The frequency of A1 is p and that of A2 is q, with p + q = 1. The following frequencies of gametes will be present at reproduction and will produce the frequencies of genotypes as follows:
The binomial expansion (p + q) (p + q) = p2 + 2pq + q2 will be maintained generation after generation unless there is an addition or loss of genes (see text).
According to their evolutionary significance, three kinds of mutations can be distinguished: beneficial, neutral, or deleterious. Individuals with genotypes that contain a beneficial new mutation will be favored by natural selection. However, since almost all conceivable beneficial mutations of a population in a stable environment have already been selected in the recent past, the occurrence of new beneficial mutations is rather rare. Mutations that do not affect the fitness of the phenotype, so-called neutral mutations, are frequent. Their evolutionary role will be discussed below. Finally, deleterious mutations will be selected against and will be eliminated in due time. If they are recessive, they may survive in a population in heterozygous condition. If they result in immediate elimination, they are called lethal. The selective value of a gene may vary depending on its interaction with the remainder of the genotype.
Even though all new genes are produced by mutation, most of the phenotypic variation in natural populations that is available for selection is the product of recombination (see below). Before the role of selection was fully understood, it was believed by many evolutionists that some evolutionary changes were due to “mutation pressure.” This is a misconception. The frequency of a gene in a population is in the long run determined by natural selection and stochastic processes, and not by the frequency of mutation.
Gene Flow
The gene content of every local population, except the most isolated ones, is strongly affected by the immigration and emigration of genes to and from other populations of the species. This exchange of genes among neighboring populations is called gene flow. Gene flow is a conservative factor that prevents the divergence of only partially isolated populations and it is a major reason for the stability of widespread species and for the stasis of populous species. The amount of gene flow differs from population to population and from species to species. Highly sedentary (philopatric) species have little gene flow, whereas those with a strong dispersal tendency may be almost panmictic.
It is important to realize that dispersal propensity seems to be highly variable among individuals of a given population. Indeed, there may be a pronounced polymorphism in that respect. Certain individuals of a population may be highly philopatric, reproducing very close to their place of birth; others may disperse relatively short distances; and a few individuals may move far away from their birthplace, sometimes up to several hundred kilometers. These latter individuals, of course, are of the greatest evolutionary significance. Most of them probably will be unsuccessful, not being optimally adapted for their new location, yet these long-distance colonizers may establish founder populations and discover suitable locations well beyond the current species range.
Some species are so successful at dispersal that they have a cosmopolitan distribution, as is true for species with spores or animal species that have wind-dispersed eggs, as found among the tardigrades and certain crustaceans. However, even relatively short dispersal can efficiently counteract any tendency for a progressive divergence of local populations. Gene flow is an extremely conservative factor in evolution.
Genetic Drift
In a small population alleles may be lost simply through errors of sampling (stochastic processes); this is known as genetic drift. Indeed, such a random loss of alleles may occur even in rather large populations. This is usually of no consequence in widespread species, because such locally lost genes will be quickly replaced by gene flow in subsequent generations. However, small founder populations, beyond the periphery of the range of a species, may have a rather unbalanced sampling of the gene pool of the parental population. This may facilitate a restructuring of the genotype of such populations (see below).
Biased Variation
Some genes (so far known only in a few species) affect the segregation of alleles during meiosis in a heterozygote such that the allele of one parental chromosome goes to the gametes in more than half of the instances. If this allele controls an unfit phenotype, it will be selected against. Only rarely is such biased variation sufficiently strong to override the eliminating power of selection.
Transposable Elements
Transposable elements (TEs) are DNA sequences (“genes”) that do not occupy fixed sites on a chromosome but can move to a new site on the same or a different chromosome. There are various kinds of TEs with various effects. When inserted in a new location on a chromosome, they may cause a mutation on an adjacent gene. They often produce short DNA sequences that replicate frequently. One of these sequences, called Alu, is highly repeated with more than 500,000 copies in an individual of many mammalian species. It constitutes about 5 percent of the human genome. No selectively valuable contributions are known for any of the TEs. Rather they often seem deleterious, but natural selection seems unable to eliminate them. Confer a genetics textbook for a detailed treatment of the manifold manifestations of transposable elements.
