
Top texture: © Laguna Design/Science Source;
Chapter 1: Genes Are DNA and Encode RNAs and Polypeptides
Edited by Esther Siegfried

Chapter Opener: © bluebay/Shutterstock, Inc.
1.1 Introduction
The hereditary basis of every living organism is its genome, a long sequence of deoxyribonucleic acid (DNA) that provides the complete set of hereditary information carried by the organism as well as its individual cells. The genome includes chromosomal DNA as well as DNA in plasmids and (in eukaryotes) organellar DNA, as found in mitochondria and chloroplasts. We use the term information because the genome does not itself perform an active role in the development of the organism. Rather, the products of expression of nucleotide sequences within the genome determine development. By a complex series of interactions, the DNA sequence directs production of all of the ribonucleic acids (RNAs) and proteins of the organism at the appropriate time and within the appropriate cells. Proteins serve a diverse series of roles in the development and functioning of an organism: they can form part of the structure of the organism; have the capacity to build the structure; perform the metabolic reactions necessary for life; and participate in regulation as transcription factors, receptors, key players in signal transduction pathways, and other molecules.
Physically, the genome can be divided into a number of different DNA molecules, or chromosomes. The ultimate definition of a genome is the sequence of the DNA of each chromosome. Functionally, the genome is divided into genes. Each gene is a sequence of DNA that encodes a single type of RNA and, in many cases, ultimately a polypeptide. Each of the discrete chromosomes comprising the genome can contain a large number of genes. Genomes for living organisms might contain as few as about 500 genes (for mycoplasma, a type of bacterium), about 20,000 for humans, or as many as about 50,000 to 60,000 for rice.
In this chapter, we explore the gene in terms of its basic molecular construction and basic function. FIGURE 1.1 summarizes the stages in the transition from the historical concept of the gene to the modern definition of the genome.

FIGURE 1.1 A brief history of genetics.
The first definition of the gene as a functional unit followed from the discovery that individual genes are responsible for the production of specific proteins. Later, the chemical differences between the DNA of the gene and its protein product led to the suggestion that a gene encodes a protein. This, in turn, led to the discovery of the complex apparatus by which the DNA sequence of a gene determines the amino acid sequence of a polypeptide.
Understanding the process by which a gene is expressed allows us to make a more rigorous definition of its nature. FIGURE 1.2 shows the basic theme of this book. A gene is a sequence of DNA that directly produces a single strand of another nucleic acid, RNA, with a sequence that is (at least initially) identical to one of the two polynucleotide strands of DNA. In many cases, the RNA is in turn used to direct production of a polypeptide. In other cases, such as ribosomal RNA (rRNA) and transfer RNA (tRNA) genes, the RNA transcribed from the gene is the functional end product. Thus, a gene is a sequence of DNA that encodes an RNA, and in protein-coding, or structural, genes, the RNA in turn encodes a polypeptide.

FIGURE 1.2 A gene encodes an RNA, which can encode a polypeptide.
The gene is the functional unit of heredity. Each gene is a sequence within the genome that functions by giving rise to a discrete product, which can be a polypeptide or an RNA. The basic pattern of inheritance of a gene was proposed by Mendel nearly 150 years ago. Summarized in his two major principles of segregation and independent assortment, the gene was recognized as a “particulate factor” that passes largely unchanged from parent to progeny. A gene can exist in alternative forms, called alleles.
In diploid organisms (having two sets of chromosomes), one of each chromosome pair is inherited from each parent. This is the same pattern of inheritance that is displayed by genes. One of the two copies of each gene is the paternal allele (inherited from the father); the other is the maternal allele (inherited from the mother). The shared pattern of inheritance of genes and chromosomes led to the discovery that chromosomes in fact carry the genes.
Each chromosome consists of a linear array of genes, and each gene resides at a particular location on the chromosome. The location is more formally called a genetic locus. The alleles of a gene are the different forms that are found at its locus. Although generally there are up to two alleles per locus in a diploid individual, a population might have many alleles of a single gene.
The key to understanding the organization of genes into chromosomes was the discovery of genetic linkage—the tendency for genes on the same chromosome to remain together in the progeny instead of assorting independently as predicted by Mendel’s principle. After the unit of recombination (reassortment) was introduced as the measure of linkage, the construction of genetic maps became possible. The recombination frequency between loci is proportional to the physical distance between the loci.
The resolution of the recombination map of a multicellular eukaryote is restricted by the small number of progeny that can be obtained from each mating. Recombination occurs so infrequently between nearby points that it is rarely observed between different variable sites in the same gene. As a result, classic linkage maps of eukaryotes can place the genes in order but cannot resolve the locations of variable sites within a gene. By using a microbial system in which a very large number of progeny can be obtained from each genetic cross, researchers could demonstrate that recombination occurs within genes and that it follows the same rules as those for recombination between genes.
Variable nucleotide sites among alleles of a gene can be arranged into a linear order, showing that the gene itself has the same linear construction as the array of genes on a chromosome. In other words, the genetic map is linear within, as well as between, loci as an unbroken sequence of nucleotides. This conclusion leads naturally to the modern view summarized in FIGURE 1.3 that the genetic material of a chromosome consists of an uninterrupted length of DNA representing many genes. Having defined the gene as an uninterrupted length of DNA, it should be noted that in eukaryotes many genes are interrupted by sequences in the DNA that are then excised from the messenger RNA (mRNA) (see the chapter titled The Interrupted Gene). Furthermore, there are regions of DNA that control the timing and pattern of expression of genes that can be located some distance from the gene itself.

FIGURE 1.3 Each chromosome consists of a single, long molecule of DNA within which are the sequences of individual genes.
From the demonstration that a gene consists of DNA, and that a chromosome consists of a long stretch of DNA representing many genes, we will move to the overall organization of the genome. In the chapter titled The Interrupted Gene, we take up in more detail the organization of the gene and its representation in proteins. In the chapter titled The Content of the Genome, we consider the total number of genes, and in the chapter titled Clusters and Repeats, we discuss other components of the genome and the maintenance of its organization.
1.2 DNA Is the Genetic Material of Bacteria and Viruses
The idea that the genetic material of organisms is DNA has its roots in the discovery of transformation by Frederick Griffith in 1928. The bacterium Streptococcus (formerly Pneumococcus) pneumoniae kills mice by causing pneumonia. The virulence of the bacterium is determined by its capsular polysaccharide, which allows the bacterium to escape destruction by its host. Several types of S. pneumoniae have different capsular polysaccharides, but they all have a smooth “S” appearance. Each of the S types can give rise to variants that fail to produce the capsular polysaccharide and therefore have a rough “R” surface (consisting of the material that was beneath the capsular polysaccharide). The R types are avirulent and do not kill the mice, because the absence of the polysaccharide capsule allows the animal’s immune system to destroy the bacteria.
When S bacteria are killed by heat treatment, they can no longer harm the animal. FIGURE 1.4, however, shows that when heat-killed S bacteria and avirulent R bacteria are jointly injected into a mouse, it dies as the result of a pneumonia infection. Virulent S bacteria can be recovered from the mouse’s blood.

FIGURE 1.4 Neither heat-killed S-type nor live R-type bacteria can kill mice, but simultaneous injection of both can kill mice just as effectively as the live S type.
In this experiment, the heat-killed S bacteria were of type III and the live R bacteria had been derived from type II. The virulent bacteria recovered from the mixed infection had the smooth coat of type III. So, some property of the dead IIIS bacteria can transform the live IIR bacteria so that they make the capsular polysaccharide and become virulent. FIGURE 1.5 shows the identification of the component of the dead bacteria responsible for transformation. This was called the transforming principle. It was purified in a cell-free system in which extracts from the dead IIIS bacteria were added to the live IIR bacteria before being plated on agar and assayed for transformation (FIGURE 1.6). Purification of the transforming principle in 1944 by Avery, MacLeod, and McCarty showed that it is DNA.

FIGURE 1.5 The DNA of S-type bacteria can transform R-type bacteria into the same S type.

FIGURE 1.6 Rough (left) and smooth (right) colonies of S. pneumoniae.
© Avery, et al., 1944. Originally published in The Journal of Experimental Medicine, 79: 137–158. Used with permission of The Rockefeller University Press.
Having shown that DNA is the genetic material of bacteria, the next step was to demonstrate that DNA is the genetic material in a quite different system. Phage T2 is a virus that infects the bacterium Escherichia coli. When phage particles are added to bacteria, they attach to the outside surface, some material enters the cell, and then approximately 20 minutes later each cell bursts open, or lyses, to release a large number of progeny phage.
FIGURE 1.7 illustrates the results of an experiment conducted in 1952 by Alfred Hershey and Martha Chase in which bacteria were infected with T2 phages that had been radioactively labeled either in their DNA component (with phosphorus-32 [32P]) or in their protein component (with sulfur-35 [35S]). The infected bacteria were agitated in a blender and two fractions were separated by centrifugation. One fraction, containing the empty phage “ghosts” that were released from the surface of the bacteria, consisted of protein and contained approximately 80% of the 35S label. The other fraction consisted of the infected bacteria themselves and contained approximately 70% of the 32P label. Previously, it had been shown that phage replication occurs intracellularly so that the genetic material of the phage would have to enter the cell during infection.

FIGURE 1.7 The genetic material of phage T2 is DNA.
Most of the 32P label was present in the fraction containing infected bacteria. The progeny phage particles produced by the infection contained approximately 30% of the original 32P label. The progeny received less than 1% of the protein contained in the original phage population. This experiment directly showed that only the DNA of the parent phages enters the bacteria and becomes part of the progeny phages, which is exactly the expected behavior of genetic material.
The phage possesses genetic material with properties analogous to those of cellular genomes: Its traits are faithfully expressed and are subject to the same rules that govern inheritance of cellular traits. The case of T2 reinforces the general conclusion that DNA is the genetic material of the genome of a cell or a virus.
1.3 DNA Is the Genetic Material of Eukaryotic Cells
When DNA is added to eukaryotic cells growing in culture, it can enter the cells, and in some of them this results in the production of new proteins. When an isolated gene is used, its incorporation leads to the production of a particular protein, as depicted in FIGURE 1.8. Although for historical reasons these experiments are described as transfection when performed with animal cells, they are analogous to bacterial transformation. The DNA that is introduced into the recipient cell becomes part of its genome and is inherited with it, and expression of the new DNA results in a new phenotype of the cells (synthesis of thymidine kinase in the example of Figure 1.8). At first, these experiments were successful only with individual cells growing in culture, but in later experiments DNA was introduced into mouse eggs by microinjection and became a stable part of the genome of the mouse. Such experiments show directly that DNA is the genetic material in eukaryotes and that it can be transferred between different species and remain functional.
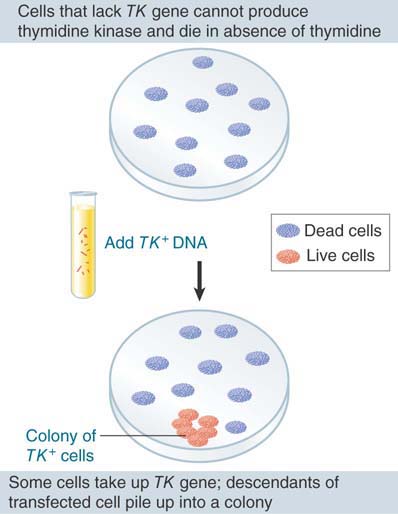
FIGURE 1.8 Eukaryotic cells can acquire a new phenotype as the result of transfection by added DNA.
The genetic material of all known organisms and many viruses is DNA. Some viruses, though, use RNA as the genetic material. As a result, the general nature of the genetic material is that it is always nucleic acid; specifically, it is DNA, except in the RNA viruses.
1.4 Polynucleotide Chains Have Nitrogenous Bases Linked to a Sugar–Phosphate Backbone
The basic building block of nucleic acids (DNA and RNA) is the nucleotide, which has three components:
A nitrogenous base
A sugar
One or more phosphates
The nitrogenous base is a purine or pyrimidine ring. The base is linked to the 1′ (“one prime”) carbon on a pentose sugar by a glycosidic bond from the N1 of pyrimidines or the N9 of purines. The pentose sugar linked to a nitrogenous base is called a nucleoside. To avoid ambiguity between the numbering systems of the heterocyclic rings and the sugar, positions on the pentose are given a prime (′).
Nucleic acids are named for the type of sugar: DNA has 2′–deoxyribose, whereas RNA has ribose. The difference is that the sugar in RNA has a hydroxyl (–OH) group on the 2′ carbon of the pentose ring. The sugar can be linked by its 5′ or 3′ carbon to a phosphate group. A nucleoside linked to a phosphate at the 5′ carbon is a nucleotide.
A polynucleotide is a long chain of nucleotides. FIGURE 1.9 shows that the backbone of the polynucleotide chain consists of an alternating series of pentose (sugar) and phosphate residues. The chain is formed by linking the 5′ carbon of one pentose ring to the 3′ carbon of the next pentose ring via a phosphate group; thus the sugar–phosphate backbone is said to consist of 5′–3′ phosphodiester linkages. Specifically, the 3′ carbon of one pentose is bonded to one oxygen of the phosphate, whereas the 5′ carbon of the other pentose is bonded to the opposite oxygen of the phosphate. The nitrogenous bases “stick out” from the backbone.

