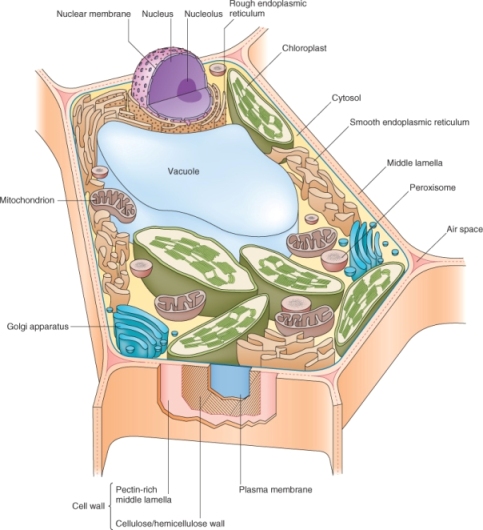
Figure 4.1 A leaf mesophyll cell. Note that the relative volume of the central vacuole shown here underestimates its actual size in a mature mesophyll cell. The plasmodesmata that connect the protoplasts of neighboring cells are not shown.
Chapter 4
Cell architecture
Living things consist of cells, distinct structural units that house the machinery that makes life possible. Plants have eukaryotic cells; a generalized plant cell is shown in Figure 4.1. Unlike animal cells, plant cells are surrounded by an extracellular coat, the cell wall. Within the cell wall lies the living protoplast, whose outer boundary is the plasma membrane that contains a variety of membrane-bound organelles, compartments with specialized functions.
Figure 4.1 A leaf mesophyll cell. Note that the relative volume of the central vacuole shown here underestimates its actual size in a mature mesophyll cell. The plasmodesmata that connect the protoplasts of neighboring cells are not shown.

Advances in our understanding of cell structure have come from developments in microscopy as well as in chemistry and molecular biology. Although electron microscopes were widely used to study cell structure in the mid-20th century, recent developments in tissue preparation have greatly expanded the scope of electron microscopy (EM). These advances include the use of rapid freezing and freeze-substitution methods that preserve cell structure at conditions nearer to those found in vivo because harsh chemical fixation procedures are avoided. Freeze substitution also preserves the antigenicity of proteins, allowing the use of antibodies to detect proteins at the EM level.
There have been recent dramatic advances in light microscopy as well, especially with laser scanning confocal microscopy which allows details of structure of living cells to be viewed at the three-dimensional level. The power of light microscopy has also been enhanced by the development of reporter molecules that allow the detection of a range of biologically important ions and macromolecules. These reporters include fluorescent dyes that are specific for a variety of molecules such as Ca2+ and H+ as well as fluorescent proteins such as green fluorescent protein (GFP) whose expression can be engineered in cells after fusion to an appropriate promoter or target gene.
Advances in chemistry have also been crucial in enhancing our understanding of plant cell structure, especially as it pertains to the cell wall. The plant cell wall is a complex of polysaccharide, protein and lignin polymers whose structures are being resolved, especially through advances in mass spectrometry. Mass spectrometry allows the identification of cell wall sugars and phenolic compounds and this information is beginning to reveal how polysaccharides and lignins are assembled.
In this chapter we will address the cellular structures that are present in most living plant cells. The chemical compositions of many of these structures, a topic integral both to plant biology and to animal and human nutrition, are discussed in Chapters 2 and 5.
A strong, flexible primary cell wall composed of complex polysaccharides and proteins surrounds each plant cell. The shape of a plant cell and eventually of the plant itself is dictated by its cell wall, and this shape is important for the cell's function (Figure 4.2). The cell wall prevents excess water uptake by living cells, allowing plants to survive in fresh water. Adjacent plant cells are glued together along their abutting walls by a layer called the middle lamella so that the cells in a plant do not normally move relative to each other (Figure 4.3). Together, all the cell walls form a network, much like a skeleton, that helps hold the plant upright and gives each plant its characteristic shape or morphology. The cell wall is dynamic, changing its structure and composition throughout the life of the cell and rapidly increasing in surface area during cell expansion. In living cells, the cell wall constrains the rate and direction of growth and exerts a profound influence on plant development and morphology. Finally, during differentiation, many cells manufacture a more rigid secondary cell wall, producing complex structures uniquely suited to the cell's function.
Figure 4.2 Scanning electron micrographs showing cell wall architecture of a selection of developing plant cells. (A) Spongy parenchyma (P) of Zinnia leaves showing large airspaces (AS) between cells that facilitate gas exchange. (B) Epidermal cells of snapdragon (Antirrhinum majus) petals that reflect light efficiently to enhance brightness. (C) Xylem tracheid with radial thickenings that strengthen the wall in these water-conducting cells which function under extreme tension.
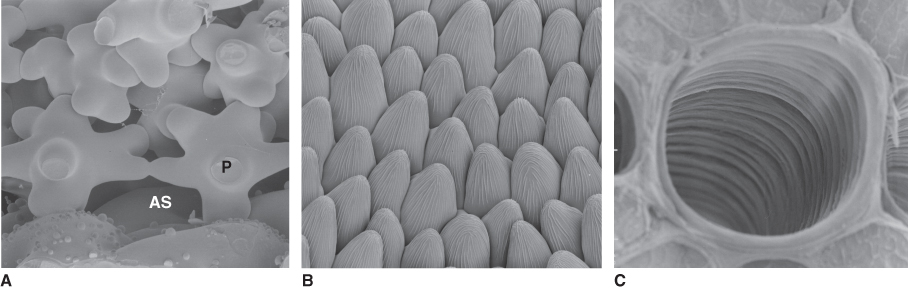
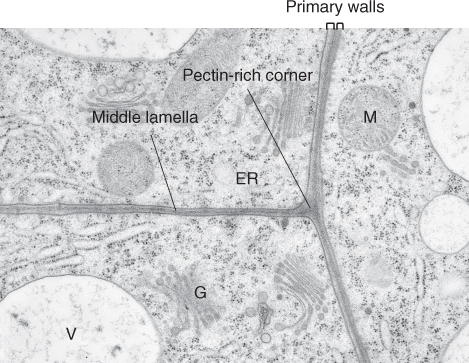
Figure 4.3 Transmission electron microscopy (TEM) image of the corners of three abutting cells. The primary walls of these cells are glued together by the middle lamella, which is formed during cell division and grows with the primary wall during cell expansion. The cell corners are often filled with pectin-rich polysaccharides; in older cells, the material in the cell corners is sometimes degraded and an airspace forms (not shown). ER, endoplasmic reticulum; G, Golgi apparatus; M, mitochondrion; V, vacuole.
The primary cell wall is made up of several structurally independent but interacting networks. The fundamental framework of the wall is polysaccharide, consisting of cellulose rods connected by cross-linking glycans. This network may be embedded in a matrix of pectic polysaccharides. A third independent, non-polysaccharide network can consist of structural proteins or phenylpropanoids. We will describe each of these networks.
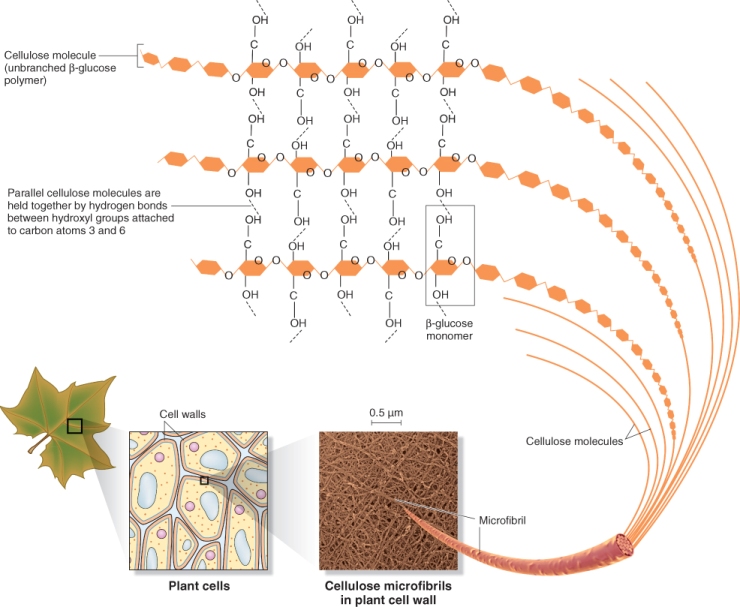
Cellulose forms the basic scaffold of cell walls. Cellulose is the most abundant plant polysaccharide, accounting for 15–30% of the dry mass of all primary cell walls and an even larger percentage of secondary walls. Cellulose molecules are linear chains of 2000–20 000 (1 → 4)-β-D-linked glucose units (see Chapter 2). In cell walls, cellulose exists as microfibrils, paracrystalline arrays of many cellulose molecules, hydrogen bonded to one another along their lengths (Figure 4.4). On average, each microfibril is 36 individual cellulose chains in diameter. Because the individual cellulose chains begin and end at different places within a microfibril, a microfibril can contain thousands of chains and be hundreds of micrometers long.
Figure 4.4 Arrangement of cellulose in cell walls. Linear cellulose molecules hydrogen bond to each other to form microfibrils. A microfibril is usually made up of about 36 cellulose molecules. Note that only the hydroxyl groups at positions 3 and 6 are shown on the glucose molecules in this figure.
Callose, another polymer of glucose, is a major component of the walls of certain types of cells and at certain times during development. It differs from cellulose because its glucose units have a (1 → 3)-β-D-linkage, which gives the molecule a helical rather than a linear shape. As a result, callose molecules do not form microfibrils because they do not hydrogen bond along their length as do linear cellulose chains. Callose is produced in the cell walls of pollen grains, in elongating pollen tubes, in the cell plates of dividing cells, and in cells that have been wounded mechanically or attacked by pathogenic fungi.
Cellulose microfibrils form a mesh that is held together by a class of cell wall polysaccharides called cross-linking glycans, which form hydrogen bonds with cellulose and with each other. They coat the cellulose microfibrils, link to other glycans, and span the distance between microfibrils to form the fundamental structural network of the cell wall. In the past, cross-linking glycans were called ‘hemicelluloses’, a term that refers to cell wall material that can be extracted with strong alkali.