Nonrandom Mating
In all species with sexual selection there may be a preference by one sexual partner for a particular phenotype of its mate. This leads to a nonrandom favoring of certain genotypes.
Some cases of sympatric speciation are best explained as products of nonrandom mating. In certain groups of fishes, particularly cichlid fishes, females seem to mate preferentially with males that prefer a certain subniche. If, for instance, in a lake in which at first species A occupies and feeds in both the benthic and the limnetic (open-water) zones and a group of females preferentially mates with benthic males, these females will select simultaneously for any visible markings that characterize males that prefer to feed in the benthic niche. Feeding and mating are no longer random and gradually two subpopulations evolve, the members of which preferentially feed and mate either benthically or limnetically. In due time the two subpopulations may evolve into two fully isolated sympatric species. In most groups of fishes this mode of sympatric speciation apparently does not occur. The same kind of process may lead to sympatric speciation in host-specific insects if mating preferentially takes place on the plant for which both mates have the same preference.
UNIPARENTAL REPRODUCTION AND EVOLUTION
Success in Darwinian evolution depends on the continuous availability of large amounts of variation. The greatest part of this phenotypic variation tends to be produced by recombination of the parental chromosomes, that is, by sexual reproduction, a process invented by the eukaryotes. However, large numbers of organisms do not have sexual reproduction; these organisms use uniparental reproduction. How do they manage to produce the variation needed to keep up with the changes in their environment?
In most forms of uniparental (“asexual”) reproduction, the offspring are genetically identical with the parent. A lineage produced by such reproduction is called a clone. How does a clone acquire new genetic variation? In higher organisms this is accomplished normally only through mutation. Any new mutation gives rise to a new mini-clone. If it is a successful mutation, the new clone will prosper and through acquiring additional mutations will gradually diverge from the parental clone. Eventually, as happened in the bdelloid rotifers, the differences between the most successful clones may become as great as those between different species of sexually reproducing species. Unsuccessful clones become extinct, and this is how the gaps between the “species” are created in asexual higher taxa.
Prokaryotes reproduce asexually. They acquire genetic variation by mutation and by unilateral exchange of genes with other clones. But as soon as sexuality had been, so to speak, “invented,” asexuality became relatively rare among the eukaryotes. Above the level of the genus there are only three higher taxa of animals that consist exclusively of uniparentally reproducing clones. Strict asexuality is rare in plants but common in some groups of fungi.
Prokaryotes reproduce asexually. All prokaryote individuals are, so to speak, of the same sex. Sexual reproduction is unknown among them. Yet sexual reproduction is now the almost universal mode of eukaryote reproduction. Every case of uniparental reproduction found in higher animals and plants is obviously a secondary (derived) condition, usually being restricted to a single species in a genus or to an isolated genus. There are only a few cases of entire families of animals being parthenogenetic (see below). It is rather obvious that in animals uniparental reproduction has been invented again and again, but the asexual clones always became extinct after a relatively short time.
Sexual versus Asexual Reproduction
What does the relative rarity of asexuality among the eukaryotes suggest? It leads to the inference that uniparental reproduction, where it is now found in higher organisms, is not primitive, but a derived condition. It has evolved independently, again and again, in unrelated groups, but soon becomes extinct. No matter what the selective advantage of sexual reproduction is, that it must have an advantage is clearly indicated by the consistent lack of success of asexuality.
And yet asexual reproduction would seem at first sight to be far more productive than sexual reproduction. Let us take a population with two kinds of females, both having the same number of 100 offspring, reduced in every generation to two survivors. Females A reproduce sexually and of its offspring 50 are males and 50 are females. Females B reproduce asexually and produce 100 females. A very simple calculation shows that in a short time the population would consist almost exclusively of the asexual B females.