FIGURE 1.9 A polynucleotide chain consists of a series of 5′–3′ sugar–phosphate links that form a backbone from which the bases protrude.
Each nucleic acid contains four types of nitrogenous bases. The same two purines, adenine (A) and guanine (G), are present in both DNA and RNA. The two pyrimidines in DNA are cytosine (C) and thymine (T); in RNA, uracil (U) is found instead of thymine. The only structural difference between uracil and thymine is the presence of a methyl group at position C5.
The terminal nucleotide at one end of the chain has a free 5′ phosphate group, whereas the terminal nucleotide at the other end has a free 3′ hydroxyl group. It is conventional to write nucleic acid sequences in the 5′ to 3′ direction—that is, from the 5′ terminus at the left to the 3′ terminus at the right.
1.5 Supercoiling Affects the Structure of DNA
The two strands of DNA are wound around each other to form a double helical structure (described in detail in the next section); the double helix can also wind around itself to change the overall conformation, or topology, of the DNA molecule in space. This is called supercoiling. The effect can be imagined like a rubber band twisted around itself. Supercoiling creates tension in the DNA; thus, it can occur only if the DNA has no free ends (otherwise the free ends can rotate to relieve the tension) or in linear DNA (FIGURE 1.10, top) if it is anchored to a protein scaffold, as in eukaryotic chromosomes. The simplest example of a DNA with no free ends is a circular molecule. The effect of supercoiling can be seen by comparing the nonsupercoiled circular DNA lying flat in Figure 1.10 (center) with the supercoiled circular molecule that forms a twisted, and therefore more condensed, shape (Figure 1.10, bottom).

FIGURE 1.10 Linear DNA is extended (top); a circular DNA remains extended if it is relaxed (nonsupercoiled; center); but a supercoiled DNA has a twisted and condensed form (bottom).
Photos courtesy of Nirupam Roy Choudhury, International Centre for Genetic Engineering and Biotechnology (ICGEB).
The consequences of supercoiling depend on whether the DNA is twisted around itself in the same direction as the two strands within the double helix (clockwise) or in the opposite direction. Twisting in the same direction produces positive supercoiling, which overwinds the DNA so that there are fewer base pairs per turn. Twisting in the opposite direction produces negative supercoiling, or underwinding, so there are more base pairs per turn. Both types of supercoiling of the double helix in space are tensions in the DNA (which is why DNA molecules with no supercoiling are said to be “relaxed”). Negative supercoiling can be thought of as creating tension in the DNA that is relieved by the unwinding of the double helix. The effect of severe negative supercoiling is to generate a region in which the two strands of DNA have separated (technically, zero base pairs per turn).
Topological manipulation of DNA is a central aspect of all of its functional activities (e.g., recombination, replication, and transcription) as well as of the organization of its higher order structure. All synthetic activities involving double-stranded DNA require the strands to separate. The strands do not simply lie side by side though; they are intertwined. Their separation therefore requires the strands to rotate about each other in space. Some possibilities for the unwinding reaction are illustrated in FIGURE 1.11.

FIGURE 1.11 Separation of the strands of a DNA double helix can be achieved in several ways.
Unwinding a short linear DNA presents no problems, because the DNA ends are free to spin around the axis of the double helix to relieve any tension. DNA in a typical chromosome, however, is not only extremely long but also coated with proteins that serve to anchor the DNA at numerous points. As a result, even a linear eukaryotic chromosome does not functionally possess free ends.
Consider the effects of separating the two strands in a molecule whose ends are not free to rotate. When two intertwined strands are pulled apart from one end, the result is to increase their winding about each other farther along the molecule, resulting in positive supercoiling elsewhere in the molecule to balance the underwinding generated in the single-stranded region. The problem can be overcome by introducing a transient nick in one strand. An internal free end allows the nicked strand to rotate about the intact strand, after which the nick can be sealed. Each repetition of the nicking and sealing reaction releases one superhelical turn.
A closed molecule of DNA can be characterized by its linking number (L), which is the number of times one strand crosses over the other in space. Closed DNA molecules of identical sequence can have different linking numbers, reflecting different degrees of supercoiling. Molecules of DNA that are the same except for their linking numbers are called topological isomers.
The linking number is made up of two components: the writhing number (W) and the twisting number (T). The twisting number, T, is a property of the double helical structure itself, representing the rotation of one strand about the other. It represents the total number of turns of the duplex and is determined by the number of base pairs per turn. For a relaxed closed circular DNA lying flat in a plane, T is the total number of base pairs divided by the number of base pairs per turn. The writhing number, W, represents the turning of the axis of the duplex in space. It corresponds to the intuitive concept of supercoiling but does not have exactly the same quantitative definition or measurement. For a relaxed molecule, W = 0, and the linking number equals the twist.
We are often concerned with the change in linking number, ΔL, given by the equation:
ΔL = ΔW + ΔT
The equation states that any change in the total number of revolutions of one DNA strand about the other can be expressed as the sum of the changes of the coiling of the duplex axis in space (ΔW) and changes in the helical repeat of the double helix itself (ΔT). In the absence of protein binding or other constraints, the twist of DNA does not tend to vary—in other words, the 10.5 base pairs per turn (bp/turn) helical repeat is a very stable conformation for DNA in solution. Thus, any ΔL is mostly likely to be expressed by a change in W; that is, by a change in supercoiling.
A decrease in linking number (that is, a change of −ΔL) corresponds to the introduction of some combination of negative supercoiling (ΔW) and/or underwinding (ΔT). An increase in linking number, measured as a change of +ΔL, corresponds to an increase in positive supercoiling and/or overwinding.
We can describe the change in state of any DNA by the specific linking difference, σ = ΔL/L0, for which L0 is the linking number when the DNA is relaxed. If all of the change in the linking number is due to change in W (that is, ΔT = 0), the specific linking difference equals the supercoiling density. In effect, σ, as defined in terms of ΔL/L0, can be assumed to correspond to supercoiling density so long as the structure of the double helix itself remains constant.
The critical feature about the use of the linking number is that this parameter is an invariant property of any individual closed DNA molecule. The linking number cannot be changed by any deformation short of one that involves the breaking and rejoining of strands. A circular molecule with a particular linking number can express the number in terms of different combinations of T and W, but it cannot change their sum so long as the strands are unbroken. (In fact, the partitioning of L between T and W prevents the assignment of fixed values for the latter parameters for a DNA molecule in solution.)
The linking number is related to the actual enzymatic events by which changes are made in the topology of DNA. The linking number of a particular closed molecule can be changed only by breaking one or both strands, using the free end to rotate one strand about the other, and rejoining the broken ends. When an enzyme performs such an action, it must change the linking number by an integer; this value can be determined as a characteristic of the reaction. The reactions to control supercoiling in the cell are performed by topoisomerase enzymes (this is explored in more detail in the chapter titled DNA Replication).
1.6 DNA Is a Double Helix
By the 1950s, the observation by Erwin Chargaff that the bases are present in different amounts in the DNAs of different species led to the concept that the sequence of bases is the form in which genetic information is carried. Given this concept, there were two remaining challenges: working out the structure of DNA, and explaining how a sequence of bases in DNA could determine the sequence of amino acids in a protein.
Three pieces of evidence contributed to the construction of the double-helix model for DNA by James Watson and Francis Crick in 1953:
X-ray diffraction data collected by Rosalind Franklin and Maurice Wilkins showed that the B-form of DNA (which is more hydrated than the A-form) is a regular helix, making a complete turn every 34 Å (3.4 nm), with a diameter of about 20 Å (2 nm). The distance between adjacent nucleotides is 3.4 Å (0.34 nm); thus, there must be 10 nucleotides per turn. (In aqueous solution, the structure averages 10.4 nucleotides per turn.)
The density of DNA suggests that the helix must contain two polynucleotide chains. The constant diameter of the helix can be explained if the bases in each chain face inward and are restricted so that a purine is always paired with a pyrimidine, avoiding partnerships of purine–purine (which would be too wide) or pyrimidine–pyrimidine (which would be too narrow).
Chargaff also observed that regardless of the absolute amounts of each base, the proportion of G is always the same as the proportion of C in DNA, and the proportion of A is always the same as that of T. Consequently, the composition of any DNA can be described by its G-C content, or the sum of the proportions of G and C bases. (The proportions of A and T bases can be determined by subtracting the G-C content from 1.) G-C content ranges from 0.26 to 0.74 among different species.
Watson and Crick proposed that the two polynucleotide chains in the double helix associate by hydrogen bonding between the nitrogenous bases. Normally, G can hydrogen-bond most stably with C, whereas A can bond most stably with T. This hydrogen bonding between bases is described as base pairing, and the paired bases (G forming three hydrogen bonds with C, or A forming two hydrogen bonds with T) are said to be complementary. Complementary base pairing occurs because of complementary shapes of the bases at the interfaces where they pair, along with the location of just the right functional groups in just the right geometry along those interfaces so that hydrogen bonds can form.
The Watson–Crick model has the two polynucleotide chains running in opposite directions, so they are said to be antiparallel, as illustrated in FIGURE 1.12. Looking in one direction along the helix, one strand runs in the 5′ to 3′ direction, whereas its complement runs 3′ to 5′.

FIGURE 1.12 The double helix maintains a constant width because purines always face pyrimidines in the complementary A-T and G-C base pairs. The sequence in the figure is T-A, C-G, A-T, G-C.
The sugar–phosphate backbones are on the outside of the double helix and carry negative charges on the phosphate groups. When DNA is in solution in vitro, the charges are neutralized by the binding of metal ions, typically Na+. In the cell, positively charged proteins provide some of the neutralizing force. These proteins play important roles in determining the organization of DNA in the cell.
The base pairs are on the inside of the double helix. They are flat and lie perpendicular to the axis of the helix. Using the analogy of the double helix as a spiral staircase, the base pairs form the steps, as illustrated schematically in FIGURE 1.13. Proceeding up the helix, bases are stacked on one another like a pile of plates.

FIGURE 1.13 Flat base pairs lie perpendicular to the sugar–phosphate backbone.
Each base pair is rotated about 36° around the axis of the helix relative to the next base pair, so approximately 10 base pairs make a complete turn of 360°. The twisting of the two strands around each other forms a double helix with a minor groove that is about 12 Å (1.2 nm) across and a major groove that is about 22 Å (2.2 nm) across, as can be seen from the scale model presented in FIGURE 1.14. In B-DNA, the double helix is said to be “right-handed”; the turns run clockwise as viewed along the helical axis. (The A-form of DNA, observed when DNA is dehydrated, is also a right-handed helix and is shorter and thicker than the B-form. A third DNA structure, Z-DNA (named for the “zig-zag” pattern of the backbone), is longer and narrower than the B-form and is a left-handed helix.
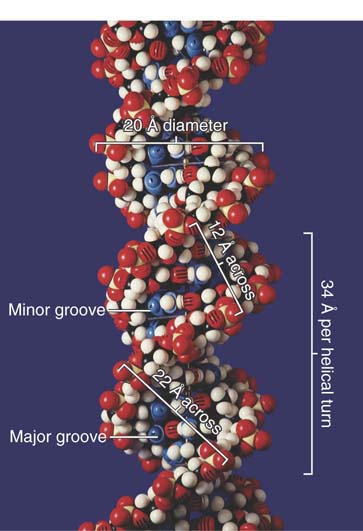
FIGURE 1.14 The two strands of DNA form a double helix. © Photodisc.
It is important to realize that the Watson–Crick model of the B-form represents an average structure and that there can be local variations in the precise structure. If DNA has more base pairs per turn, it is said to be overwound; if it has fewer base pairs per turn, it is underwound. The degree of local winding can be affected by the overall conformation of the DNA double helix or by the binding of proteins to specific sites on the DNA.
Another structural variant is bent DNA. A series of 8 to 10 adenine residues on one strand can result in intrinsic bending of the double helix. This structure allows tighter packing with consequences for nucleosome assembly (see Chapter 8, Chromatin) and gene regulation.
1.7 DNA Replication Is Semiconservative
To ensure the fidelity of genetic information, it is crucial that DNA is reproduced accurately. The two polynucleotide strands are joined only by hydrogen bonds, so they are able to separate without the breakage of covalent bonds. The specificity of base pairing suggests that both of the separated parental strands could act as template strands for the synthesis of complementary daughter strands. FIGURE 1.15 shows the principle that a new daughter strand is assembled from each parental strand. The sequence of the daughter strand is determined by the parental strand: An A in the parental strand causes a T to be placed in the daughter strand; a parental G directs incorporation of a daughter C; and so on.