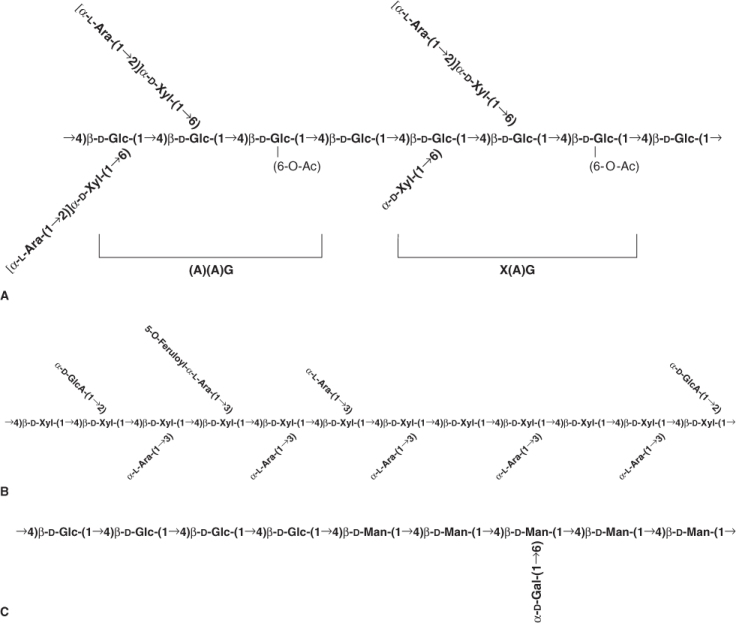
The two major cross-linking glycans of all primary cell walls of flowering plants are xyloglucans (XyGs) and glucuronoarabinoxylans (GAXs). Xyloglucans consist of linear chains of (1 → 4)-β-D-glucose with numerous α-D-xylose units linked at regular intervals at carbon 6 of the glucose units (Figure 4.5A). They may also have side linkages to other sugars. Glucuronoarabinoxylans are linear chains of (1 → 4)-β-D-xylose with side linkages to glucuronic acid and arabinose (Figure 4.5B). In addition to these major classes of cross-linking glycans, there are other less abundant polysaccharides, such as glucomannans, galactomannans and galactoglucomannans, which may interlock the microfibrils in some primary walls. These mannans are found in virtually all angiosperms examined.
Figure 4.5 Examples of cross-linking glycans from primary cell walls. (A) Solanaceous arabinoxyloglucan, a xyloglycan, with a backbone of β(1 → 4)-linked glucose and side-chains of xylose and arabinose. (B) A commelinoid glucuronoarabinoxylan with a backbone of β(1 → 4)-linked xylose and side-chains made up mostly of arabinose and galacturonic acid. (C) The mixed link glucan of grasses made up entirely of glucose linked by either β(1 → 3) or β(1 → 4) linkages.
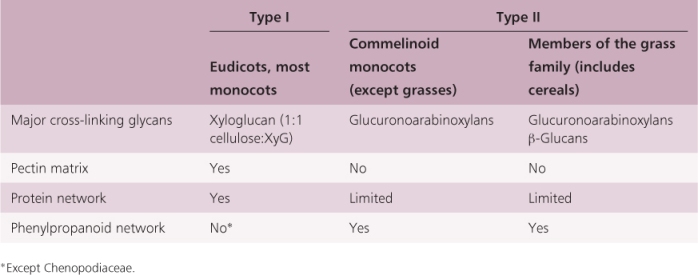
Angiosperm primary cell walls are classified into two groups based on the chemical composition of the non-cellulosic components of these walls (Table 4.1). Type I cell walls are found in all eudicot species and in about one-half of the monocots. Xyloglucans are the major cross-linking glycans in Type I walls; these walls have about an equal amount of cellulose and xyloglucan (Figure 4.6). Type II cell walls are found in commelinoid monocots (including bromeliads, palms, sedges and grasses). In Type II walls the major cross-linking glycans are glucuronoarabinoxylans (Figure 4.6). Within this latter group, the grasses, including the cereals, have additional cross-linking glycans, β-glucans, which are ‘mixed-linkage’ (1 → 3), (1 → 4)-β-D-glucans (Figure 4.5C). Mixed linkage glycans are especially abundant in the cell walls of cereal grain endosperm and make up the bulk of ‘soluble fiber’. The differences in the cross-linking glycans present in Type I and Type II cell walls may be reflected in differences in the responses of these cell walls to pH and other wall-loosening factors (see Chapter 12).
Figure 4.6 Three-dimensional models of a Type I primary cell wall (Arabidopsis) and a Type II primary cell wall (Oryza). Type I walls are found in eudicots and many monocots, while commelinoid monocots (including bromeliads, palms, sedges and grasses) have Type II walls. The Type II wall of rice, illustrated here, contains β-glucans, which is typical of grasses. These glucose-based cross-linking glycans have stretches of cellulose-like linkages (cellodextrins) that allow them to hydrogen bond together, as well as segments in which short runs of cellulose-like linkages (cellotriosyl- and cellotetraosyl-rich) are interspersed with callose-like linkages.
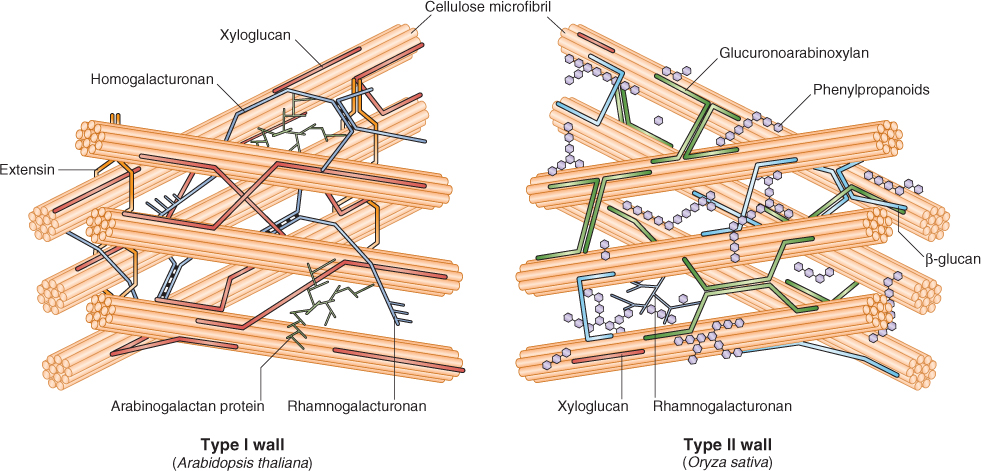
Table 4.1 Comparison of the composition of Type I and Type II primary cell walls in different taxa.
Pectins, a heterogeneous mixture of branched, and highly hydrated, polysaccharides that are rich in D-galacturonic acid, form a second network in primary cell walls. They bind Ca2+ and can be extracted from the cell wall by calcium chelators. Two fundamental constituents of pectins are homogalacturonan (HGA), a linear chain of galacturonic acid, and rhamnogalacturonan, a linear chain of alternating residues of galacturonic acid and rhamnose. Homogalacturonan chains can link with one another via Ca2+ bridges in which negatively charged galacturonic acid residues from two different HGA molecules associate with the same calcium ion.
Pectins affect the porosity of walls, modulate wall pH and ion balance, and act as recognition molecules to help plant cells detect the presence of symbiotic organisms, pathogens and insects. In addition, pectins in the middle lamella glue adjacent cells to one another (see Figure 4.3). Type I cell walls contain much more pectin than Type II walls but in both types the middle lamella is mostly pectin. Cell walls of fruits are very rich in pectins, which can be extracted (e.g. by boiling) and used as gelling agents for making jams and jellies.
Primary cell walls can also contain a third network of polymers. In Type I walls, this network is made of structural proteins. It plays a role in the control of cell wall extensibility and in pollen–stigma interactions (see Chapters 12 and 16). Five classes of these structural proteins have been identified: hydroxyproline-rich glycoproteins, proline-rich glycoproteins, glycine-rich proteins, threonine-rich proteins and arabinogalactan proteins. The synthesis and deposition of these proteins is developmentally regulated, their relative amounts varying among tissues and species. The best-studied hydroxyproline-rich glycoprotein is extensin, a rod-shaped protein (see Figure 4.6) that confers structural rigidity to the wall by cross-linking to other cell wall polymers. The proline-rich proteins are similar in composition and shape to extensin. The glycine-rich proteins are thought to form a plate-like structure at the plasma membrane–cell wall interface. The fourth group, arabinogalactan proteins, may be more than 95% sugar by mass; these proteins are anchored to the plasma membrane via a phosphatidylinositol anchor and are thought to play a role in cell signaling and incompatibility responses.
Type II walls contain less structural protein than Type I walls and lack extensin, but threonine-rich proteins that are structurally similar to extensin are found in Type II walls together with phenolic compounds, shown as phenylpropanoids in Figure 4.6. Phenolic compounds, primarily hydroxycinnamic acids, link polysaccharides to proteins or to one another. These networks render the walls rigid, locking them in place after growth has ceased.
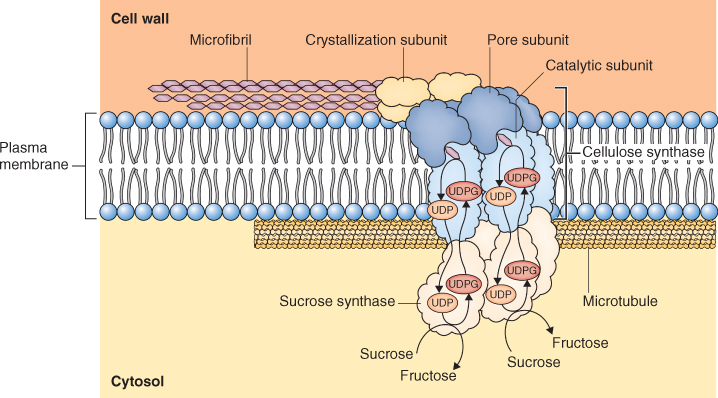
New cell walls are formed during cell division. Golgi vesicles containing glycoproteins and non-cellulosic wall polysaccharides are guided to the mid-point of the spindle where they fuse to form a plate-like, membranous organelle, the phragmosome (Figure 4.7). The phragmosome grows outward toward the lateral walls of the parent cell where it fuses with the plasma membrane to separate the contents of the daughter cells. Additional non-cellulosic components of the developing wall are delivered by secretory vesicles derived from the Golgi apparatus (see Section 4.5.5). Cellulose microfibrils are synthesized by cellulose synthase, an enzyme that is embedded in the plasma membrane. As cellulose microfibrils are synthesized, they are extruded from the outer surface of the plasma membrane into the cell wall (Figure 4.8).
Figure 4.7 High-pressure frozen and freeze-substituted TEM image of a phragmosome from Eucalyptus sieberti root tip cells. (A) The phragmosome consists of a series of flattened membranous sacs that form near the equator of two dividing cells. (B) The beginning of membrane fusion that eventually completes the process of cytokinesis and separates the two daughter cells, each of which will have its own plasma membrane.

Figure 4.8 Model of the cellulose synthase complex. Cellulose synthase is embedded in the plasma membrane and extrudes cellulose microfibrils to the exterior surface of the cell. The positioning of the cellulose synthase complex, and therefore the orientation of the microfibril, is determined by microtubules. Glucose units for cellulose synthesis are supplied by sucrose synthase which is associated with the cellulose-synthesizing complex.