An asexually reproducing female that can produce fertile eggs (parthenogenesis) does not “waste” any gametes on the production of males, and thus has twice the fertility of a sexual individual that produces both kinds of gametes. Why then does natural selection not favor parthenogenesis, the ability of females to produce eggs that do not require fertilization by males?
Since the 1880s evolutionists have argued over the selective advantage of sexual reproduction. So far, no clear-cut winner has emerged from this controversy. As often occurs in such controversies, plural answers may be the right answer. In other words, sexual reproduction has several advantages and combined they outweigh the seeming numerical advantage of asexuality. We must first grasp the entire process of sexual reproduction before we can understand why sexuality, in spite of its lower fertility, is in the long run more successful than asexual reproduction.
MEIOSIS AND RECOMBINATION
It took more than 100 years of study to achieve a full understanding of the meaning and the process of sexual reproduction. Darwin searched unsuccessfully all his life for the source of genetic variation. It required knowledge of the process of gamete formation and the difference between genotype and phenotype and their roles in natural selection, as well as an understanding of populational variation.
August Weismann and a group of cytologists found the answer. They showed that in sexual reproduction, gamete formation is preceded by two special cell divisions (see Box 5.4). During the first division, homologous maternal and paternal chromosomes attach themselves tightly to each other and then may break at one or several places. The broken chromosomes exchange parts with each other so that they now consist of a mixture of paternal and maternal chromosome pieces. This process is called crossing-over. Each new chromosome is an entirely new combination of maternal and paternal genes. In the second cell division preceding the formation of the gametes, the chromosomes do not divide, but one of each pair of homologous chromosomes goes randomly to one daughter cell and the other chromosome to the other daughter cell. As a result of this “reduction division” the “haploid” number of chromosomes in each gamete is half that of the “diploid” chromosome number of the zygote produced by the fertilized egg. This sequence of two cell divisions preceding gamete formation is called meiosis.
Box 5.4 Meiosis
Meiosis is the name for the two consecutive cell divisions that precede the formation of the haploid gametes. At the first division, sister chromatids of homologous chromosomes attach to each other. They may break at points of overlap in a process called crossing-over. A broken chromatid may join the broken end of the sister chromatid and become a composite new chromosome. In the ensuing second cell division, called the reduction division, homologous chromosomes go randomly to the opposite poles, thus producing entirely new chromosome sets. Thus at two consecutive steps an entirely new recombination of the parental genotypes is produced through crossing-over and the random movement of homologous chromosomes to opposite poles.
The gametes (spermatozoa and eggs) produced during meiosis are haploid, but diploidy is restored by fertilization. Consult a biology textbook for further details of this complex process.
Two processes during meiosis achieve a drastic recombination of the parental genotypes: (1) crossing-over during the first division and (2) the random movement of homologous chromosomes to different daughter cells (gametes) during the reduction division. The result is the production of completely new combinations of the parental genes, all of them uniquely different genotypes. These, in turn, produce unique phenotypes, providing unlimited new material for the process of natural selection.
No matter what the selective advantage of sexual reproduction may be, that it does have such an advantage in animals is clearly indicated by the consistent failure of all attempts to return to asexuality. Obligatory asexuality is not found among higher plants, but agamo-spermy, seed production without fertilization, is widespread (Grant 1981). Uniparental reproduction, however, is more frequent than sexual reproduction in certain protists, fungi, and some groups of nonva-scular plants. It is the exclusive mode of reproduction in the prokaryotes, in which unidirectional gene transfer provides genetic variation.
WHY IS THE PRODUCTION OF SUCH HIGHLY VARIABLE GENOTYPES SO FAVORED BY SELECTION?
Occasional asexual reproduction is of wide occurrence in the animal kingdom (but absent in birds and mammals). In almost every case it is restricted to a single species in an otherwise sexual genus or to an asexual genus. Only three higher taxa of animals (above the level of the genus) consist exclusively of uniparentally reproducing clones (bdelloid rotifers and some ostracods and mites). It is quite obvious that species have experimented with “buying” doubled fertility by abandoning sexuality, but the asexual clones die out sooner or later.