FIGURE 1.15 Base pairing provides the mechanism for replicating DNA.
The top part of Figure 1.15 shows an unreplicated parental duplex with the original two parental strands. The lower part shows the two daughter duplexes produced by complementary base pairing. Each of the daughter duplexes is identical in sequence to the original parent duplex, containing one parental strand and one newly synthesized strand. The structure of DNA carries the information needed for its own replication. The consequences of this mode of replication, called semiconservative replication, are illustrated in FIGURE 1.16. The parental duplex is replicated to form two daughter duplexes, each of which consists of one parental strand and one newly synthesized daughter strand. The unit conserved from one generation to the next is one of the two individual strands comprising the parental duplex.

FIGURE 1.16 Replication of DNA is semiconservative.
Figure 1.15 illustrates a prediction of this model. If the parental DNA carries a “heavy” density label because the organism has been grown in a medium containing a suitable isotope (such as 15N), its strands can be distinguished from those that are synthesized when the organism is transferred to a medium containing “light” isotopes. The parental DNA is a duplex of two “heavy” strands (red). After one generation of growth in a “light” medium, the duplex DNA is “hybrid” in density—it consists of one “heavy” parental strand (red) and one “light” daughter strand (blue). After a second generation, the two strands of each hybrid duplex have separated. Each strand gains a “light” partner so that now one half of the duplex DNA remains hybrid and the other half is entirely “light” (both strands are blue).
In this model, the individual strands of these duplexes are entirely “heavy” or entirely “light” but never some combination of “heavy” and “light.” This pattern was confirmed experimentally by Matthew Meselson and Franklin Stahl in 1958. Meselson and Stahl followed the semiconservative replication of DNA through three generations of growth of E. coli. When DNA was extracted from bacteria and separated in a density gradient by centrifugation, the DNA formed bands corresponding to its density—“heavy” for parental, hybrid for the first generation, and half hybrid and half “light” in the second generation.
1.8 Polymerases Act on Separated DNA Strands at the Replication Fork
Replication of DNA requires the two strands of the parental duplex to undergo separation, or denaturation. The disruption of the duplex, however, is transient and is reversed, or undergoes renaturation, as the daughter duplex is formed. Only a small stretch of the duplex DNA is denatured at any moment during replication. (“Denaturation” is also used to describe the loss of functional protein structure; it is a general term implying that the natural conformation of a macromolecule has been converted to some nonfunctional form.)
The helical structure of a molecule of DNA during replication is illustrated in FIGURE 1.17. The unreplicated region consists of the parental duplex opening into the replicated region where the two daughter duplexes have formed. The duplex is disrupted at the junction between the two regions, which is called the replication fork. Replication involves movement of the replication fork along the parental DNA, so that there is continuous denaturation of the parental strands and formation of daughter duplexes.

FIGURE 1.17 The replication fork is the region of DNA in which there is a transition from the unwound parental duplex to the newly replicated daughter duplexes.
The synthesis of DNA is aided by specific enzymes (called DNA polymerases) that recognize the template strand and catalyze the addition of nucleotide subunits to the polynucleotide chain that is being synthesized. They are accompanied in DNA replication by ancillary enzymes such as helicases that unwind the DNA duplex, primase that synthesizes an RNA primer required by DNA polymerase, and ligase that connects discontinuous DNA strands. Degradation of nucleic acids also requires specific enzymes: deoxyribonucleases (DNases) degrade DNA, and ribonucleases (RNases) degrade RNA. The nucleases fall into the general classes of exonucleases and endonucleases:
Endonucleases break individual phosphodiester linkages within RNA or DNA molecules, generating discrete fragments. Some DNases cleave both strands of a duplex DNA at the target site, whereas others cleave only one of the two strands. Endonucleases are involved in cutting reactions, as shown in FIGURE 1.18.

FIGURE 1.18 An endonuclease cleaves a bond within a nucleic acid. This example shows an enzyme that attacks one strand of a DNA duplex.
Exonucleases remove nucleotide residues one at a time from the end of the molecule, generating mononucleotides. They only act on a single nucleic acid strand and each exonuclease proceeds in a specific direction; that is, starting either at a 5′ or a 3′ end and proceeding toward the other end. They are involved in trimming reactions, as shown in FIGURE 1.19.

FIGURE 1.19 An exonuclease removes bases one at a time by cleaving the last bond in a polynucleotide chain.
1.9 Genetic Information Can Be Provided by DNA or RNA
The central dogma describing the expression of genetic information from DNA to RNA to polypeptide is the dominant paradigm of molecular biology. Structural genes exist as sequences of nucleic acid but function by being expressed in the form of polypeptides. Replication makes possible the inheritance of genetic information, whereas transcription and translation are responsible for its expression to another form.
FIGURE 1.20 illustrates the roles of replication, transcription, and translation in the context of the so-called central dogma:

FIGURE 1.20 The central dogma states that information in nucleic acid can be perpetuated or transferred, but the transfer of information into a polypeptide is irreversible.
Transcription of DNA by a DNA-dependent RNA polymerase generates RNA molecules. mRNAs are translated to polypeptides. Other types of RNA, such as rRNAs and tRNAs, are functional themselves and are not translated.
A genetic system might involve either DNA or RNA as the genetic material. Cells use only DNA. Some viruses use RNA, and replication of viral RNA by an RNA-dependent RNA polymerase occurs in cells infected by these viruses.
The expression of cellular genetic information is usually unidirectional. Transcription of DNA generates RNA molecules; the exception is the reverse transcription of retroviral RNA to DNA that occurs when retroviruses infect cells (discussed shortly). Generally, polypeptides cannot be retrieved for use as genetic information; translation of RNA into polypeptide is always irreversible.
These mechanisms are equally effective for the cellular genetic information of prokaryotes or eukaryotes and for the information carried by viruses. The genomes of all living organisms consist of duplex DNA. Viruses have genomes that consist of DNA or RNA, and there are examples of each type that are double-stranded (dsDNA or dsRNA) or single-stranded (ssDNA or ssRNA). Details of the mechanism used to replicate the nucleic acid vary among viruses, but the principle of replication via synthesis of complementary strands remains the same, as illustrated in FIGURE 1.21.

FIGURE 1.21 Double-stranded and single-stranded nucleic acids both replicate by synthesis of complementary strands governed by the rules of base pairing.
Cellular genomes reproduce DNA by the mechanism of semiconservative replication. Double-stranded viral genomes, whether DNA or RNA, also replicate by using the individual strands of the duplex as templates to synthesize complementary strands.
Viruses with single-stranded genomes use the single strand as a template to synthesize a complementary strand; this complementary strand in turn is used to synthesize its complement (which is, of course, identical to the original strand). Replication might involve the formation of stable double-stranded intermediates or use double-stranded nucleic acid only as a transient stage.
The restriction of a unidirectional transfer of information from DNA to RNA in cells is not absolute. The restriction is violated by the retroviruses, which have genomes consisting of a single-stranded RNA molecule. During the retroviral cycle of infection, the RNA is converted into a single-stranded DNA by the process of reverse transcription, which is accomplished by the enzyme reverse transcriptase, an RNA-dependent DNA polymerase. The resulting ssDNA is in turn converted into a dsDNA. This duplex DNA becomes part of the genome of the host cell and is inherited like any other gene. Thus, reverse transcription allows a sequence of RNA to be retrieved and used as DNA in a cell.
The existence of RNA replication and reverse transcription establishes the general principle that information in the form of either type of nucleic acid sequence can be converted into the other type. In the usual course of events, however, the cell relies on the processes of DNA replication (to copy DNA from DNA), transcription (to copy RNA from DNA), and translation (to use mRNA to direct the synthesis of a polypeptide). On rare occasions though (possibly mediated by an RNA virus), information from a cellular RNA is converted into DNA and inserted into the genome. Although retroviral reverse transcription is not necessary for the regular operations of the cell, it becomes a mechanism of potential importance when we consider the evolution of the genome.
The same principles for the perpetuation of genetic information apply to the massive genomes of plants or amphibians as well as the tiny genomes of mycoplasma and the even smaller genomes of DNA or RNA viruses. TABLE 1.1 presents some examples that illustrate the range of genome types and sizes. The reasons for such variation in genome size and gene number are explored in the chapters titled The Content of the Genome and Genome Sequences and Evolution.
TABLE 1.1 The amount of nucleic acid in the genome varies greatly.
| Genome | Number of Genes | Number of Base Pairs |
|---|---|---|
| Organism | ||
| Plants | <50,000 | <1011 |
| Mammals | 30,000 | ~3 × 109 |
| Worms | 14,000 | ~108 |
| Flies | 12,000 | 1.6 × 108 |
| Fungi | 6,000 | 1.3 × 107 |
| Bacteria | 2–4,000 | <107 |
| Mycoplasma | 500 | <108 |
| dsDNA Viruses | ||
| Vaccinia | <300 | 187,000 |
| Papova (SV40) | ~6 | 5,226 |
| Phage T4 | ~200 | 165,000 |
| ssDNA Viruses | ||
| Parvovirus | 5 | 5,000 |
| Phage fX174 | 11 | 5,387 |
| dsRNA Viruses | ||
| Reovirus | 22 | 23,000 |
| ssRNA Viruses | ||
| Ciribavirus | 7 | 20,000 |
| Influenza | 12 | 13,500 |
| TMV | 4 | 6,400 |
| Phage MS2 | 4 | 3,569 |
| STNV | 1 | 1,300 |
| Viroids | ||
| PSTV RNA | 0 | 359 |
Note: TMV=tobacco mosaic virus; STNV=satellite tobacco necrosis virus; PSTV=potato spindle tuber viroid. |
||
Among the various living organisms, with genomes varying in size over a 100,000-fold range, a common principle prevails: The DNA encodes all of the proteins that the cell(s) of the organism must synthesize and the proteins in turn (directly or indirectly) provide the functions needed for survival. A similar principle describes the function of the genetic information of viruses, whether DNA or RNA: The nucleic acid encodes the protein(s) needed to package the genome and for any other functions in addition to those provided by the host cell that are needed to reproduce the virus. (The smallest virus—the satellite tobacco necrosis virus [STNV]—cannot replicate independently. It requires the presence of a “helper” virus—the tobacco necrosis virus [TNV], which is itself a normally infectious virus.)
1.10 Nucleic Acids Hybridize by Base Pairing
A crucial property of the double helix is the capacity to separate the two strands without disrupting the covalent bonds that form the polynucleotides and at the (very rapid) rates needed to sustain genetic functions. The specificity of the processes of denaturation and renaturation is determined by complementary base pairing.
The concept of base pairing is central to all processes involving nucleic acids. Disruption of the base pairs is crucial to the function of a double-stranded nucleic acid, whereas the ability to form base pairs is essential for the activity of a single-stranded nucleic acid. FIGURE 1.22 shows that base pairing enables complementary single-stranded nucleic acids to form a duplex:

FIGURE 1.22 Base pairing occurs in duplex DNA and also in intra- and intermolecular interactions in single-stranded RNA (or DNA).
An intramolecular duplex region can form by base pairing between two complementary sequences that are part of a single-stranded nucleic acid.
A single-stranded nucleic acid can base pair with an independent, complementary single-stranded nucleic acid to form an intermolecular duplex.
Formation of duplex regions from single-stranded nucleic acids is most important for RNA, but it is also important for single-stranded viral DNA genomes. Base pairing between independent complementary single strands is not restricted to DNA–DNA or RNA–RNA; it also can occur between DNA and RNA.
The lack of covalent bonds between complementary strands makes it possible to manipulate DNA in vitro. The hydrogen bonds that stabilize the double helix are disrupted by heating or by low salt concentration. The two strands of a double helix separate entirely when all of the hydrogen bonds between them are broken.
Denaturation of DNA occurs over a narrow temperature range and results in striking changes in many of its physical properties. The midpoint of the temperature range over which the strands of DNA separate is called the melting temperature (Tm) and it depends on the G-C content of the duplex. Each G-C base pair has three hydrogen bonds; as a result, it is more stable than an A-T base pair, which has only two hydrogen bonds. The more G-C base pairs in a DNA, the greater the energy that is needed to separate the two strands. In solution under physiological conditions, a DNA that is 40% G-C (a value typical of mammalian genomes) denatures with a Tm of about 87°C, so duplex DNA is stable at the temperature of the cell.
The denaturation of DNA is reversible under appropriate conditions. Renaturation depends on specific base pairing between the complementary strands. FIGURE 1.23 shows that the reaction takes place in two stages. First, single strands of DNA in the solution encounter one another by chance; if their sequences are complementary, the two strands base pair to generate a short, double-stranded region. This region of base pairing then extends along the molecule, much like a zipper, to form a lengthy duplex. Complete renaturation restores the properties of the original double helix. The property of renaturation applies to any two complementary nucleic acid sequences. This is sometimes called annealing, but the reaction is more generally called hybridization whenever nucleic acids from different sources are involved, as in the case when DNA hybridizes to RNA. The ability of two nucleic acids to hybridize constitutes a precise test for their complementarity because only complementary sequences can form a duplex.