Plant cells expand 20–50-fold in length during their development. Primary cell walls must also increase in size during cell expansion. Existing wall must be loosened and new wall material inserted in a coordinated process that does not compromise the structural integrity of the existing wall. This process is discussed in detail in Chapter 12.
When cells stop enlarging, the primary wall is cross-linked into its final shape. At this point, deposition of the secondary wall on the inner surface of the primary wall may begin (Figure 4.9). The composition and pattern of deposition of the secondary wall vary from cell to cell (Figure 4.10). For example, the secondary walls of cotton fiber cells are nearly 98% cellulose, whereas the secondary walls in some seed tissues are made of mostly non-cellulosic polysaccharides. The secondary walls of cereal endosperm cells are composed of β-glucans, whereas those of the date endosperm are mostly mannans, polymers of mannose. In these cases, secondary wall polysaccharides are used as a food source by the embryo during seed germination and early seedling growth. The β-glucans in cereal endosperm are important in human diets as well. They provide ‘soluble fiber’, thought to be important in lowering blood cholesterol levels. Because the yeast used in beer making does not easily breakdown these glycans, they accumulate in beer causing a cloudy ‘haze’ (see Chapter 6).
Figure 4.9 Secondary cell walls. Once they have achieved their final size and shape, some cells elaborate a multilayered secondary wall. In the diagram (A), a living protoplast has generated several distinct layers of secondary walls (S1 through S3, with the innermost S3 being laid down last), whereas the micrograph (B) shows secondary wall layers surrounding the empty lumen of a dead fiber cell in the young stem of a locust tree. In both panels, the original primary wall (CW1) and the middle lamella (ML) form the outermost layers of the wall.
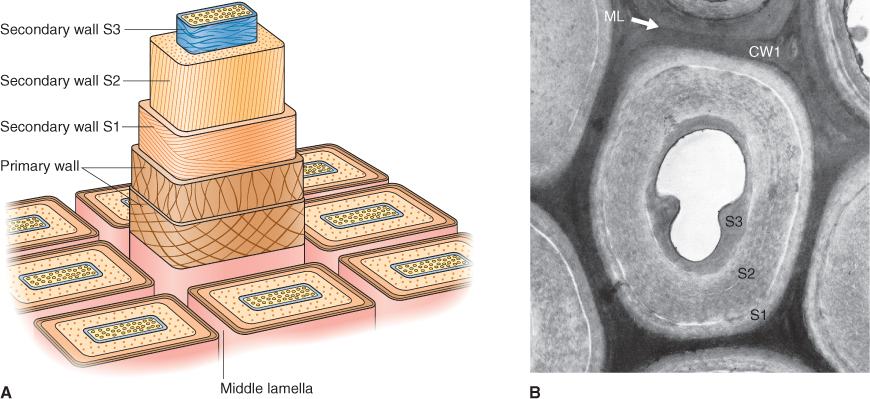
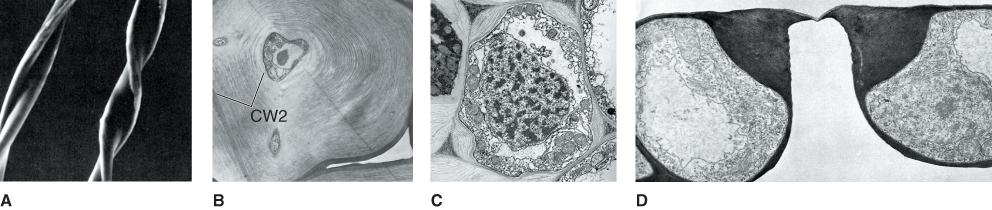
Figure 4.10 Secondary walls may differ in composition in different cell types. They serve a number of different functions. (A) The thick secondary wall of almost pure cellulose gives the flattened helical form to a cotton fiber. (B) The secondary wall (CW2) nearly fills the lumen of a pear fruit stone cell. (C) A collenchyma cell has reinforced thickenings only in the cell corners. (D) The thickened and elaborated inner wall of a guard cell pair provides the physical form needed to control formation of the stoma.
Many secondary walls contain lignins, complex networks of aromatic compounds called phenylpropanoids. In the water-conducting cells of the xylem, lignified secondary walls are deposited in distinctive patterns, such as annular or helical coils or reticulate and pitted sheets (see Figure 1.28). Lignins make the wall rigid and waterproof. In trees, 15–25% of the dry mass of wood is lignin; lignified secondary walls give wood its strength and make up the skeleton that keeps 100 m tree trunks upright. When wood is processed to make paper, the lignins must be extracted by harsh chemical treatments.
Secondary deposition of suberin and cutin can render cell walls impermeable to water. Suberin is found in cell walls in specific cell types, notably stem and older root epidermal cells, cork cells of bark, the surfaces of wounded cells and parts of endodermal and bundle sheath cell walls. The core of suberin is lignin-like, and the attachment of the long-chain hydrocarbons imparts a strongly hydrophobic characteristic to suberin that prevents water movement. Cutin, a polymer of ester-linked fatty acids, and its associated waxes are also found on leaf and stem surfaces, providing a barrier to the diffusion of water vapor. Waxes are generally esters of long-chain fatty acids and alcohols that are synthesized in the ER.
Inside the cell wall is the living protoplast enclosed by the plasma membrane. The plasma membrane creates and maintains an electrochemical environment within the cell that is different from the outside environment (see Chapter 5). Within the protoplast are many organelles whose membranes maintain distinctive electrochemical environments within them. The functioning of these organelles depends on the integrity of their membranes and their embedded transporters. Plant cells contain at least 14 distinct membrane types (Table 4.2). We will discuss the features common to all cellular membranes and then look specifically at the plasma membrane.
Table 4.2 Membrane types found in plant cells.
| Endomembrane system | Other membranes |
| Plasma membrane | Peroxisomal membrane |
| Nuclear membrane | Plastid envelope membranes (inner and outer) |
| Endoplasmic reticulum | Thylakoid membranes of the plastid |
| Golgi cisternae (cis, medial and trans types) | Mitochondrial membranes (inner and outer) |
| Trans-Golgi network/partially-coated reticulum | |
| Clathrin- and COP-coated transport vesicles | |
| Endocytic vesicle membrane | |
| Endosomal membrane | |
| Multivesicular body/autophagic vacuole membranes | |
| Tonoplast (vacuole membrane) |
COP, coat protein.
Almost all biological membranes consist of a bilayer of polar lipid molecules in association with a variety of proteins. Most cellular membranes have equal masses of protein and lipid components. Virtually all membrane molecules are able to diffuse freely within the planes of the membrane, permitting membranes to change shape and membrane molecules to rearrange rapidly.
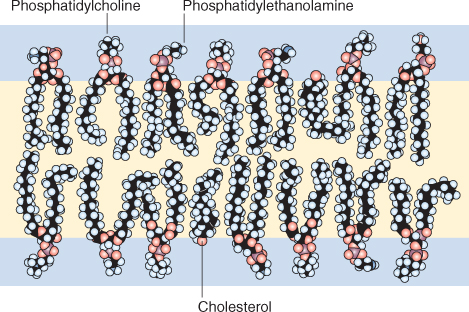
The primary components of the polar lipid bilayer of cellular membranes are phospholipids and glycolipids (see Section 2.2.7). These molecules are amphipathic because they have polar or charged hydrophilic (‘water-loving’) heads and hydrophobic (‘water-hating’) tails that consist of two fatty acids that contain between 14 and 24 carbon atoms. At least one of the fatty acids has one or more double bonds that form a kink in the otherwise straight fatty acid chain (see Figures 2.9 and 2.10). The hydrophilic head of phospholipids contains a phosphate group to which other polar molecules may be attached, while the hydrophilic head of glycolipids contains a sugar. In water, these molecules spontaneously form bilayers so that their hydrophilic heads interact with the water and their hydrophobic tails exclude water and interact with each other (Figure 4.11). Another class of membrane lipids is that of sterols; these molecules have a hydrocarbon skeleton that is hydrophobic, and a single, hydrophilic hydroxyl group. Cholesterol is one sterol commonly found in membranes. The ratio of lipid classes varies between different membranes within a cell.
Figure 4.11 Organization of amphipathic lipid molecules in a membrane bilayer. The hydrophilic heads of the molecules interact with water; their hydrophobic tails repel water and interact with each other in the center of the bilayer.
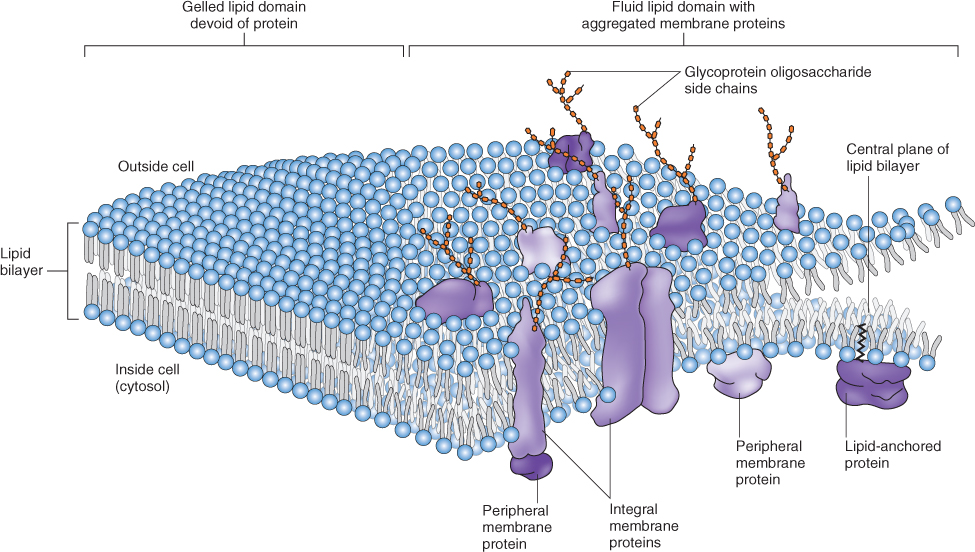
Membrane proteins may be associated with the lipid bilayer in a number of different ways (Figure 4.12). Integral proteins are embedded in the bilayer and, like membrane lipids, have both hydrophilic and hydrophobic domains. Many integral membrane proteins are decorated with chains of sugars. Water-soluble peripheral proteins interact with membrane lipids or proteins through salt bridges, hydrogen bonds, electrostatic reactions or a combination thereof. Unlike integral proteins, they can be removed by using treatments that do not disrupt the membrane itself. Anchored proteins attach to the bilayer via lipid anchors that are covalently bound to the proteins.