For more than a century evolutionists have speculated about the nature of sexuality’s powerful advantage, but until now no unanimity has been reached. Surely when a population suddenly encounters an extremely adverse situation, the more genetically diverse it is, the greater is the chance that it contains genotypes that can better cope with the environmental demands, compared to a uniform clone or a group of closely related clones.
A considerable number of solutions have been proposed for the mechanism by which sexuality (recombination) is favored by selection. They all have in common a greater survival of beneficial mutations and a faster elimination of deleterious mutations in sexual populations than in asexual ones. Pathogens (new diseases), for example, are best coped with by the origin of new resistant genotypes. The genotype, consisting of nucleic acids, is not directly exposed to natural selection, but is translated during the development of the fertilized egg into the proteins and other constituents of the phenotype (see Chapter 6). The phenotype is the result of the interaction of the environment with the genotype.
The process of sexual reproduction makes far more new phenotypes available for natural selection than does mutation or any other process. It is the major source of the variation found in populations of sexual species. This capacity for the production of large amounts of variation would seem to be the major selective advantage of sexual reproduction (see the special section “The Evolution of Sex,” Science 281(1988): 1979–2008). It is this capacity for recombination that gives sexual reproduction its enormous evolutionary importance.
Recombination
A member of a population in a sexually reproducing species mates with another member of its population and they produce in their offspring an entirely new recombination of the parental genes. The phrase “gene pool” for the genes found in a population is somewhat misleading. The genes are not independently swimming in a “pool,” but are linearly arranged on the chromosomes, each individual in a sexually reproducing diploid species carrying on its chromosomes one haploid set of paternal and one haploid set of maternal genes. This is the Sutton-Boveri theory, first put forth at the turn of the twentieth century, and later confirmed by T. H. Morgan. This diploid combination of the parental genetic material is called the genotype. Each individual is a unique combination of the two sets of parental genes, and it is the phenotype, the product of the genotype (the recombined set of genes), that is ordinarily the actual target of selection (see below). Recombination in a population is the major source of the phenotypic variation available for effective natural selection.
Lateral Transfer
There is no sexual reproduction in the prokaryotes and thus no replenishment of genetic variation by recombination. Instead, genetic variation in bacteria is renewed by a process called unidirectional lateral transfer, in which a bacterium attaches itself to another one and transfers some of its genes. There is little information on the types of genes transferred by this process. It is probably limited to certain classes of genes, since the major types of bacteria, such as gram-negative, gram-positive, and cyanobacteria, are not fused by this process. Even the archaebacteria exchange genes with other families of bacteria.
What happened to lateral transfer after the origin of sexual reproduction? Until the 1940s, it was assumed that this process had disappeared among sexually reproducing organisms. However, Barbara McClintock then discovered in maize the transposons, or genes that move from their position on one chromosome to another chromosome. This is such a new and unexpected discovery that it is not yet clear whether the phenomenon is widespread. There are also nucleic acid entities (e.g., plasmids) that are largely independent of the chromosomes. These genetic elements are of particular importance among the asexually reproducing prokaryotes. Whenever they affect the phenotype, they are subject to natural selection.
Gene Interaction
How the phenotype is produced by the action of the genes is the subject of physiological or developmental genetics. For the sake of simplicity, it was traditionally assumed that each gene acted independently of all others. This is not correct. Indeed, there are numerous interactions among the genes. Many genes, for example, may affect simultaneously several aspects of the phenotype. Such genes are called pleiotropic. Pleiotropy is most conspicuously demonstrated by deleterious genes, like the genes for sickle cell anemia (see Box 6.3), cystic fibrosis, and similar mutations, which affect some basic tissue activity that manifests itself in numerous different organs. On the other hand, a particular aspect of the phenotype may be affected by several different genes. Such inheritance is called polygenic inheritance. Pleiotropy and polygeny contribute to the cohesion of the genotype; the multiple interactions of genes is referred to as epistasis.