FIGURE 1.23 Denatured single strands of DNA can renature to give the duplex form.
Experimentally, the hybridization reaction is used to combine two single-stranded nucleic acids in solution and then to measure the amount of double-stranded material that forms. FIGURE 1.24 illustrates a procedure in which a DNA preparation is denatured and the single strands are linked to a filter. A second denatured DNA (or RNA) preparation is then added. The filter is treated so that the second preparation of nucleic acid can attach to it only if it is able to base-pair with the DNA that was originally linked to the filter. Usually the second preparation is labeled so that the hybridization reaction can be measured as the amount of label retained by the filter. Alternatively, hybridization in solution can be measured as the change in UV absorbance of a nucleic acid solution at 260 nm as detected via spectrophotometry. As DNA denatures to single strands with increasing temperature, UV absorbance of the DNA solution increases; UV absorbance consequently decreases as ssDNA hybridizes to complementary DNA or RNA with decreasing temperature.

FIGURE 1.24 Filter hybridization establishes whether a solution of denatured DNA (or RNA) contains sequences complementary to the strands immobilized on the filter.
The extent of hybridization between two single-stranded nucleic acids is determined by their complementarity. Two sequences need not be perfectly complementary to hybridize under the appropriate conditions. If they are similar but not identical, an imperfect duplex is formed in which base pairing is interrupted at positions where the two single strands are not complementary.
1.11 Mutations Change the Sequence of DNA
Mutations provide decisive evidence that DNA is the genetic material. When a change in the sequence of DNA causes an alteration in the sequence of a protein, we can conclude that the DNA encodes that protein. Furthermore, a corresponding change in the phenotype of the organism can allow us to identify the function of that protein. The existence of many mutations in a gene might allow many variant forms of a protein to be compared, and a detailed analysis can be used to identify regions of the protein responsible for individual enzymatic or other functions.
All organisms experience a certain number of mutations as the result of normal cellular operations or random interactions with the environment. These are called spontaneous mutations, and the rate at which they occur (the “background level”) is different among species, and can be different among tissue types within the same species. Mutations are rare events, and, of course, those that have deleterious effects are selected against during evolution. It is therefore difficult to observe large numbers of spontaneous mutants from natural populations.
The occurrence of mutations can be increased by treatment with certain compounds. These are called mutagens, and the changes they cause are called induced mutations. Most mutagens either modify a particular base of DNA or become incorporated into the nucleic acid. The potency of a mutagen is judged by how much it increases the rate of mutation above background. By using mutagens, it becomes possible to induce many changes in any gene or genome.
Researchers can measure mutation rates at several levels of resolution: mutation across the entire genome (as the rate per genome per generation), mutation in a gene (as the rate per locus per generation), or mutation at a specific nucleotide site (as the rate per base pair per generation). These rates correspondingly decrease as a smaller unit is observed.
Spontaneous mutations that inactivate gene function occur in bacteriophages and bacteria at a relatively constant rate of 3–4 × 10−3 per genome per generation. Given the large variation in genome sizes between bacteriophages and bacteria (about 103), this corresponds to great differences in the mutation rate per base pair.
This suggests that the overall rate of mutation has been subject to selective forces that have balanced the deleterious effects of most mutations against the advantageous effects of some mutations. Such a conclusion is strengthened by the observation that an archaean that lives under harsh conditions of high temperature and acidity (which are expected to damage DNA) does not show an elevated mutation rate, but in fact has an overall mutation rate just below the average range. FIGURE 1.25 shows that in bacteria, the mutation rate corresponds to about 10−6 events per locus per generation or to an average rate of change per base pair of 10−9–10−10 per generation. The rate at individual base pairs varies very widely, over a 10,000-fold range. We have no accurate measurement of the rate of mutation in eukaryotes, although usually it is thought to be somewhat similar to that of bacteria on a per-locus, per-generation basis. Each human infant is estimated to carry about 35 new mutations.

FIGURE 1.25 A base pair is mutated at a rate of 10−9–10−10 per generation, a gene of 1,000 bp is mutated at about 10−6 per generation, and a bacterial genome is mutated at 3 × 10−3 per generation.
1.12 Mutations Can Affect Single Base Pairs or Longer Sequences
Any base pair of DNA can be mutated. A point mutation changes only a single base pair and can be caused by either of two types of event:
Chemical modification of DNA directly changes one base into a different base.
An error during the replication of DNA causes the wrong base to be inserted into a polynucleotide.
Point mutations can be divided into two types, depending on the nature of the base substitution:
The most common class is the transition, which results from the substitution of one pyrimidine by the other, or of one purine by the other. This replaces a G-C pair with an A-T pair, or vice versa.
The less common class is the transversion, in which a purine is replaced by a pyrimidine, or vice versa, so that an A-T pair becomes a T-A or C-G pair.
As shown in FIGURE 1.26, the mutagen nitrous acid performs an oxidative deamination that converts cytosine into uracil, resulting in a transition. In the replication cycle following the transition, the U pairs with an A, instead of the G with which the original C would have paired. So the C-G pair is replaced by a T-A pair when the A pairs with the T in the next replication cycle. (Nitrous acid can also deaminate adenine, causing the reverse transition from A-T to G-C.)
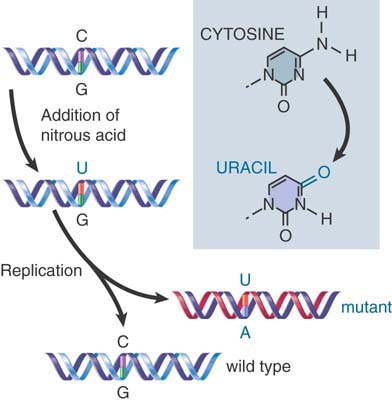
FIGURE 1.26 Mutations can be induced by chemical modification of a base.
Transitions are also caused by base mispairing, which occurs when noncomplementary bases pair instead of the conventional G-C and A-T base pairs. Base mispairing usually occurs as an aberration resulting from the incorporation into DNA of an abnormal base that has flexible pairing properties. FIGURE 1.27 shows the example of the mutagen bromouracil (BrdU), an analog of thymine that contains a bromine atom in place of thymine’s methyl group and can be incorporated into DNA in place of thymine. BrdU has flexible pairing properties, though, because the presence of the bromine atom allows a tautomeric shift from a keto (=O) form to an enol (–OH) form. The enol form of BrdU can pair with guanine, which after replication leads to substitution of the original A-T pair by a G-C pair.

FIGURE 1.27 Mutations can be induced by the incorporation of base analogs into DNA.
The mistaken pairing can occur either during the original incorporation of the base or in a subsequent replication cycle. The transition is induced with a certain probability in each replication cycle, so the incorporation of BrdU has continuing effects on the sequence of DNA.
Point mutations were thought for a long time to be the principal means of change in individual genes. We now know, though, that insertions of short sequences are quite frequent. Often, the insertions are the result of transposable elements, which are sequences of DNA with the ability to move from one site to another (see the chapter titled Transposable Elements and Retroviruses). An insertion within a coding region usually abolishes the activity of the gene because it can alter the reading frame; such an insertion is a frameshift mutation. (Similarly, a deletion within a coding region is usually a frameshift mutation.) Insertions of transposable elements can subsequently result in deletion of part or all of the inserted material, and sometimes of the adjacent regions.
A significant difference between point mutations and insertions is that mutagens can increase the frequency of point mutations, but do not affect the frequency of transposition. Both insertions and deletions of short sequences (often called indels) can occur by other mechanisms, however—for example, those involving errors during replication or recombination. In addition, a class of mutagens called the acridines introduces very small insertions and deletions.
1.13 The Effects of Mutations Can Be Reversed
FIGURE 1.28 shows that the possibility of reversion mutations, or revertants, is an important characteristic that distinguishes point mutations and insertions from deletions:

FIGURE 1.28 Point mutations and insertions can revert, but deletions cannot revert.
A point mutation can revert either by restoring the original sequence or by gaining a compensatory mutation elsewhere in the gene.
An insertion can revert by deletion of the inserted sequence.
A deletion of a sequence cannot revert in the absence of some mechanism to restore the lost sequence.
Mutations that inactivate a gene are called forward mutations. Their effects are reversed by back mutations, which are of two types: true reversions and second-site reversions. An exact reversal of the original mutation is called a true reversion. Consequently, if an A-T pair has been replaced by a G-C pair, another mutation to restore the A-T pair will exactly regenerate the original sequence. The exact removal of a transposable element following its insertion is another example of a true reversion. The second type of back mutation, second-site reversion, can occur elsewhere in the gene, and its effects compensate for the first mutation. For example, one amino acid change in a protein can abolish gene function, but a second alteration can compensate for the first and restore protein activity.
A forward mutation results from any change that alters the function of a gene product, whereas a back mutation must restore the original function to the altered gene product. The possibilities for back mutations are thus much more restricted than those for forward mutations. The rate of back mutations is correspondingly lower than that of forward mutations, typically by a factor of about 10.
Mutations in other genes can also occur to circumvent the effects of mutation in the original gene. This is called a suppression mutation. A locus in which a mutation suppresses the effect of a mutation in another unlinked locus is called a suppressor. For example, a point mutation might cause an amino acid substitution in a polypeptide, whereas a second mutation in a tRNA gene might cause it to recognize the mutated codon, and as a result insert the original amino acid during translation. (Note that this suppresses the original mutation but causes errors during translation of other mRNAs.)
1.14 Mutations Are Concentrated at Hotspots
So far, we have dealt with mutations in terms of individual changes in the sequence of DNA that influence the activity of the DNA in which they occur. When we consider mutations in terms of the alteration of function of the gene, most genes within a species show more or less similar rates of mutation relative to their size. This suggests that the gene can be regarded as a target for mutation, and that damage to any part of it can alter its function. As a result, susceptibility to mutation is roughly proportional to the size of the gene. Are all base pairs in a gene equally susceptible, though, or are some more likely to be mutated than others?
What happens when we isolate a large number of independent mutations in the same gene? Each is the result of an individual mutational event. Most mutations will occur at different sites, but some will occur at the same position. Two independently isolated mutations at the same site can constitute exactly the same change in DNA (in which case the same mutation has happened more than once), or they can constitute different changes (three different point mutations are possible at each base pair).
The histogram in FIGURE 1.29 shows the frequency with which mutations are found at each base pair in the lacI gene of E. coli. The statistical probability that more than one mutation occurs at a particular site is given by random-hit kinetics (as seen in the Poisson distribution). Some sites will gain one, two, or three mutations, whereas others will not gain any. Some sites gain far more than the number of mutations expected from a random distribution; they might have 10× or even 100× more mutations than predicted by random hits. These sites are called hotspots. Spontaneous mutations can occur at hotspots, and different mutagens can have different hotspots.