Figure 4.12 The fluid–mosaic membrane model. Integral, peripheral and lipid-anchored membrane proteins are shown. Note that the drawing is not to scale.
The lipid bilayer of cellular membranes serves as a permeability barrier to many molecules. While non-polar molecules and gases can readily diffuse across the hydrophobic core of the bilayer, hydrophilic polar molecules and ions cannot. An important exception to this rule is water, which, although polar, can freely cross the lipid bilayer. Membrane proteins serve as highly specific transporters allowing cells to take up or excrete ions and molecules (see Chapter 5). Membrane proteins perform a wide array of other functions including signal transduction, enzymatic catalysis and structural roles. What's crucial to remember is that membrane proteins define the specificity of each membrane system.
All living cells have a plasma membrane. The lipids, proteins and their associated carbohydrate side-chains that make up plant plasma membranes occur in a molecular ratio of about 2:2:1. The most prevalent lipids of plasma membranes may be phospholipids or sterols. The ratio of different lipids may vary in the plasma membranes of cells in different organs or of cells in the same organ type in different species. Proteins in the plasma membrane participate in transmembrane transport and signaling, catalyze reactions such as the synthesis of cellulose (see Section 4.2.5), and connect both the membrane and its attached cytoskeletal elements to the cell wall.
Most cells in a plant are connected by numerous narrow (40–50 nm) cytoplasmic channels lined by plasma membrane. These plasmodesmata (singular = plasmodesma) span adjoining cell walls (Figure 4.13). Thus most cells in a plant share a physically continuous plasma membrane. Notable exceptions to this rule are stomatal guard cells and cells of the embryo sac. The structure of plasmodesmata is fairly complex (see Figures 4.13 and 14.7); ions and small molecules up to about 800 Da in mass can freely diffuse from cell to cell through plasmodesmata. Some cells have the ability to increase the size of plasmodesmata so that larger molecules may pass through; in certain cases, molecules up to 10 kDa have been shown to move via this pathway. The network of interconnected protoplasts in a plant is called the symplasm and the parts of the cell outside the plasma membrane, the cell wall and airspaces, collectively are called the apoplast (see Chapter 14).
Figure 4.13 Plasmodesmata. (A) TEM image of Azolla root tip cells showing the many plasmodesmata (box) interconnecting neighboring cells. (B) Higher magnification image showing a side view of plasmodesmata from a Vicia faba extrafloral nectary. (C) TEM image of a face view of plasmodesmata in an Abutilon nectary trichome. Note that a plasmodesma is bounded by the plasma membrane (PM) and the core is filled with a piece of tightly rolled endoplasmic reticulum that forms the desmotubule (DT).
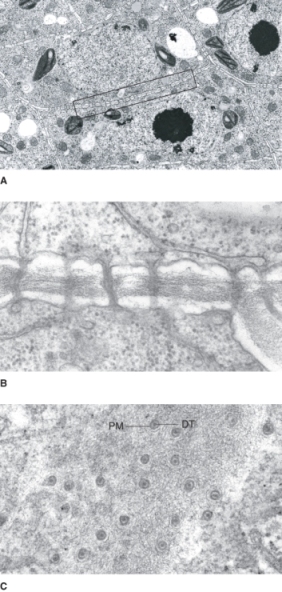
A prominent organelle in most living plant cells is the nucleus. One exception is the sugar-conducting sieve tube elements of the phloem, which lack nuclei at maturity. Nuclei are 3–10 μm in diameter, contain most of the cell's genetic information, and serve as the center of regulatory activity (Figure 4.14). The nucleus is bounded by a nuclear envelope that consists of two bilayer membranes separated by a lumen, the perinuclear space. This envelope contains complex nuclear pores through which RNA and ribosome subunits, synthesized in the nucleus, are exported to the cytosol, and proteins, synthesized in the cytosol, are imported into the nucleus (Figure 4.15). Molecules smaller than 40 kDa can pass through nuclear pores by diffusion but those greater than 40 kDa move by energy-dependent transporters. The outer membrane of the nuclear envelope has ribosomes on its cytoplasmic face. It is continuous with the membranes of the endoplasmic reticulum and therefore may be considered to be part of the endomembrane system (see below).
Figure 4.14 TEM image showing the nucleus (N) of a bean (Phaseolus vulgaris) root tip cell. Note the large, darkly stained central nucleolus (NU); the nuclear envelope (NE) consists of two membranes.
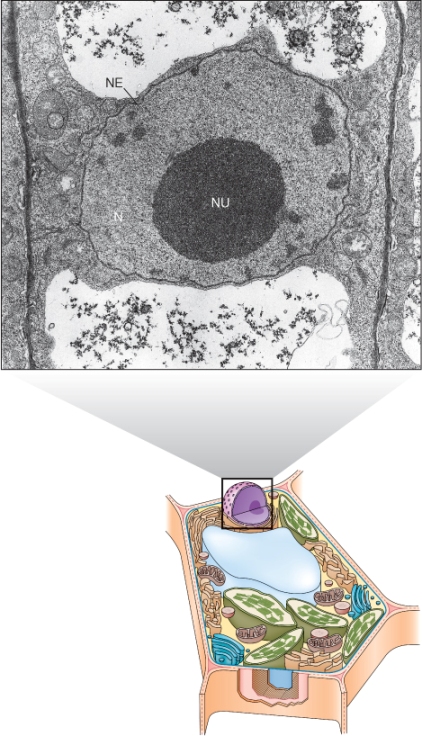
Figure 4.15 (A) TEM image of a freeze-fractured nuclear envelope (NE) with nuclear pores (NP). The continuity of the inner and outer membranes becomes apparent when the membranes are seen in cross-section. (B) Transport of small molecules into the nucleus via the nuclear pores takes place by diffusion, whereas import or export of larger molecules requires active (energy-consuming) transport. ER, endoplasmic reticulum.
Chromosomes are made up of DNA–protein complexes called chromatin (see Chapter 3). Chromatin forms a network in the nucleoplasm during interphase when gene transcription and DNA synthesis are occurring. Individual chromosomes occupy discrete nuclear domains throughout this part of the cell cycle.
A typical interphase nucleus contains one to several nucleoli (singular = nucleolus), distinctive regions that stain densely in transmission electron micrographs (see Figure 4.14). These are the sites where ribosomal RNA is synthesized, processed and assembled with ribosomal proteins to form ribosomal subunits. The subunits then move through nuclear pores to the cytosol.
The endomembrane system of a plant cell is an extensive, interconnected series of organelles that are responsible for the synthesis, processing and storage of a wide variety of macromolecules. Ten of the 14 cellular membrane types listed in Table 4.2 are part of the endomembrane system. Extensive vesicle traffic between these compartments not only transports secreted molecules to the cell surface and vacuolar proteins to the storage vacuoles, but also distributes membrane proteins and membrane lipids from their sites of synthesis to other parts of the endomembrane system (Figure 4.16). A large number of sorting, targeting and retrieval systems regulate traffic between different compartments, ensuring delivery of molecules to the correct organelle and the maintenance of organelle identity (see Chapter 5).
Figure 4.16 Diagram of the plant endomembrane system. Note the 16 different domains of the ER that are separately numbered in this diagram.
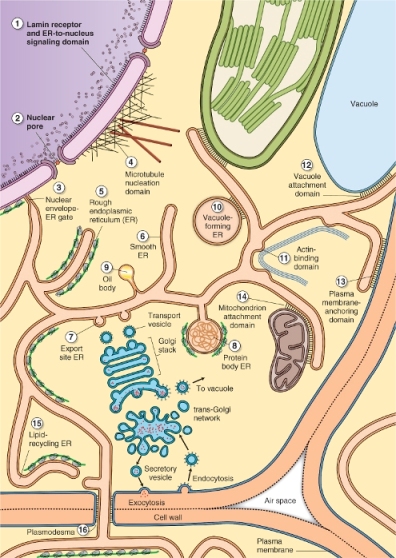
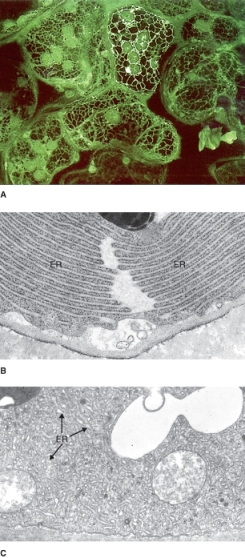
The endoplasmic reticulum (ER) is the most extensive, versatile and adaptable organelle in eukaryotic cells. It consists of a three-dimensional network of continuous tubules and flattened sacs that connect to the nuclear envelope, course through the cytoplasm and underlie the plasma membrane (Figures 4.16 and 4.17). In plants, the principal functions of ER include the synthesis, processing and sorting of proteins that will be targeted to various membranes, to vacuoles or to the secretory pathway. Also, a diverse array of lipid molecules is synthesized within the ER. In addition to its synthetic functions, the ER provides anchoring sites for actin filament bundles that drive cytoplasmic streaming. It also plays a critical role in regulating the cytosolic concentration of calcium that in turn influences many other cellular activities. Three types of ER membranes are recognized: rough ER, which is studded with ribosomes and is a site of protein synthesis (Figure 4.17B), smooth ER (Figure 4.17C), which is free of ribosomes and is associated with lipid synthesis, and the nuclear envelope. In addition, many more morphologically distinct subdomains of ER that perform various different functions are found in plant cells (see Figure 4.16).
Figure 4.17 Confocal (A) and TEM images (B, C) of endoplasmic reticulum (ER). (A) Fluorescently labeled ER forming a net within Nicotiana tabacum (tobacco) leaf mesophyll cells. (B) Rough ER in a Coleus gland cell and (C) smooth tubular ER in a cap cell of a Primula malacoides farina gland (a surface structure that secretes a floury white wax).
Translation of all nuclear messenger RNAs (mRNAs) begins in the cytosol on free ribosomes. The synthesis of proteins that are destined to be secreted or are targeted to organelles of the endomembrane system is completed on the ER. As soluble proteins are synthesized, they are translocated across the ER membrane into the lumen; integral membrane proteins are incorporated directly into the ER membrane (see Chapter 5). Many of these proteins undergo further processing within the ER, modifications that affect their final three-dimensional conformation. For example, monomeric proteins may assemble into polymeric protein complexes. In addition, some proteins are glycosylated by enzymes that add branched sugar chains to the amide nitrogen of asparagine residues. Once ER-localized protein processing is complete, most proteins are transported to the Golgi apparatus in vesicles that bud from the ER and fuse with the Golgi cisternae (see Figure 4.16 and Section 4.5.4). Budding and transport of vesicles from the ER is aided by coat proteins (COPs).