These interactions of genes are the least understood properties of the genotype. They will be referred to again in later chapters in connection with phenomena such as evolutionary stasis, bursts of evolutionary change, and mosaic evolution. The so-called “cohesion of the genotype” is one of the aspects of these interactions (see below). The study of the structure of the genotype is the most challenging of all future tasks of evolutionary biology.
Genome Size
If the production of new genes would parallel evolutionary advance one would expect that the organisms that are highest on the phylogenetic tree would have the largest genome. Up to a point this is indeed true. Genome size is measured in terms of the number of base pairs, although for practical reasons the units are megabases (1,000 base pairs, abbreviated Mb). The genome of humans is about 3500 Mb. In a bacterium it may be only 4 Mb. Very large figures were found in salamanders and lungfishes. An equally great variation was found in plants.
Why should there be such enormous variation and, in particular, such great differences among closely related organisms? The answer is that there are two kinds of DNA, that active in development (coding genes) and that not active (noncoding DNA) (see Box 5.5). The great differences in the Mb numbers are almost completely due to the presence of smaller or greater amounts of noncoding genes, often referred to as “junk.” There are numerous mechanisms by which noncoding genes are produced and multiplied, particularly by retro-transposable elements. There are also mechanisms by which junk DNA is eliminated, and different species differ in the efficiency of their elimination mechanisms. Research on the factors that control genome size still has a long way to go before full understanding is achieved. The size of the active genome is not only much smaller, but also far less variable than these numbers suggest.
Box 5.5 Noncoding DNA
A remarkably high proportion of the DNA in the chromosomes seems not to perform an obvious function such as coding for RNAs and proteins. Such DNA, sometimes probably incorrectly referred to as “junk,” is estimated for humans to be as much as 97 percent of the total DNA. This portion of our genome includes introns, repetitive sequences such as microsatellite DNA, and various kinds of “interspersed elements” such as Alu sequences. There is a widespread belief among Darwinians that such apparently unnecessary DNA would have been eliminated long ago by natural selection if it did not have some, as of yet undiscovered, function. Indeed the introns have a recognized function, to keep the exons apart prior to the activation of a gene (translation of the DNA message into proteins). During the translation process the introns are excised prior to the translation of a gene into proteins. Introns also contain many regulatory elements (DNA motifs that serve as binding sites for transcription regulation genes) and are thought to enhance eukaryotic genetic complexity via alternative splicing through both cis- and trans-acting elements.
THE ORIGIN OF NEW GENES
A bacterium has about 1,000 genes. A human has perhaps 30,000 functional genes. Where did all these new genes come from? They originate by duplication, with the duplicated gene inserted in tandem in the genome next to the sister gene. Such a new gene is called a paralogous gene. At first, it will have the same function as its sister gene. However, it will usually evolve by having its own mutations and in due time it may acquire functions that differ from those of its sister gene. The original gene, however, will also evolve, and such direct descendants of the original gene are called orthologous genes. In homology studies only orthologous genes may be compared.
Additions to the genome come not only by the duplication of single genes, but sometimes through the duplication of groups of genes, whole chromosomes, and entire chromosome sets. For instance, a special mechanism, involving the kinetochores, can lead to a duplication of chromosome sets in certain orders of mammals, leading to highly variable chromosome numbers in these orders. Lateral transfer is another way for addition to the genome.
Kinds of Genes
Molecular biology has discovered that there are many kinds of genes. Some directly control the production of organic material (via enzymes) and others control the activity of the material that produces genes. No mutation in 8,000 of the 12,000 genes of the Drosophila genome seems to have an effect on the phenotype. Changes in these genes have been referred to as neutral evolution (see below).