FIGURE 1.29 Spontaneous mutations occur throughout the lacI gene of E. coli, but are concentrated at a hotspot.
1.15 Many Hotspots Result from Modified Bases
A major cause of spontaneous mutation is the presence of an unusual base in the DNA. In addition to the four standard bases of DNA, modified bases are sometimes found. The name reflects their origin; they are produced by chemical modification of one of the four standard bases. The most common modified base is 5-methylcytosine, which is generated when a methyltransferase enzyme adds a methyl group to cytosine residues at specific sites in the DNA. Sites containing 5-methylcytosine are hotspots for spontaneous point mutation in E. coli. In each case, the mutation is a G-C to A-T transition. The hotspots are not found in mutant strains of E. coli that cannot methylate cytosine.
The reason for the existence of these hotspots is that cytosine bases suffer a higher frequency of spontaneous deamination. In this reaction, the amino group is replaced by a keto group. Recall that deamination of cytosine generates uracil (see Figure 1.26). FIGURE 1.30 compares this reaction with the deamination of 5-methylcytosine where deamination generates thymine. The effect is to generate the mismatched base pairs G-U and G-T, respectively.
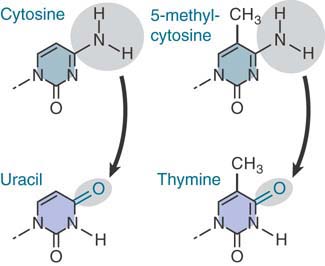
FIGURE 1.30 Deamination of cytosine produces uracil, whereas deamination of 5-methylcytosine produces thymine.
All organisms have repair systems that correct mismatched base pairs by removing and replacing one of the bases (see Chapter 14, Repair Systems). The operation of these systems determines whether mismatched pairs such as G-U and G-T persist into the next round of DNA replication and thereby result in mutations.
FIGURE 1.31 shows that the consequences of deamination are different for 5-methylcytosine and cytosine. Deaminating the (rare) 5-methylcytosine causes a mutation, whereas deaminating cytosine does not have this effect. This happens because the DNA repair systems are much more effective in accurately repairing G-U than G-T base pairs.

FIGURE 1.31 The deamination of 5-methylcytosine produces thymine (by C-G to T-A transitions), whereas the deamination of cytosine produces uracil (which usually is removed and then replaced by cytosine).
E. coli contain an enzyme, uracil-DNA-glycosidase, that removes uracil residues from DNA. This action leaves an unpaired G residue, and a repair system then inserts a complementary C base. The net result of these reactions is to restore the original sequence of the DNA. Thus, this system protects DNA against the consequences of spontaneous deamination of cytosine. (This system is not, however, efficient enough to prevent the effects of the increased deamination caused by nitrous acid; see Figure 1.26.)
Note that the deamination of 5-methylcytosine creates thymine and results in a mismatched base pair, G-T. If the mismatch is not corrected before the next replication cycle, a mutation results. The bases in the mispaired G-T first separate and then pair with the correct complements to produce the wild-type G-C in one daughter DNA and the mutant A-T in the other.
Deamination of 5-methylcytosine is the most common cause of mismatched G-T pairs in DNA. Repair systems that act on G-T mismatches have a bias toward replacing the T with a C (rather than the alternative of replacing the G with an A), which helps to reduce the rate of mutation (see the chapter titled Repair Systems). However, these systems are not as effective as those that remove U from G-U mismatches. As a result, deamination of 5-methylcytosine leads to mutation much more often than does deamination of cytosine.
Additionally, 5-methylcytosine creates hotspots in eukaryotic DNA. It is common in CpG dinucleotide repeats that are concentrated in regions called CpG islands (see the chapter titled Epigenetics I Effects Are Inherited). Although 5-methylcytosine accounts for about 1% of the bases in human DNA, sites containing the modified base account for about 30% of all point mutations.
The importance of repair systems in reducing the rate of mutation is emphasized by the effects of eliminating the mouse enzyme MBD4, a glycosylase that can remove T (or U) from mismatches with G. The result is to increase the mutation rate at CpG sites by a factor of 3. The reason the effect is not greater is that MBD4 is only one of several systems that act on G-T mismatches; most likely the elimination of all the systems would increase the mutation rate much more.
The operation of these systems casts an interesting light on the use of T in DNA as compared to U in RNA. It might relate to the need for stability of DNA sequences; the use of T means that any deaminations of C are immediately recognized because they generate a base (U) that is not usually present in the DNA. This greatly increases the efficiency with which repair systems can function (compared with the situation when they have to recognize G-T mismatches, which can also be produced by situations in which removing the T would not be the appropriate correction). In addition, the phosphodiester bond of the backbone is more easily broken when the base is U.
Another type of hotspot, though not often found in coding regions, is the “slippery sequence”—a homopolymer run, or region where a very short sequence (one or a few nucleotides) is repeated many times in tandem. During replication, a DNA polymerase can skip one repeat or replicate the same repeat twice, leading to a decrease or increase in repeat number.
1.16 Some Hereditary Agents Are Extremely Small
Viroids (or subviral pathogens) are infectious agents that cause diseases in some plants. They are very small circular molecules of RNA. Unlike viruses—for which the infectious agent consists of a virion, a genome encapsulated in a protein coat—the viroid RNA is itself the infectious agent. The viroid consists solely of the RNA molecule, which is extensively folded by imperfect base pairing, forming a characteristic rod as shown in FIGURE 1.32. Mutations that interfere with the structure of this rod reduce the infectivity of the viroid.

FIGURE 1.32 PSTV RNA is a circular molecule that forms an extensive double-stranded structure, interrupted by many interior loops. The severe and mild forms of PSTV have RNAs that differ at three sites.
A viroid RNA consists of a single molecule that is replicated autonomously and accurately in infected cells. Viroids are categorized into several groups. A particular viroid is assigned to a group according to sequence similarity with other members of the group. For example, four viroids in the potato spindle tuber viroid (PSTV) group have 70%–83% sequence similarity with PSTV. Different isolates of a particular viroid strain vary from one another in sequence, which can result in phenotypic differences among infected cells. For example, the “mild” and “severe” strains of PSTV differ by three nucleotide substitutions.
Viroids are similar to viruses in that they have heritable nucleic acid genomes, but differ from viruses in both structure and function. Viroid RNA does not appear to be translated into polypeptide, so it cannot itself encode the functions needed for its survival. This situation poses two as yet unanswered questions: How does viroid RNA replicate, and how does it affect the phenotype of the infected plant cell?
Replication must be carried out by enzymes of the host cell. The heritability of the viroid sequence indicates that viroid RNA is the template for replication.
Viroids are presumably pathogenic because they interfere with normal cellular processes. They might do this in a relatively random way—for example, by taking control of an essential enzyme for their own replication or by interfering with the production of necessary cellular RNAs. Alternatively, they might behave as abnormal regulatory molecules, with particular effects upon the expression of individual host cell genes.
An even more unusual agent is the cause of scrapie, a degenerative neurological disease of sheep and goats. The disease is similar to the human diseases of kuru and Creutzfeldt–Jakob disease, which affect brain function. The infectious agent of scrapie does not contain nucleic acid. This extraordinary agent is called a prion (proteinaceous infectious agent). It is a 28 kD hydrophobic glycoprotein, PrP. PrP is encoded by a cellular gene (conserved among the mammals) that is expressed in normal brain cells. The protein exists in two forms: The version found in normal brain cells is called PrPc and is entirely degraded by proteases; the version found in infected brains is called PrPsc and is extremely resistant to degradation by proteases. PrPc is converted to PrPsc by a conformational change that confers protease-resistance and that has yet to be fully defined.
As the infectious agent of scrapie, PrPsc must in some way modify the synthesis of its normal cellular counterpart so that it becomes infectious instead of harmless (see the chapters titled Epigenetics I and Epigenetics II). Mice that lack a PrP gene cannot develop scrapie, which demonstrates that PrP is essential for development of the disease.
1.17 Most Genes Encode Polypeptides
The first systematic attempt to associate genes with enzymes, carried out by Beadle and Tatum in the 1940s, showed that each stage in a metabolic pathway is catalyzed by a single enzyme and can be blocked by mutation in a single gene. This led to the one gene–one enzyme hypothesis. A mutation in a gene alters the activity of the protein enzyme it encodes.
A modification in the hypothesis is needed to apply to proteins that consist of more than one polypeptide subunit. If the subunits are all the same, the protein is a homomultimer and is encoded by a single gene. If the subunits are different, the protein is a heteromultimer, and each different subunit can be encoded by a different gene. Stated as a more general rule applicable to any heteromultimeric protein, the one gene–one enzyme hypothesis becomes more precisely expressed as the one gene–one polypeptide hypothesis. (Even this modification is not completely descriptive of the relationship between genes and proteins, because many genes encode alternate versions of a polypeptide; this concept can be explored further under the topic of alternative splicing in multicellular eukaryotes in the chapter titled RNA Splicing and Processing.)
Identifying the biochemical effects of a particular mutation can be a protracted task. The mutation responsible for Mendel’s wrinkled-pea phenotype was identified only in 1990 as an alteration that inactivates the gene for a starch-debranching enzyme!
It is important to remember that a gene does not directly generate a polypeptide: A gene encodes an RNA, which can in turn encode a polypeptide. Most genes are structural genes that encode messenger RNAs, which in turn direct the synthesis of polypeptides, but some genes encode RNAs that are not translated to polypeptides. These RNAs might be structural components of the protein synthesis machinery or might have roles in regulating gene expression (see the chapter titled Regulatory RNA). The basic principle is that the gene is a sequence of DNA that specifies the sequence of an independent product. The process of gene expression might terminate in a product that is either RNA or polypeptide.
A mutation in a coding region is generally a random event with regard to the structure and function of the gene; mutations can have little or no effect (as in the case of neutral mutations), or they can damage or even abolish gene function. Most mutations that affect gene function are recessive: They result in an absence of function, because the mutant gene does not produce its usual polypeptide. FIGURE 1.33 illustrates the relationship between mutant recessive and wild-type alleles. When a heterozygote contains one wild-type allele and one mutant allele, the wild-type allele is able to direct production of the enzyme and is therefore dominant. (This assumes that an adequate amount of product is made by the single wild-type allele. When this is not true, the smaller amount made by one allele as compared to two alleles results in the intermediate phenotype of a partially dominant allele in a heterozygote.)

FIGURE 1.33 Genes encode proteins; dominance is explained by the properties of mutant proteins. A recessive allele does not contribute to the phenotype because it produces no protein (or protein that is nonfunctional).
1.18 Mutations in the Same Gene Cannot Complement
How do we determine whether two mutations that cause a similar phenotype have occurred in the same gene? If they map to positions that are very close together (i.e., they recombine very rarely), they might be alleles. However, in the absence of information about their relative positions, they could also represent mutations in two different genes whose proteins are involved in the same function. The complementation test is used to determine whether two recessive mutations are alleles of the same gene or in different genes. The test consists of generating a heterozygote for the two mutations (by mating parents homozygous for each mutation) and observing its phenotype.
If the mutations are alleles of the same gene, the parental genotypes can be represented as follows:

The first parent provides an m1 mutant allele and the second parent provides an m2 allele, so that the heterozygote progeny have the genotype:

No wild-type allele is present, so the heterozygotes have mutant phenotypes and the alleles fail to complement. If the mutations lie in different linked genes, the parental genotypes can be represented as:

Each chromosome has one wild-type allele at one locus (represented by the plus sign [+]) and one mutant allele at the other locus. Then, the heterozygote progeny have the genotype:

in which the two parents between them have provided a wild-type allele from each gene. The heterozygotes have wild-type phenotypes because they are heterozygous for both mutant alleles, and thus the two genes are said to complement.
The complementation test is shown in more detail in FIGURE 1.34. The basic test consists of the comparison shown in the top part of the figure. If two mutations are alleles of the same gene, we see a difference in the phenotypes of the trans configuration (both mutations are not in the same allele) and the cis configuration (both mutations are in the same allele). The trans configuration (where the mutations lie on the same DNA molecule) is mutant because each allele has a (different) mutation, whereas the cis configuration (where the mutations lie on different DNA molecules) is wild-type because one allele has two mutations and the other allele has no mutations. The lower part of the figure shows that if the two mutations are in different genes, we always see a wild phenotype. There is always one wild-type and one mutant allele of each gene in both the cis and trans configurations. “Failure to complement” means that two mutations occurred in the same gene. Mutations that do not complement one another are said to comprise part of the same complementation group. Another term used to describe the unit defined by the complementation test is the cistron, which is the same as the gene. Basically these three terms all describe a stretch of DNA that functions as a unit to give rise to an RNA or polypeptide product. The properties of the gene with regard to complementation are explained by the fact that this product is a single molecule that behaves as a functional unit.