Some storage proteins that are synthesized during seed development are notable exceptions to the general rule of ER-to-Golgi transport. Prolamins, the alcohol-soluble storage proteins of cereal grains, are synthesized in specific ER regions and either aggregate in the lumen of the ER or are transported directly to vacuoles by vesicles that by-pass the Golgi (see Figure 4.16 and Chapter 6).
The smooth ER membrane is the site of many reactions involved in lipid synthesis. Some of the 16- and 18-carbon fatty acids that are synthesized in the plastids are transported to the ER. There they are combined with glycerol-3-phosphate to make phosphatidic acid, which is further modified to make the phospholipid molecules from which most cellular membranes are made. Triacylglycerols (oils) that are produced in developing seeds and pollen grains (see Chapter 6) are also synthesized in the ER. As they are synthesized, triacylglycerols accumulate between the inner and outer leaflets of the lipid bilayer of the ER membrane in regions of the membrane that contain proteins called oleosins. Mature oil bodies (oleosomes) are therefore enclosed by a ‘half-unit’ membrane, a lipid monolayer that contains oleosins and other proteins that are important for the integrity of this organelle (Figure 4.18).
Figure 4.18 Formation of oil bodies. (A) Triglycerides (oil) accumulate between the two lipid monolayers of the ER membrane and bud to form oil bodies at sites defined by the presence of molecules of the protein oleosin. (B) TEM iamge of oil bodies (OB) showing their close proximity to peroxisomes (P).

Finally, the monomers that make up the waterproof waxes that coat the above-ground surfaces of plants (see Section 4.2.6) are synthesized by the ER of epidermal cells (Figure 4.19). Elongase enzymes located in ER membranes elongate fatty acids that are produced in the chloroplast. The resulting very-long-chain fatty acids are the monomers of waxes and cutin. It is not known how these monomers are transported to the exterior of the cell, although one hypothesis is that vesicles transport monomers to the plasma membrane where ‘flippase’ enzymes bring about their relocation to the cell's outer surface (Figure 4.19).
Figure 4.19 Hypothetical scheme for the transport of fatty acid precursors for cutin and wax synthesis to the site of polymerization in the cell wall. It is proposed that fatty acids move from the endoplasmic reticulum (ER) to the plasma membrane in the membranes of vesicles. A ‘flippase’ transfers cutin monomers from the inner side of the plasma membrane to the outer leaflet, where they can be extracted from the membrane by a water-soluble, lipid-transfer protein. The lipid-transfer protein diffuses in the solution phase of the cell wall matrix and transfers cutin monomers to enzymes that polymerize them into waxes near the surface of epidermal cell walls.

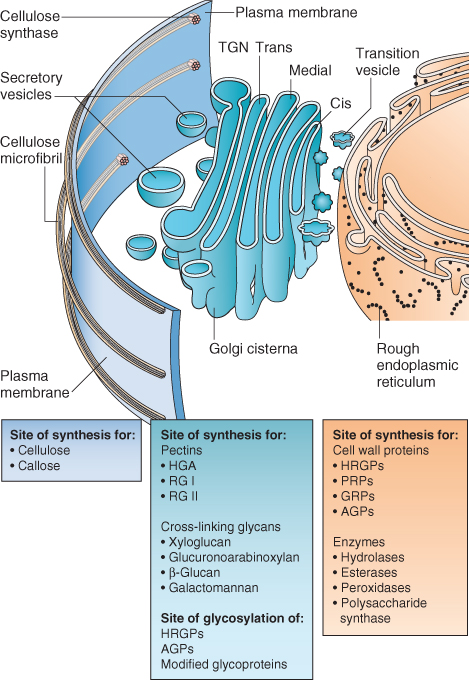
The Golgi apparatus occupies a central position in the secretory pathway. It is responsible for the synthesis, modification, packaging and sorting of a variety of macromolecules destined for export to the cell wall or for delivery to other organelles. The Golgi apparatus receives newly synthesized glycoproteins and glycolipids from the ER and modifies their sugar side-chains, creating a wide variety of oligosaccharide groups. The sugar groups on proteins may specify plasma membrane–cell wall interactions, prevent the premature activation of lectins, or contribute to protein folding or the assembly of multiprotein complexes. In addition, the Golgi is responsible for the synthesis of cross-linking glycans and other non-cellulosic cell wall polysaccharides (Figure 4.20).
Figure 4.20 Cell wall biosynthesis requires the coordination of multiple compartments of the endomembrane system. Cellulose microfibrils are synthesized at the plasma membrane surface (see Figure 4.8).Structural proteins and wall-modifying enzymes are translated on the rough ER and transported to the Golgi apparatus, where some are glycosylated before being packaged in secretory vesicles for delivery to the cell exterior (AGP, arabinogalactan protein; GRP, glycine-rich protein; HRGP, hydroxyproline-rich protein; PRP, proline-rich protein). The Golgi also synthesizes and packages for secretion the various non-cellulose cell wall polysaccharides (HGA, homogalacturonan; RG I and RG I, rhamnogalacturonan I and II). At the cell surface, vesicle contents are integrated with the newly synthesized microfibrils. Assembly of the new wall is estimated to begin when the cellulose chains are no more than ten glucose residues in length. TGN, trans-Golgi network.
The Golgi apparatus is a membranous organelle that has several components: the Golgi stack, about 1 μm in diameter and comprised of five to eight membrane-enclosed flattened sacs (cisternae); the trans-Golgi network (TGN), a tubular structure associated with one side of the Golgi stack; and the Golgi matrix, a filamentous cage that surrounds the Golgi stack and its TGN (Figure 4.21). The number of Golgi stacks per cell varies widely depending on cell type and function. For example, small, shoot apical meristem cells of willow-herb (Epilobium) contain 20 Golgi stacks, the larger onion (Allium cepa) root apical meristem cells have about 400 stacks, and the giant fiber cells of cotton (Gossypium hirsutum) contain more than 10 000.
Figure 4.21 Structural features of the Golgi apparatus. (A) The spatial relationship of a Golgi stack with its associated trans-Golgi network (TGN). Two types of vesicles bud from the TGN: vesicles coated with clathrin protein are targeted to the vacuoles, while a different type of protein-coated vesicle moves products to the plasma membrane (PM) or cell wall. (B) TEM image of a Golgi stack and its TGN in a green alga, Micrasterias. Note the fuzzy protein coats on vesicles on the cis face (upper) and around the margins of the cis-cisternae and around the TGN cell.

Each Golgi stack is asymmetrical. Transport vesicles from the ER deliver their contents by fusing with the cis face of the Golgi stack. ER-derived products move sequentially via transport vesicles through the cis-, medial-, and trans-cisternae of the Golgi stack and then to the TGN. In addition, different compartments of the Golgi apparatus participate in the synthesis of the various classes of cell wall polysaccharides. Vesicles containing the non-cellulosic polysaccharide components of the cell wall bud from the TGN and move to the plasma membrane for delivery to the cell wall. Vesicles involved in intra-Golgi and Golgi-to-TGN transport bud from cisternae with the aid of a COP.
Within the TGN, materials are sorted for distribution to vacuoles or the plasma membrane, or for transport to the cell exterior. Proteins destined for vacuoles contain specific amino acid sequences that target them for transport to this organelle and are transported via the multivesicular body (MVB) (see Chapter 5). All other proteins are packaged for transport to the plasma membrane. Vesicles from the Golgi destined for vacuoles are coated by a protein called clathrin, which differs from the coat protein of vesicles bound for the plasma membrane (Figure 4.21).
When a transport vesicle from the Golgi apparatus reaches the plasma membrane, the vesicle membrane fuses with the plasma membrane and its contents are discharged into the cell wall, a process called exocytosis. Exocytosis allows the delivery of proteins and polysaccharides to the cell wall and thus the cell exterior. In addition, it allows the addition of new membrane proteins and lipids to the plasma membrane.
A second mechanism, endocytosis, allows plant cells to retrieve plasma membrane components for recycling or degradation. This process also may allow a plant cell to take up extracellular molecules. Endocytosis involves the import of plasma membrane-derived vesicles that are coated in clathrin. Models for endocytosis in plants propose that imported materials move from the clathrin-coated vesicles through the MVB before arriving at vacuoles, where they are broken down and their component parts are recycled into new molecules (see Figures 4.16 and 5.30).
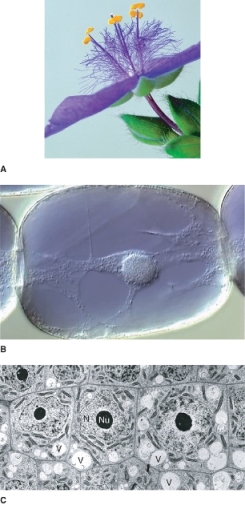
Vacuoles, fluid-filled compartments enclosed in a single membrane, the tonoplast, can occupy 30–90% of the volume of a plant cell (see Figures 4.1 and 4.22). Large, mature plant cells have a large central vacuole, while meristematic cells often contain numerous small vacuoles (Figure 4.22C) that coalesce into one or a few larger vacuoles as the cell matures and expands. Vacuoles typically originate as provacuoles that bud from the ER.
Figure 4.22 Vacuoles. (A) Flower of Tradescantia. Note the purple stamen hairs on the filaments of the stamens; each hair is a chain of cells. (B) Light micrograph of a single stamen hair cell. Vacuole contents are purple due to the presence of anthocyanin pigment. Note a very thin layer of cytoplasm around the large central vacuole; strands of cytoplasm cross the vacuole, and one strand contains the nucleus. (C) TEM iamge of Hydrocharis root tip cells showing early stages of vacuolation. N, nucleus; Nu, nucleolus; V, vacuole.
Vacuoles serve several functions. In mature plant cells, the large central vacuole occupies most of the volume of a cell and confines the metabolically-active cytoplasm to a thin layer, increasing its surface area and maximizing gas and nutrient exchange. The vacuole also allows plants to produce large cells without having to produce very much protein-rich cytoplasm.
Plant cells also use vacuoles for short- and long-term storage of a variety of ions and molecules. Vacuoles act as reservoirs of protons, organic acids and metabolically important ions, such as calcium; they also serve as storage organelles for polysaccharides called fructans in grasses (see Chapter 17). In addition vacuoles may contain lytic enzymes that can be used in digestion of stored reserves as well as in the breakdown and recycling of cellular components. Vacuoles may also store heavy metals, and plant defense compounds such as tannins, oxalic acid crystals, proteases and nucleases, and, in the case of one marine alga, sulfuric acid. In some cells, water-soluble pigments, such as the anthocyanins that give leaves and flowers a red, pink or blue color (see Chapter 15), are found in vacuoles (Figure 4.22A, B). In seeds, there are specialized, protein-storing vacuoles.