Genes that do not code for proteins have long been considered to be “junk.” However, they may play an important but not yet understood role in the regulation of other genes. The explanation of the role of the noncoding DNA may provide the solution to some of the open questions about the structure of the genotype. There are several different kinds of noncoding genetic material, including introns, pseudogenes, and highly repetitive DNA (Li 1997). At least some noncoding DNA definitely has a function: introns keep the exons separate. What is particularly difficult to understand is the great amount of noncoding DNA. According to some estimates, 95 percent of the human DNA is “junk.” A Darwinian finds it difficult to believe that selection would not have been able to get rid of it if it was indeed totally useless. After all, the production of this DNA is expensive.
Homeobox Genes, Regulatory Genes
All living animals belong to a limited number of basic designs: radially symmetrical, bilaterally symmetrical, segmented (metamerical), and characteristic subdivisions of these basic patterns. The great German morphologists have referred to such a basic design as Bauplan, which was translated into English (not quite correctly) as “body plan.” In German, the syllable plan in Bauplan means “map” or “blueprint” —not something that someone had planned. It is not a metaphysical concept.
Until a few years ago it was a complete riddle how a set of genes could determine what in the development of the zygote should become the anterior or the posterior end of the embryo, or the dorsal or the ventral side, and in a metameric organism which segment should bear what appendages. However, developmental genetics has now provided many explanations. In addition to the substrate-producing “structural” genes, there are regulatory genes that produce proteins able to determine front or rear, ventral or dorsal, and so on (Hox genes), or the construction of special organs, like the eye (pax gene). Sponges have only a single Hox gene, arthropods have 8, and mammals have 4 Hox clusters with 38 genes. Mice and flies share 6 Hox genes, which the common ancestor of Protostomia and Deuterostomia already must have had (see Box 5.6).
Everything indicates that the basic regulatory systems are very ancient and were later coopted for additional functions when these were acquired (Erwin et al. 1997). Such specialized developmental genes are largely independent of the action of other genes and permit the independent development of different parts and structures of the developing embryo. For example, the development of wings in a bat can take place with minimal disturbance of the other developmental pathways. This explains why so-called mosaic evolution is such a widespread phenomenon.
THE NATURE OF VARIATION
In Darwin’s day, the nature of variation in populations was not yet understood. This understanding was possible only following developments in the late nineteenth and twentieth centuries. What Darwin did know as a naturalist, taxonomist, and student of natural populations was that variation in natural populations seemed to be virtually inexhaustible. It provides abundant material for natural selection in all organisms, at least in sexually reproducing species of animals and plants. The visible characteristics of an organism, its phenotype, are due to instruction during development by their genes and by the genotype interacting with the environment.
Box 5.6 Hox Genes
Developmental as well as evolutionary biologists aim to better understand the evolution of complexity and the origin of morphological novelties in evolution through the analyses of the expression patterns of Hox genes during the ontogeny of organisms. It is suspected that these genes might play a pivotal role in specifying regional identity in body plans. Hox genes are arranged in genomic clusters and code for a class of transcription factors (genes that control the expression of other genes), and, importantly, their expression takes place in a spatially and temporally colinear fashion. Anterior genes in the Hox gene clusters are expressed earlier in development and more anteriorly in the embryo, whereas posterior genes are switched on later in development and in more distal portions of the body.
It has been suggested that increasing complexity of body plans during evolution might be causally correlated with increasing complexity of the Hox gene complexes. Invertebrates have only a single Hox gene cluster, and the common ancestor of all chordates probably also had only a single set of 13 Hox genes. During the evolution of chordates from relatively simply and rather segmentally organized cephalochordates like Amphioxus to more complex organisms like mice and humans, who have four Hox gene complexes, the single ancestral cluster probably duplicated twice for a complete set of 52 Hox genes in four clusters. These duplications from one to two and then to four clusters (A–D) occurred either as individual chromosome duplications, rather than through tandem duplications, because the clusters are each on different chromosomes, or through the duplications of entire genomes. Later in evolution, individual Hox genes on these clusters were lost on particular evolutionary lineages, yet mice and humans have the same set of 39 Hox genes distributed on four Hox clusters. None of these clusters retained their original set of 13 genes and each contains a unique combination of genes.