FIGURE 1.34 The cistron is defined by the complementation test. Genes are represented by DNA helices; red stars identify sites of mutation.
1.19 Mutations May Cause Loss of Function or Gain of Function
The various possible effects of mutation in a gene are summarized in FIGURE 1.35. In principle, when a gene has been identified, insight into its function can be gained by generating a mutant organism that entirely lacks the gene. A mutation that completely eliminates gene function—usually because the gene has been deleted—is called a null mutation. If a gene is essential to the organism’s survival, a null mutation is lethal when homozygous or hemizygous. Many null mutations might not be lethal but nonetheless disrupt some aspect of the form, growth, or development of the organism, resulting in a specific phenotype.

FIGURE 1.35 Mutations that do not affect protein sequence or function are silent. Mutations that abolish all protein activity are null. Point mutations that cause loss of function are recessive; those that cause gain of function are dominant.
To determine how a gene affects the phenotype, it is essential to characterize the effect of a null mutation. Generally, if a null mutant fails to affect a phenotype, we can safely conclude that the gene function is not essential. Some genes are duplicated or have overlapping functions, though, and loss of function of one of the genes is not sufficient to significantly affect the phenotype. Null mutations, or other mutations that impede gene function (but do not necessarily abolish it entirely), are called loss-of-function mutations. A loss-of-function mutation is recessive (as in the example of Figure 1.33). Loss-of-function mutations that affect protein activity but retain sufficient activity so that the phenotype is not altered are referred to as leaky mutations. Sometimes, a mutation has the opposite effect and causes a protein to acquire a new function or expression pattern; such a change is called a gain-of-function mutation. A gain-of-function mutation is dominant.
Not all mutations in protein-coding genes lead to a detectable change in the phenotype. Mutations without apparent phenotypic effect are called silent mutations. They fall into two categories: (1) base changes in DNA that do not cause any change in the amino acid in the resulting polypeptide (called synonymous mutations); and (2) base changes in DNA that change the amino acid, but the replacement in the polypeptide does not affect its activity (called neutral substitutions).
1.20 A Locus Can Have Many Different Mutant Alleles
If a recessive mutation is produced by every change in a gene that prevents the production of an active protein, there should be a large number of such mutations for any one gene. Many amino acid replacements can change the structure of the protein sufficiently to impede its function.
Different variants of the same gene are called multiple alleles, and their existence makes it possible to generate heterozygotes with two mutant alleles. The relationships between these multiple alleles can take various forms.
In the simplest case, a wild-type allele encodes a polypeptide product that is functional, whereas a mutant allele(s) encodes polypeptides that are nonfunctional. However, there are often cases in which a series of loss-of-function mutant alleles have different, variable phenotypes. For example, wild-type function of the X-linked white locus of Drosophila melanogaster is required for development of the normal red color of the eye. The locus is named for the effect of null mutations that, in homozygous females or hemizygous males, cause the fly to have white eyes.
The wild-type allele is indicated as w+ or just +, and the phenotype is red eyes. An entirely defective form of the gene (white eye phenotype) might be indicated by a “minus” superscript (w–). To distinguish among a variety of mutant alleles with different effects, other superscripts can be introduced, such as wi (ivory eye color) or wa (apricot eye color). Although some alleles produce no visible pigment, and therefore the eyes are white, many alleles produce some color. Therefore, each of these mutant alleles must represent a different mutation of the gene, many of which do not eliminate its function entirely but leave a residual activity that produces a characteristic phenotype.
The w+ allele is dominant over any other allele in heterozygotes and there are many different mutant alleles for this locus. TABLE 1.2 shows a small sample. These alleles are named for the color of the eye in a homozygous female or hemizygous male. (Most w alleles affect the quantity of pigment in the eye. The list of white alleles in the figure is arranged in roughly declining amount of color in the eye pigment, but others, such as wsp, affect the pattern in which pigment is deposited.)
TABLE 1.2 The w locus in Drosophila melanogaster has an extensive series of alleles whose phenotypes extend from wild-type (red) color to complete lack of pigment.
| Allele | Phenotype of Homozygote |
|---|---|
| w+ | Red eye (wild type) |
| wbl | Blood |
| wch | Cherry |
| wbf | Buff |
| wh | Honey |
| wa | Apricot |
| we | Eosin |
| wl | Ivory |
| wz | Zeste (lemon-yellow) |
| wsp | Mottled, color varies |
| w1 | White (no color) |
When multiple alleles exist, an organism might be a heterozygote that carries two different mutant alleles. The phenotype of such a heterozygote depends on the nature of the residual activity of each allele. The relationship between two mutant alleles is, in principle, no different from that between wild-type and mutant alleles: One allele might be dominant, there might be partial dominance, or there might be codominance.
1.21 A Locus Can Have More Than One Wild-Type Allele
In some instances, such as the gene that controls the human ABO blood group system, there is not necessarily a unique wild-type allele for a particular locus. Lack of function is represented by the null, or O, allele. However, the functional alleles A and B are codominant with one another and dominant to the O allele. The basis for this relationship is illustrated in FIGURE 1.36.

FIGURE 1.36 The ABO human blood group locus encodes a galactosyltransferase whose specificity determines the blood group.
The H antigen is generated in all individuals and consists of a particular carbohydrate group that is added to proteins and lipids. The ABO locus encodes a galactosyltransferase enzyme that puts an additional sugar group on the H antigen. The specificity of this enzyme determines the blood group. The A allele produces an enzyme that uses the modified sugar UDP-N-acetylgalactose to form the A antigen. The B allele produces an enzyme that uses the modified sugar UDP-galactose to form the B antigen. The A and B versions of the transferase enzyme differ in four amino acids that presumably affect its ability to catalyze the addition of specific sugars. The O allele has a small deletion that eliminates the activity of the transferase, so no modification of the H antigen occurs.
This explains why A and B alleles are dominant in the AO and BO heterozygotes: The corresponding transferase activity forms the A or B antigen. The A and B alleles are codominant in AB heterozygotes because both transferase activities are expressed. The OO homozygote is a null that has neither activity and therefore lacks both A and B antigens.
Neither A nor B alleles can be regarded as uniquely wild type because they represent alternative activities rather than loss or gain of function. A situation such as this—that is, there are multiple functional alleles in a population—is described as a polymorphism (see the chapter titled The Content of the Genome).
1.22 Recombination Occurs by Physical Exchange of DNA
The term genetic recombination describes the generation of new combinations of alleles at each generation in diploid organisms. This arises because the two homologous copies of each chromosome might have different alleles at some loci. By the exchange of corresponding segments between the homologs, called crossing over, recombinant chromosomes that are different from the parental chromosomes can be generated.
Recombination results from a physical exchange of chromosomal material. For example, recombination might result from the crossing over that occurs when homologous chromosomes align during meiosis (the specialized division that produces haploid gametes). Meiosis begins with a cell that has duplicated its chromosomes so that it has four copies of each chromatid (the two homologous chromosomes and their identical [sister] copies that remain joined after duplication). Early in meiosis, all four chromatids are closely associated (synapsed) in a structure called a bivalent and, later, a tetrad. At this point, pairwise exchanges of material between two nonidentical (nonsister) chromatids (of the four total) can occur.
The point of synapsis between homologs is called a chiasma; this is illustrated diagrammatically in FIGURE 1.37. A chiasma represents a site at which one DNA strand in each of two nonsister chromatids in a tetrad has been broken and exchanged. If during the resolution of the chiasma the previously unbroken strands are also broken and exchanged, recombinant chromatids will be generated. Each recombinant chromatid consists of material derived from one chromatid on one side of the chiasma, with material from the other chromatid on the opposite side. The two recombinant chromatids have reciprocal structures. The event is described as a “breakage and reunion.” Because each individual crossing-over event involves only two of the four associated chromatids, a single recombination event can produce only 50% recombinants.

FIGURE 1.37 Chiasma formation at Prophase I of meiosis is responsible for generating recombinant chromosomes.
The complementarity of the two strands of DNA is essential for the recombination process. Each of the chromatids shown in Figure 1.36 consists of a very long duplex of DNA. For them to be broken and reconnected without any loss of material requires a mechanism to recognize and align at exactly corresponding positions; this mechanism is complementary base pairing.
Recombination results from a process in which the single strands in the region of the crossover exchange their partners, resulting in a branch that might migrate for some distance in either direction. FIGURE 1.38 shows that this creates a stretch of heteroduplex DNA in which the single strand of one duplex is paired with its complement from the other duplex. Each duplex DNA corresponds to one of the chromatids involved in recombination in Figure 1.37. The mechanism, of course, involves other stages in which strands must be broken and religated, which we discuss in more detail in the chapter titled Homologous and Site-Specific Recombination, but the crucial feature that makes precise recombination possible is the complementarity of DNA strands. Figure 1.38 shows only some stages of the reaction, but we see that a stretch of heteroduplex DNA forms in the recombination intermediate when a single strand crosses over from one duplex to the other. Each recombinant consists of one parental duplex DNA at the left, which is connected by a stretch of heteroduplex DNA to the other parental duplex at the right.

FIGURE 1.38 Recombination involves pairing between complementary strands of the two parental duplex DNAs.
The formation of heteroduplex DNA requires the sequences of the two recombining duplexes to be close enough to allow pairing between the complementary strands. If there are no differences between the two parental genomes in this region, formation of heteroduplex DNA will be perfect. However, pairing can still occur even when there are small differences. In this case, the heteroduplex DNA has points of mismatch, at which a base in one strand is paired with a base in the other strand that is not complementary to it. The correction of such mismatches is another feature of genetic recombination (see the chapter titled Repair Systems).
Over chromosomal distances, recombination events occur more or less at random with a characteristic frequency. The probability that a crossover will occur within any specific region of the chromosome is more or less proportional to the length of the region, up to a saturation point. For example, a large human chromosome usually has three or four crossover events per meiosis, whereas a small chromosome might have only one on average.
FIGURE 1.39 compares recombination frequencies in three situations: two genes on different chromosomes, two genes that are far apart on the same chromosome, and two genes that are close together on the same chromosome. Genes on different chromosomes segregate independently according to Mendel’s principles, resulting in the production of 50% “parental” types and 50% “recombinant” types during meiosis. When genes are sufficiently far apart on the same chromosome, the probability of at least one crossover in the region between them becomes so high that their association is the same as that of genes on different chromosomes and they show 50% recombination.