Plastids are found in the cells of all autotrophic eukaryotes; in plants they are present in almost all cells. They produce sugar in photosynthesis (see Chapter 9) and store carbohydrate as starch. They are involved in the biosynthesis of other essential metabolites, including chlorophylls, carotenoids, purines, pyrimidines, amino acids, fatty acids and hormones, e.g. gibberellins and abscisic acid. They also reduce nitrite and sulfate, facilitating the incorporation of nitrogen and sulfur into organic compounds.
Plastids are enclosed by two membranes, the outer and inner membranes, and contain internal membrane systems that vary in complexity according to the plastid's developmental stage and function. The outer membrane contains non-specific protein pores that allow ions and molecules up to 10 kDa in mass to diffuse into the intermembrane space between the two membranes. The enzymes involved in galactolipid metabolism are localized in the outer membrane. The inner membrane, which encloses an aqueous compartment called the stroma, is less permeable. Gases and small, uncharged molecules can cross the inner membrane unassisted, but most metabolites enter or leave the stroma by way of specific transporters. The inner membrane invaginates to produce the internal thylakoid membrane system of the plastid. In plants, de novo synthesis of fatty acids takes place exclusively in the stroma of plastids.
Plastids contain the machinery required to synthesize some of their own proteins, including a naked DNA genome (discussed in more detail in Chapter 3) and ribosomes that resemble those of prokaryotes more closely than they do the linear nuclear chromosomes, and cytoplasmic ribosomes of eukaryotic cells. Morphological, biochemical and nucleic acid sequence evidence indicates that plastids originated as prokaryotic endosymbionts related to cyanobacteria. However, plastids have since lost many genes and must import about 75% of their proteins, which are encoded in the nucleus and transcribed on free ribosomes in the cytosol.
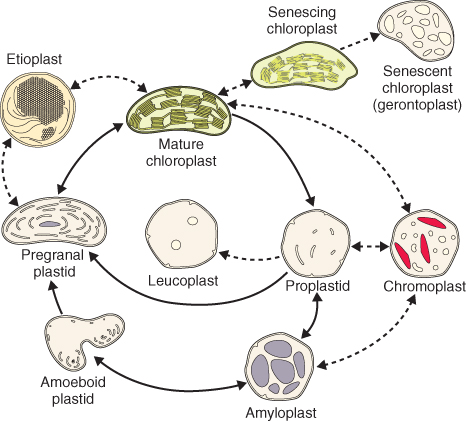
All plastid types are related to the undifferentiated proplastids that are present in meristematic cells. Plastids possess a remarkable capacity to differentiate, dedifferentiate and redifferentiate (Figure 4.23); differentiated plastids can convert from one type to another in response to developmental or environmental conditions.
Figure 4.23 Plastid developmental cycle and interconversion of various plastid types. Solid arrows depict normal steps of chloroplast development; dotted arrows show plastid interconversions that occur under certain environmental or developmental conditions. The gray structures seen in the amyloplast are starch granules.
Proplastids are spherical or ovoid and range in diameter from about 0.2 to 1.0 μm. A proplastid contains few ribosomes, and its internal membrane system is poorly developed, consisting only of a few invaginations of the inner envelope and a small number of flattened sacs called lamellae (Figure 4.24A).
Figure 4.24 A range of plastid types found in plant cells. (A) TEM image showing a proplastid (left) adjacent to a mitochondrion in a bean (Phaseolus vulgaris) root cell. The electron-dense particles near the proplastid's periphery are lipid droplets. Beneath the large, round protein deposit in the middle of the proplastid can be seen the flattened lamella of an internal membrane. (B) TEM image of amyloplasts in a soybean (Glycine max) root cap columella cell containing many starch granules (S). (C) TEM image showing a leucoplast in an actively secreting glandular trichome of peppermint (Mentha spp.). The black, electron-dense globules are lipid droplets. Note the presence of extensive tubular smooth endoplasmic reticulum cisternae in the surrounding cytosol. (D) TEM image of a chromoplast in a cell of a ripe Jerusalem cherry (Solanum pseudocapsicum) fruit. The electron-dense bodies in the plastid contain the carotenoids.

Chloroplasts are the site of photosynthesis and contain the green pigment chlorophyll. They are commonly hemispherical or lens-shaped in vascular plants (Figure 4.25A). They range in diameter from 3 to 10 μm and from 0.5 to 1.0 μm in thickness. Within each chloroplast the thylakoids form an extensive continuous network that encloses a single, branched chamber, the thylakoid lumen. The thylakoid network contains two types of membrane domains: stacks of thylakoids called grana (singular = granum) are connected by unstacked stroma thylakoids (Figure 4.25B). The components of the light-energy-capture reactions, including the photosynthetic pigments, are associated with the thylakoid membranes (see Chapter 9). In a mature chloroplast the number of thylakoids in a granum may vary from several to 40 or more depending on the plant species and light conditions. For example, plants grown in shade generally have not only more numerous grana, but also more thylakoids per granum, than individuals of the same species growing in bright light.
Figure 4.25 Chloroplast ultrastructure. (A) TEM image showing a chloroplast with stacked grana (GT) and unstacked stroma (ST) thylakoids in a maize leaf mesophyll cell. The dark granules are lipid droplets. (B) High-magnification TEM iamge of stacked grana and unstacked stroma thylakoids in a chloroplast from a leaf of timothy grass (Phleum pratense). The membranes, which are stacked in one area and unstacked in another, enclose a single lumen.

Chloroplasts contain numerous ribosomes that can account for up to half the ribosomes present in photosynthetic tissues. The chloroplast enzyme, rubisco, makes up about half the soluble protein in leaves and may be Earth's most abundant protein. Rubisco's large subunit is encoded by chloroplast DNA and is synthesized on chloroplast ribosomes. In contrast, the small subunit of rubisco is encoded by nuclear DNA and is synthesized on cytoplasmic ribosomes (see Chapter 9). This is an example of the importance of coordinated nuclear and plastid gene expression during plastid development.
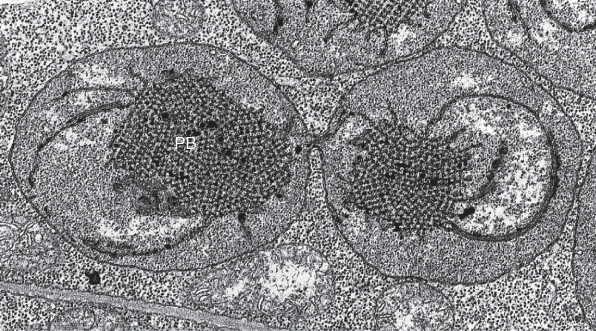
Etioplasts are plastids whose development from proplastids to chloroplasts has been arrested by the absence of light or by very low light; they are 2–3 μm in length. Etioplasts lack chlorophyll, but produce protochlorophyllide, a precursor of chlorophyll. A prominent structure in the etioplast is the prolamellar body (Figure 4.26), a lattice-like inclusion that consists of membrane lipids, protochlorophyllide and the light-requiring enzyme protochlorophyllide oxidoreductase (see Chapter 8). Protochlorophyllide is converted to chlorophyll, chlorophyll–protein stable complexes are produced, and lipids from the prolamellar body are incorporated into developing thylakoid membranes.
Figure 4.26 TEM image of an etioplast from Phaseolus vulgaris undergoing binary fission. Note the prominent prolamellar body (PB) in each etioplast and the constriction between the two daughter plastids.
Amyloplasts (see Figure 4.24B) are unpigmented plastids that synthesize and store starch; they are about the same size as chloroplasts. Their name is derived from amylose, a major component of starch (see Chapter 6). Amyloplasts are especially common organelles in storage tissue of roots and stems and the endosperm of cereal grains. In potato tubers, starch grains commonly fill the entire plastid except for a thin layer of stroma lining the inner envelope. Amyloplasts occasionally redifferentiate into chloroplasts, as happens when potato tubers are left in the light. Note that amyloplasts in the cereal endosperm cannot redifferentiate as this tissue is dead at functional maturity.
Leucoplasts (see Figure 4.24C) are colorless plastids that synthesize and store monoterpenes, volatile compounds contained in essential oils (see Chapter 15). They have few internal membranes or ribosomes and contain lipid droplets. They are found in specialized secretory gland cells associated with leaf and stem trichomes (hairs), the nectaries of flowers, and secretory cavities such as those found in citrus peel.
Chromoplasts (see Figure 4.24D) are plastids that store carotenoids; they may be yellow, orange or red, depending on the particular combination of carotenes and xanthophylls present. They are responsible for the colors of many fruits (e.g. tomatoes, oranges), flowers (e.g. buttercups, marigolds), roots (e.g. carrots) and tubers (e.g. sweet potatoes). Chromoplasts have an irregular shape ranging from round to amoeboid to elongate; in some cases they contain large crystals. Chromoplasts can develop directly from proplastids or by the dedifferentiation of chloroplasts, as happens in ripening tomato fruits. The plastids of senescing leaves are sometimes called gerontoplasts and resemble chromoplasts in accumulating carotenoids that account for the bright yellow and orange colors of foliage in fall (see Chapter 18). Occasionally chromoplasts will redifferentiate into chloroplasts, as they do in some yellow and orange citrus fruits that regreen under the appropriate conditions, or in carrot roots exposed to light.
Plastids arise by binary fission of existing plastids (see Figure 4.26). The division of proplastids, etioplasts and young chloroplasts is most common, but fully developed chloroplasts can also divide. In root and shoot meristems, proplastid division keeps pace with cell division, so that mother and daughter cells both possess approximately the same number of plastids—about 20. As cells expand, the number of plastids per cell increases due to continued plastid division.
Cytoplasmic organelles can be passed from one generation to the next by way of the egg (maternal inheritance), the sperm (paternal inheritance) or both (biparental inheritance). Genetic evidence indicates that in most angiosperms, the plastids and mitochondria are inherited maternally. These organelles are either excluded from angiosperm sperm cells or are degraded during male gametophyte development or after fertilization. This is discussed further in Chapter 16. In gymnosperms, plastids are usually passed to the next generation through sperm. Biparental inheritance of plastids and mitochondria has been documented both cytologically and genetically in a few flowering plant genera. Whether from egg, or sperm, plastids are in the proplastid stage at the time they are inherited.
Mitochondria, often called the powerhouses of the cell, are found in nearly all plant cells. Mitochondria are the site of aerobic respiration (see Chapter 7) and participate in gluconeogenesis (see Chapter 6) and photorespiration (see Chapter 9). Electron transport in mitochondria is a key source of ATP, and mitochondrial metabolic pathways supply important intermediates such as organic acids and amino acids that are used in biosynthesis elsewhere in the cell. Like plastids, mitochondria have their own DNA and prokaryote-like ribosomes, and divide by fission. Sequence data from mitochondrial ribosomal DNA indicates that mitochondria originated as endosymbionts related to the α-proteobacteria.