Differences in the gene content and expression patterns of Hox genes are assumed to be at least partially responsible for the different body plans that differentiate phyla of animals. The function of many Hox genes is paradoxically often extremely conserved in evolution, permitting remarkable experiments that demonstrated that Hox genes from, for example, Amphioxus can rescue the function of homologous genes in mice that had been experimentally removed from these mice. It remains an open question how new body plans are specified and evolved in light of, or in spite of, the remarkably conserved genomic architecture of Hox gene clusters and their highly conserved function in evolution.
The Impact of the Molecular Revolution
Although the basic principles of inheritance were worked out between 1900 and the 1930s, the real understanding of the nature of inheritance was achieved only through the molecular revolution. It began in 1944 (Avery et al.) when it was established that the genetic material consisted not of proteins but of nucleic acids. In 1953 Watson and Crick discovered the structure of DNA, and after this one major discovery followed the other, culminating in the discovery of the genetic code by Nirenberg in 1961 (Kay 2000). Finally, every step in the translation of the genetic information in the course of the developing organism was understood in principle. Unexpectedly, the basic Darwinian concepts of variation and selection were not affected in any way. Not even the replacement of proteins by nucleic acids as the carriers of genetic information required a change in the evolutionary theory. On the contrary, an understanding of the nature of genetic variation greatly strengthened Darwinism, for it confirmed the finding of the geneticists that an inheritance of acquired characters is impossible.
Molecular biology’s greatest contribution to evolutionary biology was the creation of the field of developmental genetics. Developmental biology, which had so long resisted the evolutionary synthesis, now adopted Darwinian thinking and analyzed the functional role of the genotype. This led to the discovery of regulatory genes (box, pax, etc.) and thus vastly enlarged our understanding of the evolutionary aspects of development.
Evolutionary Developmental Biology
One of the most important discoveries of molecular genetics was that some genes are very old. This means that the same gene (essentially the same sequence of base pairs) is found in organisms that are only very distantly related, say in Drosophila and mammals. A second discovery was that certain genes, often referred to as regulatory genes, control such basic developmental processes as the determination of anterior vs. posterior or of dorsal vs. ventral. These findings shed considerable light not only on previously completely puzzling developmental processes, but also on the causation of fundamental events (branching points) in phylogeny.
Scientists had always assumed that the same gene, no matter where found, always had the same phenotypic effect. But developmental geneticists have now shown that this is not necessarily so. The same gene may have rather different expressions in annelids (polychaetes) and arthropods (crustaceans). Selection seems to be able to recruit genes in new developmental processes that previously had seemed to have other functions.
It had been shown by morphological-phylogenetic research that photoreceptor organs (eyes) had developed at least 40 times independently during the evolution of animal diversity. A developmental geneticist, however, showed that all animals with eyes have the same regulatory gene, Pax 6, which organizes the construction of the eye. It was therefore at first concluded that all eyes were derived from a single ancestral eye with the Pax 6 gene. But then the geneticist also found Pax 6 in species without eyes, and proposed that they must have descended from ancestors with eyes. However, this scenario turned out to be quite improbable and the wide distribution of Pax 6 required a different explanation. It is now believed that Pax 6, even before the origin of eyes, had an unknown function in eyeless organisms, and was subsequently recruited for its role as an eye organizer.
CONCLUSIONS
It is shown in this chapter that Darwin, by making biopopulations the foundation of his evolutionary theorizing, rather than Platonic types, had found an entirely novel solution for the explanation of evolution. He postulated that the inexhaustible genetic variation of a population, together with selection (elimination), is the key to evolutionary change. To understand how this is implemented one must understand inheritance, and much of this chapter is therefore devoted to an explanation of the genetic basis of variation. The genetic material is constant and does not permit an inheritance of acquired characteristics. The genotype, interacting with the environment, produces the phenotype during development. Mutations continually replenish the variability of the gene pool. However, the variation of the phenotypes that provide the material for selection is produced by recombination in meiosis, a process of restructuring and reassorting the chromosomes.