FIGURE 1.39 Genes on different chromosomes segregate independently so that all possible combinations of alleles are produced in equal proportions. Crossing over occurs so frequently between genes that are far apart on the same chromosome that they effectively segregate independently. But recombination is reduced when genes are closer together, and for adjacent genes it might hardly ever occur.
When genes are close together, though, the probability of a crossover between them is reduced, and recombination occurs only in some proportion of meioses. For example, if it occurs in one-quarter of the meioses, the overall rate of recombination is 12.5% (because a single recombination event produces 50% recombination, and this occurs in 25% of meioses). When genes are very close together, as shown in the bottom panel of Figure 1.39, recombination between them might never be observed in phenotypes of multicellular eukaryotes (because they produce few offspring).
This leads us to the concept that a chromosome is an array of many genes. Each protein-coding gene is an independent unit of expression and is represented in one or more polypeptide chains. The properties of a gene can be changed by mutation. The allelic combinations present on a chromosome can be changed by recombination. We can now ask, “What is the relationship between the sequence of a gene and the sequence of the polypeptide chain it encodes?”
1.23 The Genetic Code Is Triplet
Each protein-coding gene encodes a particular polypeptide chain (or chains). The concept that each polypeptide consists of a particular series of amino acids dates from Sanger’s characterization of insulin in the 1950s. The discovery that a gene consists of DNA presents us with the issue of how a sequence of nucleotides in DNA is used to construct a sequence of amino acids in protein.
The sequence of nucleotides in DNA is important not because of its structure per se, but because it encodes the sequence of amino acids that constitutes the corresponding polypeptide. The relationship between a sequence of DNA and the sequence of the corresponding polypeptide is called the genetic code.
The structure and/or enzymatic activity of each protein follows from its primary sequence of amino acids and its overall conformation, which is determined by interactions between the amino acids. By determining the sequence of amino acids in each protein, the gene is able to carry all the information needed to specify an active polypeptide chain. In this way, the thousands of genes found in the genome of a complex organism are able to direct the synthesis of many thousands of polypeptide types in a cell.
Together, the various proteins of a cell undertake the catalytic and structural activities that are responsible for establishing its phenotype. Of course, in addition to sequences that encode proteins, DNA also contains certain control sequences that are recognized by regulator molecules, usually proteins. Here, the function of the DNA is determined by its sequence directly, not via any intermediary molecule. Both types of sequence—genes expressed as proteins and sequences recognized by proteins—constitute genetic information.
The coding region of a gene is deciphered by a complex apparatus that interprets the nucleic acid sequence; this apparatus is essential if the information carried in DNA is to have meaning. The initial step in the interpretation of the genetic code is to copy DNA into RNA. In any particular region it is usually the case that only one of the two strands of DNA encodes a functional RNA, so we write the genetic code as a sequence of bases (rather than base pairs). (Recent evidence suggests that both strands are transcribed in some regions, but in most cases it is not clear that both resulting transcripts have functional importance.)
A coding sequence is read in groups of three nucleotides, each group representing one amino acid. Each trinucleotide sequence is called a codon. A gene includes a series of codons that is read sequentially from a starting point at one end to a termination point at the other end. Written in the conventional 5′ to 3′ direction, the nucleotide sequence of the DNA strand that encodes a polypeptide corresponds to the amino acid sequence of the polypeptide written in the direction from N-terminus to C-terminus.
A coding sequence is read in nonoverlapping triplets from a fixed starting point:
Nonoverlapping implies that each codon consists of three nucleotides and that successive codons are represented by successive trinucleotides. An individual nucleotide is part of only one codon.
The use of a fixed starting point means that assembly of a polypeptide must begin at one end and work to the other, so that different parts of the coding sequence cannot be read independently.
The nature of the code predicts that two types of mutations, base substitution and base insertion/deletion, will have different effects. If a particular sequence is read sequentially, such as
UUU AAA GGG CCC (codons)
aa1 aa2 aa3 aa4 (amino acids; the number reflects different types of amino acids, not position)
a nucleotide substitution, or point mutation, will affect only one amino acid. For example, the substitution of an A by some other base (X) causes aa2 to be replaced by aa5
UUU AAX GGG CCC
aa1 aa5 aa3 aa4
because only the second codon has been changed.
However, a mutation that inserts or deletes a single nucleotide will change the triplet sets for the entire subsequent sequence. A change of this sort is called a frameshift. An insertion might take the following form:
UUU AAX AGG GCC C
aa1 aa5 aa6 aa7
Because the new sequence of triplets is completely different from the old one, the entire amino acid sequence of the polypeptide is altered downstream from the site of mutation, so the function of the protein is likely to be lost completely.
Frameshift mutations are induced by the acridines, compounds that bind to DNA and distort the structure of the double helix, causing additional bases to be incorporated or omitted during replication. Each mutagenic event in the presence of an acridine results in the addition or removal of a single base pair.
If an acridine mutant is produced by, say, the addition of a nucleotide, it should revert to wild type by deletion of the nucleotide. However, reversion also can be caused by deletion of a different base at a site close to the first. Combinations of such mutations provided revealing evidence about the nature of the genetic code, as is discussed in a moment.
FIGURE 1.40 illustrates the properties of frameshift mutations. An insertion or deletion changes the entire polypeptide sequence following the site of mutation. However, the combination of an insertion and a deletion of the same number of nucleotides causes the code to be read incorrectly only between the two sites of mutation; reading in the original frame resumes after the second site.

FIGURE 1.40 Frameshift mutations show that the genetic code is read in triplets from a fixed starting point.
In a 1961 experiment by Francis Crick, Leslie Barnett, Sydney Brenner, and R. J. Watts-Tobin, genetic analysis of acridine mutations in the rII region of the phage T4 showed that all the mutations could be classified into one of two sets, described as (+) and (−). Either type of mutation by itself causes a frameshift: the (+) type by virtue of a base addition, and the (−) type by virtue of a base deletion. Double mutant combinations of the types (+ +) and (− −) continue to show mutant behavior. However, combinations of the types (+ −) and (− +) suppress one another so that one mutation is described as a frameshift suppressor of the other. (In the context of this work, “suppressor” is used in an unusual sense because the second mutation is in the same gene as the first; in fact, these are second-site reversions.)
These results show that the genetic code must be read as a sequence that is fixed by the starting point. Therefore, a single nucleotide addition and deletion compensate for each other, whereas double additions or double deletions remain mutant. However, these observations do not suggest how many nucleotides make up each codon.
When triple mutants are constructed, only (+ + +) and (− − −) combinations show the wild-type phenotype, whereas other combinations remain mutant. If we take three single nucleotide additions or three deletions to correspond respectively to the addition or omission overall of a single amino acid, this implies that the code is read in triplets. An incorrect amino acid sequence is found between the two outside sites of mutation and the sequence on either side remains wild type, as indicated in Figure 1.40.
1.24 Every Coding Sequence Has Three Possible Reading Frames
If the genetic code is read in nonoverlapping triplets, there are three possible ways of translating any nucleotide sequence into polypeptide, depending on the starting point. These are called reading frames. For the sequence
A C G A C G A C G A C G A C G A C G
the three possible reading frames are
ACG ACG ACG ACG ACG ACG ACG
CGA CGA CGA CGA CGA CGA CGA
GAC GAC GAC GAC GAC GAC GAC
A reading frame that consists exclusively of triplets encoding amino acids is called an open reading frame (ORF). A sequence that is translated into polypeptide has a reading frame that begins with a special initiation codon (AUG) and then extends through a series of triplets encoding amino acids until it ends at one of three termination codons (UAA, UAG, or UGA).
A reading frame that cannot be read into polypeptide because termination codons occur frequently is said to be closed, or blocked. If a sequence is closed in all three reading frames, it cannot have the function of encoding polypeptide.
When the sequence of a DNA region of unknown function is obtained, each possible reading frame can be analyzed to determine whether it is open or closed. Usually no more than one of the three possible reading frames is open in any single stretch of DNA. FIGURE 1.41 shows an example of a sequence that can be read in only one reading frame because the alternative reading frames are closed by frequent termination codons. A long ORF is unlikely to exist by chance; if it had not been translated into polypeptide, there would have been no selective pressure to prevent the accumulation of termination codons. Therefore, the identification of a lengthy open reading frame is taken to be prima facie evidence that the sequence is (or until recently has been) translated into a polypeptide in that frame. An ORF for which no protein product has been identified is sometimes called an unidentified reading frame (URF).
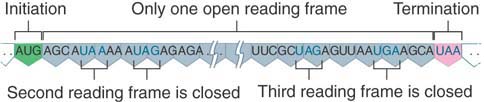
FIGURE 1.41 An open reading frame starts with AUG and continues in triplets to a termination codon. Closed reading frames can be interrupted frequently by termination codons.
1.25 Bacterial Genes Are Colinear with Their Products
By comparing the nucleotide sequence of a gene with the amino acid sequence of its polypeptide product, we can determine whether the gene and the polypeptide are colinear—that is, whether the sequence of nucleotides in the gene exactly corresponds to the sequence of amino acids in the polypeptide. In bacteria and their viruses, genes and their products are colinear. Each gene is a continuous stretch of DNA with a coding region that is three times the number of amino acids in the polypeptide that it encodes (due to the triplet nature of the genetic code). In other words, if a polypeptide contains N amino acids, the gene encoding that polypeptide contains 3N nucleotides.
The equivalence of the bacterial gene and its product means that a physical map of DNA will exactly match an amino acid map of the polypeptide. How well do these maps match the recombination map?
The colinearity of gene and polypeptide was originally investigated in the tryptophan synthetase gene of E. coli. Genetic distance was measured by the percentage of recombination between variable sites in the DNA; amino acid distance was measured as the number of amino acids separating sites of amino acid replacement. FIGURE 1.42 compares the two maps; the wild-type protein sequence is illustrated on top, highlighting the seven amino acids that were replaced in the mutant protein (shown below). The order of seven variable sites is the same as the order of the corresponding sites of amino acid replacement, and the recombination distances are roughly similar to the actual distances in the protein. The recombination map expands the distances between some variable sites, but otherwise there is little distortion of the recombination map relative to the physical map.

FIGURE 1.42 The recombination map of the tryptophan synthetase gene corresponds with the amino acid sequence of the polypeptide.
The recombination map leads to two further general points about the organization of the gene. Different mutations can cause a wild-type amino acid to be replaced with different alternatives. If two such mutations cannot recombine, they must involve different point mutations at the same position in DNA. If the mutations can be separated on the genetic map but affect the same amino acid on the upper map (the connecting lines converge in the figure), they must involve point mutations at different positions in the same codon. This happens because the unit of genetic recombination (1 bp) is smaller than the unit encoding the amino acid (3 bp).
1.26 Several Processes Are Required to Express the Product of a Gene
In comparing a gene and its polypeptide product, we are restricted to the sequence of DNA that lies between the points corresponding to the N-terminus and C-terminus of the polypeptide. However, a gene is not directly translated into polypeptide but is expressed via the production of a messenger RNA (mRNA), a nucleic acid intermediate actually used to synthesize a polypeptide (as we see in detail in the chapter titled Translation).
Messenger RNA is synthesized by the same process of complementary base pairing used to replicate DNA, with the important difference that it corresponds to only one strand of the DNA double helix. FIGURE 1.43 shows that the sequence of mRNA is complementary to the sequence of one strand of DNA—called the antisense (or template) strand—and is identical (apart from the replacement of T with U) to the other strand of DNA—called the coding (or sense) strand. The convention for writing DNA sequences is that the top strand is the coding strand and runs 5′ to 3′.

FIGURE 1.43 RNA is synthesized by using one strand of DNA as a template for complementary base pairing.
The process by which information from a gene is used to synthesize an RNA or polypeptide product is called gene expression. In bacteria, expression of a structural gene consists of two stages. The first stage is transcription, when an mRNA copy of the coding strand of the DNA is produced. The second stage is translation of the mRNA into a polypeptide. This is the process by which the sequence of an mRNA is read in triplets to give the series of amino acids that make the corresponding polypeptide.
An mRNA includes a sequence of nucleotides that contain the codons for the amino acids in the polypeptide. This part of the nucleic acid is called the coding region. However, the mRNA includes additional sequences on either end that do not encode amino acids. The 5′ untranslated region is called the leader, or 5′ UTR, and the 3′ untranslated region is called the trailer, or 3′ UTR. These UTRs are important for mRNA stability and translation.
The gene includes the entire sequence represented in mRNA, including the UTRs. Sometimes, mutations impeding gene function are found in the additional, noncoding regions, confirming the view that these comprise a legitimate part of the genetic unit. FIGURE 1.44 illustrates this situation, in which the gene is considered to comprise a continuous stretch of DNA needed to produce a particular polypeptide, including the 5′ UTR, the coding region, and the 3′ UTR.

FIGURE 1.44 The gene is usually longer than the sequence encoding the polypeptide.
A bacterial cell has only a single compartment, so transcription and translation occur in the same place and are concurrent, as illustrated in FIGURE 1.45. In eukaryotes, transcription occurs in the nucleus, but the mRNA product must be transported to the cytoplasm in order to be translated. This results in a spatial separation between transcription (in the nucleus) and translation (in the cytoplasm). However, for eukaryotic genes, the primary transcript of the gene is a pre-mRNA that requires processing to generate the mature mRNA. The basic stages of gene expression in a eukaryote are outlined in FIGURE 1.46.

FIGURE 1.45 Transcription and translation take place in the same compartment in bacteria.