In electron micrographs, plant mitochondria appear spherical to ellipsoid, generally about 1 μm wide and 1–3 μm long (Figure 4.27). Viewed in living cells, however, these organelles change shape frequently from round to oval to dumbbell-shaped and back. A plant cell may contain hundreds or thousands of mitochondria, depending on the cell type and its stage of development. The cells of the maize root cap have about 200 mitochondria when young and 2000–3000 when fully enlarged and mature.
Figure 4.27 Mitochondrion morphology and structure. (A) The three-dimensional structure of a mitochondrion and the distribution of ATP synthase molecules in the inner membrane. (B) TEM image of a mitochondrion in an Avena (oat) leaf mesophyll cell.

Mitochondria are bounded by two membranes, which are separated by the intermembrane space. The outer membrane contains few enzymes. The inner membrane encloses the mitochondrial matrix, which contains soluble enzymes, mitochondrial DNA and ribosomes. The inner membrane is highly infolded producing long, tubular cristae extending into the matrix (Figure 4.27). The inner membrane (including cristae) is the site of the respiratory electron transport chain and contains ATP synthase that makes ATP.
As in plastids, the outer membrane of a mitochondrion is highly permeable to small molecules, whereas the inner membrane has a very low permeability. Protein complexes, known as porins, mediate the transport of small molecules across the outer membrane. Each complex creates an aqueous channel through which ions and molecules move freely. In contrast, solute-specific transporters control solute movement across the inner membrane.
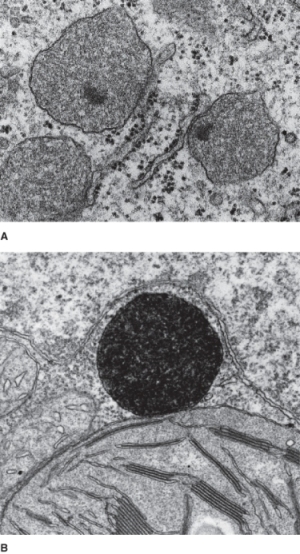
Peroxisomes are roughly spherical organelles, about 0.2–2.0 μm in diameter, surrounded by a single membrane (Figure 4.28). These organelles grow by importing lipids, membrane proteins and matrix proteins from the cytosol, and periodically divide. Unlike plastids and mitochondria, peroxisomes contain neither DNA nor ribosomes. All proteins in these organelles are encoded by nuclear genes, translated on free polysomes in the cytosol and targeted to the organelle.
Figure 4.28 Peroxisomes. (A) TEM image showing three unspecialized peroxisomes closely associated with endoplasmic reticulum in a bean (Phaseolus vulgaris) root cell. (B) TEM image of a peroxisome adjacent to a chloroplast in a tobacco (Nicotiana tabacum) leaf cell that has been stained with diaminobenzidine, which becomes electron-dense where the enzyme catalase is active. In photosynthetic tissues, peroxisomes engage in photorespiration.
While their function varies in different tissues and organs, peroxisomes contain oxidases that generate hydrogen peroxide, a toxic by-product of metabolism, and catalase, an enzyme that breaks down hydrogen peroxide to form oxygen and water. In leaves, peroxisomes participate in the glycolate pathway, also known as photorespiration (see Chapter 9). In nitrogen-fixing root nodules of some legumes, peroxisomes are involved in the conversion of fixed nitrogen into nitrogen-rich organic compounds. Peroxisomes are the site of the β-oxidation of fatty acids and the conversion of fats to sugars (see Chapter 6). Peroxisomes are especially prominent is seeds that store fats and oils; in this case they have often been called glyoxysomes. Peroxisomes are implicated in the production of the plant hormone jasmonic acid.
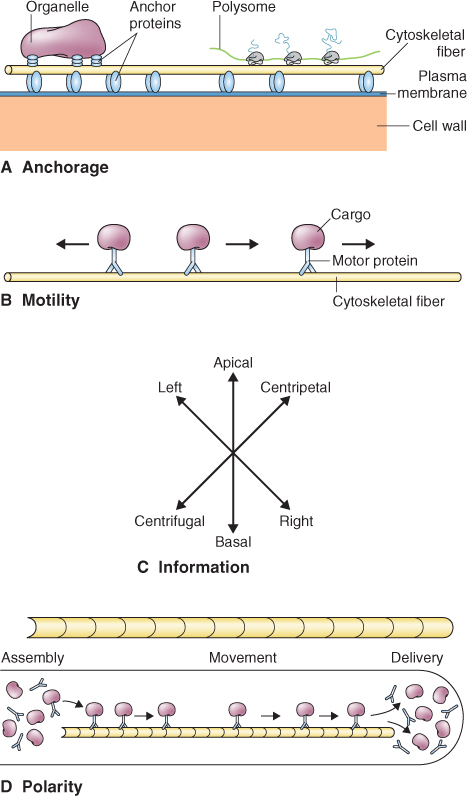
Spatial organization within the eukaryotic cell and directed movements of the cell or its contents are mediated by the cytoskeleton, a network of proteinaceous fibers that permeates the cytosol (Figure 4.29). The cytoskeleton includes three major families of proteins: intermediate filaments, actin and tubulin. Associated with these fibers are accessory proteins that link, move and modify the network. The cytoskeleton provides structural stability to cytoplasm, anchoring proteins and other macromolecules, and supporting organelles during and after their assembly (Figure 4.29A). The cytoskeleton moves cellular components during cytoplasmic streaming and plays a key role in cell wall deposition as well as in the orientation of the new cell wall during cell division. It can confer polarity to a cell, for example, by determining the direction in which organelles move (Figure 4.29B–D).
Figure 4.29 Different functions of the cytoskeleton: (A) anchorage, (B) motility, (C) information and (D) polarity.
The cytoskeleton evolved before plants diverged from animals and the main features of the cytoskeleton have been conserved in both. However, the plant cytoskeleton has also evolved unique functions and many of these are related to properties of the cell wall and the way in which the plant cell divides.
In plant cells, three types of cytoskeletal filaments have been identified: intermediate filaments, actin filaments or microfilaments, and microtubules. Each type of filament is composed of self-assembling, asymmetrical protein subunits. Non-covalent bonds link these subunits; therefore they assemble and disassemble in response to factors that affect protein–protein interactions, such as ionic strength, pH and temperature.
Intermediate filaments are 10–15 nm diameter protein filaments that are thinner than microtubules but thicker than actin filaments. Plants lack the various classes of intermediate filament proteins found in other eukaryotes, e.g. lamins, keratins, vimentins and neurofilaments. Plants also lack a nuclear lamina, a sheet-like structure lying just inside the nuclear envelope that stabilizes the nuclear envelope and maintains the shape of the organelle. Although homologs of the intermediate filament proteins of other eukaryotes are absent from plants, several groups of plant proteins that form coiled-coil interactions typical of intermediate filament proteins have been identified. Among these are a group of proteins known as filament-like proteins (FLPs). FLPs show coiled-coil interactions with partner filaments that allow the formation of rope-like, 10–15 nm long intermediate filaments (Figure 4.30). The coiled-coil proteins identified in plants may be functional homologs of the intermediate filament proteins found in other eukaryotes. These proteins diverged over evolutionary time and now share limited sequence homology.
Figure 4.30 Assembly of intermediate filaments. Identical monomers pair through coiled-coil interactions to form a dimer. Dimers associate laterally to form tetramers. Tetrameters polymerize to form protofilaments. Finally, protofilaments associate laterally to form a rope-like filament.
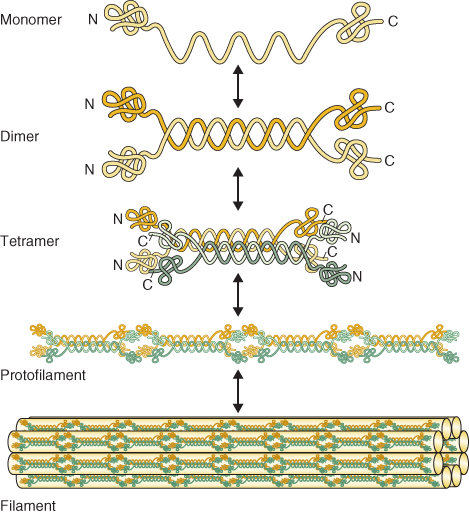
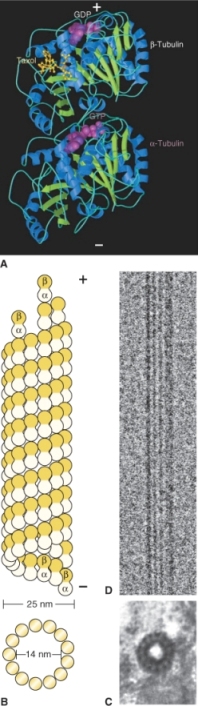
Microtubules are made of heterodimeric subunits of the globular proteins α- and β-tubulin (Figure 4.31). The dimers associate to form a hollow structure 25 nm in diameter. Tubulin dimers bind together at their sides and at their ends. The dimers lie in straight columns called protofilaments. Most microtubules have 13 protofilaments, but the number can vary from 11 to 16.
Figure 4.31 Structure of microtubules. (A) Ribbon diagram of the tubulin dimer within a microtubule. Regions of β-sheet structure are shown in green and the α-helix in blue. (B) Diagrams of a microtubule in cross-section and side view. (C) Electron micrograph of a cross-section of microtubule from a plant cell showing 13 protofilaments. (D) TEM image of a microtubule assembled from purified tubulin and viewed from the side. Unlike (C), this side view was prepared using cryotechniques, without fixation or staining with heavy metals.
Actin filaments or microfilaments are made of the protein actin. Soluble actin is a globular protein of 375 amino acids (sometimes termed ‘G-actin’ in contrast to the filamentous form called ‘F-actin’). Subunits polymerize into a tightly helical filament, about 8 nm wide (Figure 4.32A).
Figure 4.32 Structure and assembly of actin filaments. (A) (i) Ribbon diagram of G-actin monomers in the open configuration with bound ATP shown in yellow. (ii) Sequence showing open G-actin, an F-actin trimer in the closed configuration following hydrolysis of ATP, and a growing actin filament showing staggering of the monomers that give the filament its helical configuration. (B) (i) Actin filaments and microtubules form by the addition of polarized subunits to the plus and minus ends. (ii) Actin and tubulin monomers bind nucleoside triphosphates (NTPs) and the NTP is hydrolyzed when the monomer becomes incorporated into the growing polymer.

Both actin and tubulin occur in all eukaryotes. Actin and tubulin are highly conserved proteins that appear to have arisen from single-copy genes present before multicellular eukaryotes diverged. In plants, α- and β-tubulin genes are present in similar numbers (four to nine copies), whereas the number of actin genes present varies widely, e.g. Arabidopsis contains ten actin genes whereas as Petunia has more than 100.