FIGURE 1.46 In eukaryotes, transcription occurs in the nucleus and translation occurs in the cytoplasm.
The most important stage in RNA processing is splicing. Many genes in eukaryotes (and a majority in multicellular eukaryotes) contain regions of noncoding sequence embedded in coding sequence; these internal DNA sequences are initially transcribed but are excised and are not present in the mature mRNA. These excised sequences are referred to as introns. The remaining sequences are joined together. The sequences that are transcribed, retained, and joined in the mature mRNA are called exons. Other processing events that occur at this stage involve the modification of the 5′ and 3′ ends of the pre-mRNA.
Translation of the mature mRNA into a polypeptide is accomplished by a complex apparatus that includes both protein and RNA components. The actual “machine” that undertakes the process is the ribosome, a large complex that includes some large RNAs—ribosomal RNAs (rRNAs)—and many small proteins. The process of recognizing which amino acid corresponds to a particular nucleotide triplet requires an intermediate transfer RNA (tRNA); there is at least one tRNA species for every amino acid. Many ancillary proteins are involved. We describe translation in the chapter titled Translation, but note for now that the ribosomes are the large structures in Figure 1.45 that translate the mRNA.
It is an important point to note that the process of gene expression involves RNA not only as the essential substrate but also in providing components of the apparatus. The rRNA and tRNA components are encoded by genes and are generated by the process of transcription (like mRNA), but they are not translated to polypeptide. In addition, there are RNAs (e.g., snRNA and microRNAs) that do not encode polypeptides but are nonetheless essential for gene expression.
1.27 Proteins Are trans-Acting but Sites on DNA Are cis-Acting
A crucial progression in the definition of the gene was the realization that all of its parts must be present on one contiguous stretch of DNA. In genetic terminology, sites that are located on the same DNA are said to be in cis. Sites that are located on two different molecules of DNA are described as being in trans. So two mutations might be in cis (on the same DNA) or in trans (on different DNAs). The complementation test uses this concept to determine whether two mutations are in the same gene (see the section Mutations in the Same Gene Cannot Complement earlier in this chapter). We can now extend the concept of the difference between cis and trans effects from defining the coding region of a gene to describing the interaction between a gene and its regulatory elements.
Suppose that the ability of a gene to be expressed is controlled by a protein that binds to the DNA close to the coding region. In the example depicted in FIGURE 1.47, RNA can be synthesized only when the protein is bound to a control site on the DNA. Now, suppose that a mutation occurs in the control site so that the protein can no longer bind to it. As a result, the gene can no longer be expressed.

FIGURE 1.47 Control sites in DNA provide binding sites for proteins; coding regions are expressed via the synthesis of RNA.
Gene expression can be inactivated either by a mutation in a control site or by a mutation in a coding region. The mutations cannot be distinguished genetically because both have the property of acting only on the DNA sequence of the single allele in which they occur. They have identical properties in the complementation test, so a mutation in a control region is defined as comprising part of the gene in the same way as a mutation in the coding region.
FIGURE 1.48 shows that a deficiency in the control site affects only the coding region to which it is connected; it does not affect the ability of the homologous allele to be expressed. A mutation that acts solely by affecting the properties of the contiguous sequence of DNA is called cis-acting. It should be noted that in many eukaryotes the control region can influence the expression of DNA at some distance, but nonetheless the control region is on the same DNA molecule as the coding sequence.

FIGURE 1.48 A cis-acting site controls expression of the adjacent DNA but does not influence the homologous allele.
We can contrast the behavior of the cis-acting mutation shown in Figure 1.47 with the result of a mutation in the gene encoding the regulatory protein. FIGURE 1.49 shows that the absence of regulatory protein would prevent both alleles from being expressed. A mutation of this sort is said to be trans-acting.

FIGURE 1.49 A trans-acting mutation in a gene for a regulatory protein affects both alleles of a gene that it controls.
Reversing the argument, if a mutation is trans-acting, we know that its effects must be exerted through some diffusible product (either a protein or a regulatory RNA) that acts on multiple targets within a cell. However, if a mutation is cis-acting, it must function by directly affecting the properties of the contiguous DNA, which means that it is not expressed in the form of RNA or protein but instead is some alteration in the DNA of the control region itself.
Summary
Two classic experiments provided strong evidence that DNA is the genetic material of bacteria, viruses, and eukaryotic cells. DNA isolated from one strain of Pneumococcus bacteria can confer properties of that strain upon another strain. In addition, DNA is the only component that is inherited by progeny phages from parental phages. We can use DNA to transfect new properties into eukaryotic cells.
DNA is a double helix consisting of anti-parallel strands in which the nucleotide units are linked by 5′ to 3′ phosphodiester bonds. The backbone is on the exterior; purine and pyrimidine bases are stacked in the interior in pairs in which A is complementary to T, and G is complementary to C. In semiconservative replication, the two strands separate and both are used as templates for the assembly of daughter strands by complementary base pairing. Complementary base pairing is also used to transcribe an RNA from one strand of a DNA duplex.
A stretch of DNA can encode a polypeptide. The genetic code describes the relationship between the sequence of DNA and the sequence of the polypeptide. In general, only one of the two strands of DNA encodes a polypeptide.
A mutation consists of a change in the sequence of A-T and G-C base pairs in DNA. A mutation in a coding sequence can change the sequence of amino acids in the corresponding polypeptide. Point mutations can be reverted by back mutation of the original mutation. Insertions can revert by loss of the inserted material, but deletions cannot revert. Mutations can also be suppressed indirectly when a mutation in a different gene counters the original defect.
The natural incidence of mutations is increased by mutagens. Mutations can be concentrated at hotspots. A type of hotspot responsible for some point mutations is caused by deamination of the modified base 5-methylcytosine. Forward mutations occur at a rate of about 10−6 per locus per generation; back mutations are rarer.
Although all genetic information in cells is carried by DNA, viruses have genomes of double-stranded or single-stranded DNA or RNA. Viroids are subviral pathogens that consist solely of small molecules of RNA with no protective packaging. The RNA does not code for protein and its mode of perpetuation and of pathogenesis is unknown. Scrapie results from a proteinaceous infectious agent, or prion.
A chromosome consists of an uninterrupted length of duplex DNA that contains many genes. Each gene (or cistron) is transcribed into an RNA product, which in turn is translated into a polypeptide sequence if it is a structural gene. An RNA or protein product of a gene is said to be trans-acting. A gene is defined as a unit of a single stretch of DNA by the complementation test. A site on DNA that regulates the activity of an adjacent gene is said to be cis-acting.
When a gene encodes a polypeptide, the relationship between the sequence of DNA and sequence of the polypeptide is given by the genetic code. Only one of the two strands of DNA encodes polypeptide. A codon consists of three nucleotides that represent a single amino acid. A coding sequence of DNA consists of a series of codons, read from a fixed starting point and nonoverlapping. Usually only one of the three possible reading frames can be translated into polypeptide.
A gene can have multiple alleles. Recessive alleles are caused by loss-of-function mutations that interfere with the function of the protein. A null allele has total loss of function. Dominant alleles are caused by gain-of-function mutations that create a new property in the protein.
References
1.1 Introduction
Reviews
Cairns, J., Stent, G., and Watson, J. D. 1966. Phage and the Origins of Molecular Biology. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
Judson, H. 1978. The Eighth Day of Creation. Knopf, New York.
Olby, R. 1974. The Path to the Double Helix. Macmillan, London.
1.2 DNA Is the Genetic Material of Bacteria and Viruses
Research
Avery, O. T., MacLeod, C. M., and McCarty, M. (1944). Studies on the chemical nature of the substance inducing transformation of pneumococcal types. J. Exp. Med. 98, 451–460.
Griffith, F. (1928). The significance of pneumococcal types. J. Hyg. 27, 113–159.
Hershey, A. D., and Chase, M. (1952). Independent functions of viral protein and nucleic acid in growth of bacteriophage. J. Gen. Physiol. 36, 39–56.
1.3 DNA Is the Genetic Material of Eukaryotic Cells
Research
Pellicer, A., Wigler, M., Axel, R., and Silverstein, S. (1978). The transfer and stable integration of the HSV thymidine kinase gene into mouse cells. Cell 14, 133–141.
1.6 DNA Is a Double Helix
Review
Watson, J. D. 1981. The Double Helix: A Personal Account of the Discovery of the Structure of DNA (Norton Critical Editions). W. W. Norton, New York.
Research
Franklin, R. E., and Gosling, R. G. (1953). Molecular configuration in sodium thymonucleate. Nature 171, 740–741.
Watson, J. D., and Crick, F. H. C. (1953). A structure for DNA. Nature 171, 737–738.
Watson, J. D., and Crick, F. H. C. (1953). Genetic implications of the structure of DNA. Nature 171, 964–967.
Wilkins, M. F. H., Stokes, A. R., and Wilson, H. R. (1953). Molecular structure of deoxypentose nucleic acids. Nature 171, 738–740.
1.7 DNA Replication Is Semiconservative
Review
Holmes, F. (2001). Meselson, Stahl, and the Replication of DNA: A History of the Most Beautiful Experiment in Biology. Yale University Press, New Haven, CT.
Research
Meselson, M., and Stahl, F. W. (1958). The replication of DNA in E. coli. Proc. Natl. Acad. Sci. USA. 44, 671–682.
1.11 Mutations Change the Sequence of DNA
Reviews
Drake, J. W. (1991). A constant rate of spontaneous mutation in DNA-based microbes. Proc. Natl. Acad. Sci. USA. 88, 7160–7164.
Drake, J. W., and Balz, R. H. (1976). The biochemistry of mutagenesis. Annu. Rev. Biochem. 45, 11–37.
Research
Drake, J. W., Charlesworth, B., Charlesworth, D., and Crow, J. F. (1998). Rates of spontaneous mutation. Genetics 148, 1667–1686.
Grogan, D. W., Carver, G. T., and Drake, J. W. (2001). Genetic fidelity under harsh conditions: analysis of spontaneous mutation in the thermoacidophilic archaeon Sulfolobus acidocaldarius. Proc. Natl. Acad. Sci. USA. 98, 7928–7933.
1.12 Mutations Can Affect Single Base Pairs or Longer Sequences
Review
Maki, H. (2002). Origins of spontaneous mutations: specificity and directionality of base-substitution, frameshift, and sequence-substitution mutageneses. Annu. Rev. Genet. 36, 279–303.
1.14 Mutations Are Concentrated at Hotspots
Research
Coulondre, C., et al. (1978). Molecular basis of base substitution hotspots in E. coli. Nature 274, 775–780.
Millar, C. B., Guy, J., Sansom, O. J., Selfridge, J., MacDougall, E., Hendrich, B., Keightley, P. D., Bishop, S. M., Clarke, A. R., and Bird, A. (2002). Enhanced CpG mutability and tumorigenesis in MBD4-deficient mice. Science 297, 403–405.
1.16 Some Hereditary Agents Are Extremely Small
Reviews
Diener, T. O. (1986). Viroid processing: a model involving the central conserved region and hairpin. Proc. Natl. Acad. Sci. USA 83, 58–62.
Diener, T. O. (1999). Viroids and the nature of viroid diseases. Arch. Virol. Suppl. 15, 203–220.
Prusiner, S. B. (1998). Prions. Proc. Natl. Acad. Sci. USA 95, 13363–13383.
Research
Bueler, H., et al. (1993). Mice devoid of PrP are resistant to scrapie. Cell 73, 1339–1347.
McKinley, M. P., Bolton, D. C., and Prusiner, S. B. (1983). A protease-resistant protein is a structural component of the scrapie prion. Cell 35, 57–62.
1.23 The Genetic Code Is Triplet
Review
Roth, J. R. (1974). Frameshift mutations. Annu. Rev. Genet. 8, 319–346.
Research
Benzer, S., and Champe, S. P. (1961). Ambivalent rII mutants of phage T4. Proc. Natl. Acad. Sci. USA 47, 403–416.
Crick, F. H. C., Barnett, L., Brenner, S., and Watts-Tobin, R. J. (1961). General nature of the genetic code for proteins. Nature 192, 1227–1232.
1.25 Bacterial Genes Are Colinear with Their Products
Research
Yanofsky, C., Drapeau, G. R., Guest, J. R., and Carlton, B. C. (1967). The complete amino acid sequence of the tryptophan synthetase A protein (μ subunit) and its colinear relationship with the genetic map of the A gene. Proc. Natl. Acad. Sci. USA 57, 2966–2968.
Yanofsky, C., et al. (1964). On the colinearity of gene structure and protein structure. Proc. Natl. Acad. Sci. USA 51, 266–272.