Actin filaments and microtubules are polar structures because their protein subunits are asymmetrical. In polymer assembly, the asymmetrical subunits line up end to end with a uniform orientation (Figure 4.32B). The polarity thus conferred to the polymer means that each end has a different biochemical character and therefore each end of the polymer may have different rate constants for assembly and disassembly reactions. In addition, each end of the polymer can be recognized specifically so that cells may build polymer arrays with uniform polarity. The more dynamic end of the polymer is designated ‘plus’ and the less active end ‘minus’.
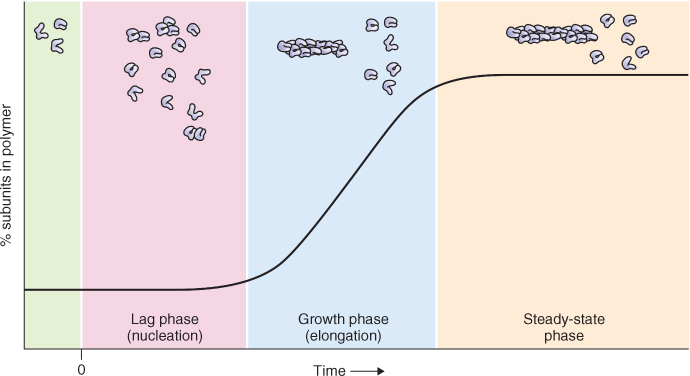
Actin filaments and microtubules assemble by similar processes that share several characteristics (Figure 4.33). The first stage of assembly is called nucleation and results in the formation of a template. During the second stage, elongation, templates formed during nucleation grow by the endwise addition of subunits. The rate of elongation represents the difference between the rate of subunit addition and the rate of subunit loss. The third stage is a steady state in which a constant length of fiber is maintained over time. The steady state is dynamic, the fiber gaining and losing subunits constantly. However, at the steady state, the rate of subunit addition is balanced exactly by the rate of subunit loss.
Figure 4.33 Graph showing the process and kinetics of polymerization. At time zero, a solution of individual subunits is induced to polymerize. During nucleation (pink panel), subunits must associate to create a stable template that serves as a platform for further elongation. In this example, using actin filaments, templates are formed when three subunits associate, generating a trimer. During elongation (growth phase, blue panel), subunits are rapidly added to the ends of growing polymers. A steady-state phase (yellow panel) is reached when the rate of subunit addition is balanced exactly by the rate of subunit loss.
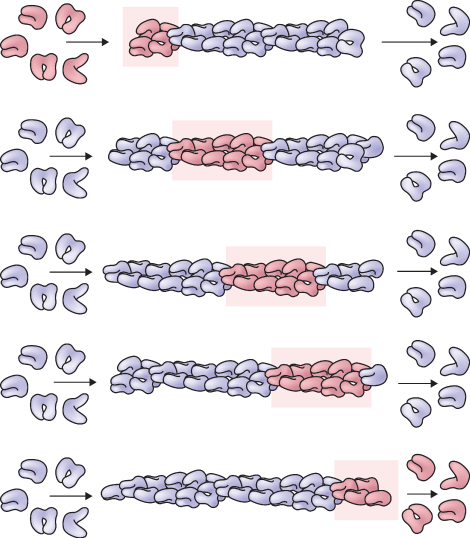
The assembly of microtubules and microfilaments requires the binding and hydrolysis of nucleoside triphosphate (NTP) by their respective protein subunits. G-actin binds ATP and α- and β-tubulin bind guanosine triphosphate (GTP). The γ-phosphate bond of the NTP is hydrolyzed only after a subunit has bound to the growing structure. Nucleotide hydrolysis has different consequences for the assembly of actin and tubulin. In the case of actin, the presence of ATP at the plus end of the filament results in an assembly rate far exceeding that at the minus end, where the actin subunits contain ADP. This difference underlies a type of dynamic behavior called treadmilling, which occurs when the concentration of free subunits supports growth at the plus end that is balanced by shrinkage at the minus end. The net rate of change in polymer length can be zero, but subunits incorporated at the plus end will ‘treadmill’ through the polymer and are eventually released from the minus end (Figure 4.34). Because the difference in assembly rate at the plus and minus ends is much larger for actin than for microtubules, treadmilling is presumed to be more common among actin assemblies. However, both polymers demonstrate this behavior.
Figure 4.34 Treadmilling, a dynamic behavior observed in cytoskeletal polymers. Subunits (dark pink) added at one end (top row) ‘treadmill’ through the polymer and are released from the other end (bottom row). Although the diagram illustrates an actin filament, treadmilling also occurs in microtubules.
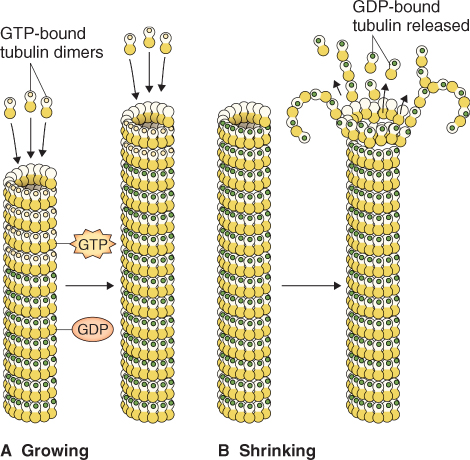
Nucleotide hydrolysis also gives rise to a different kind of behavior in microtubules, called dynamic instability. Here, energy released by GTP hydrolysis accelerates the rate of depolymerization. Growing microtubules add GTP-bound subunits. Because hydrolysis lags slightly behind assembly, the growing ends of a microtubule will have a cap of subunits with bound GTP. This cap stabilizes the structure and favors further growth (Figure 4.35A, B). However, when the supply of tubulin dimers runs low, or when the rate of GTP hydrolysis accelerates, the end of the microtubule may contain a majority of GDP-bound subunits. When freed of the GTP cap, the protofilaments peel apart and initiate a catastrophic disassembly, hundreds of times faster than the rate of growth.
Figure 4.35 Dynamic instability of microtubules. (A) Diagram of growing microtubules (GTP is shown by yellow dots and GDP by green dots). Since hydrolysis of GTP usually lags behind polymerization of new subunits, the growing ends of microtubules are rich in subunits in which β-tubulin monomers bind GTP. Such microtubules are said to have a ‘GTP cap’. (B) Hydrolysis of GTP causes a conformational change that tends to bend protofilaments outward, weakening lateral contacts between dimers shown with green dots in adjacent protofilaments.
Accessory proteins interact with and regulate the different functions of both actin and tubulin. These accessory proteins fall into two general classes, those that influence the assembly and organization of cytoskeletal proteins, and those, called motor proteins, that convert chemical energy into motion.
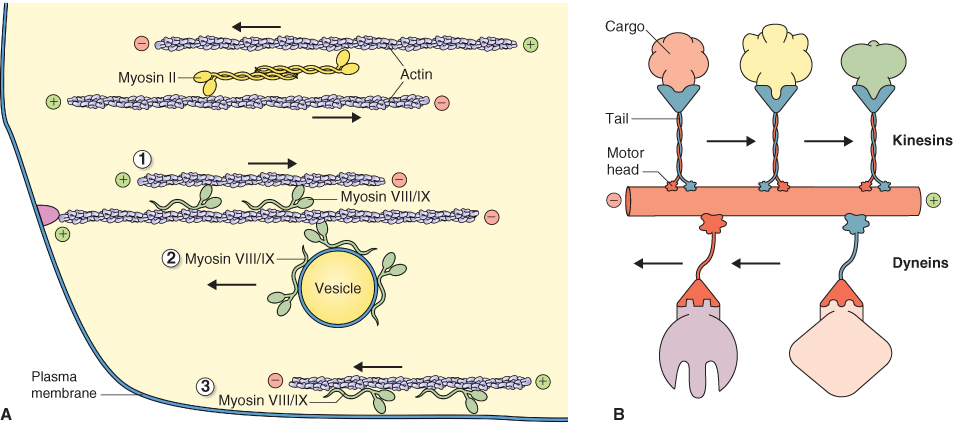
Motor proteins fall into three families: myosins, dyneins and kinesins. All motor proteins contain a globular, force-producing ‘head’ domain that binds a cytoskeletal polymer and a rod-shaped ‘tail’ domain that binds cargo (Figure 4.36). Each also uses ATP as a source of energy. Myosin powers many types of cellular motility involving actin filaments (Figure 4.37A), whereas dynein and kinesin interact with microtubules (Figure 4.37B). Dynein transports cargo to the minus end of a microtubule while kinesin moves cargo towards the plus end. Kinesins are implicated in vesicle traffic and the formation of mitotic spindles. All three families of accessory proteins are found in plants but their relative abundance differs from that in animals. In plants, axonemal dynein is only found in those phyla that have motile sperm; angiosperms appear to lack this motor protein. In contrast, plants have more kinds of kinesin than animals, including forms that are plant-specific.
Figure 4.36 TEM images of highly purified motor proteins. (A) The flagellar form of dynein has three spherical head domains. (B) The cytoplasmic form of dynein has two head domains. (C) Myosin II; the asterisk-shaped structure in the lower right of the image is an IgM antibody used to purify the myosin. (D) Kinesin, showing two small motor domains at one end of the rod-shaped protein while the light chains at the other end are not distinguishable from each other.
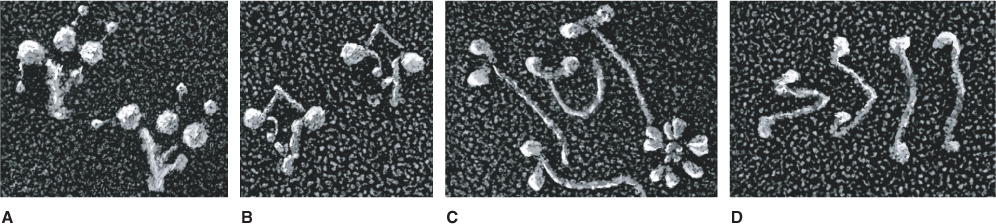
Figure 4.37 Motor proteins and their interactions with actin filaments. (A) Myosin proteins interact with actin filaments, moving from the minus (pointed) end to the plus (barbed) end of the polymer. Myosin II proteins, such as those in muscle, have long rod-like domains that promote assembly into bipolar (head-to-tail) filaments. Myosin VIII/IX motors have short tail domains that can bind an actin filament or a membrane. Force transduction by the myosin VIII/IX head may then (1) move one actin filament relative to another, (2) move a vesicle along an actin filament, or (3) move an actin filament along a membrane. (B) Diagram of the microtubule-associated motor proteins dynein and kinesin. Dyneins move cargo toward the minus end of a microtubule, whereas most kinesins move cargo toward the plus end.
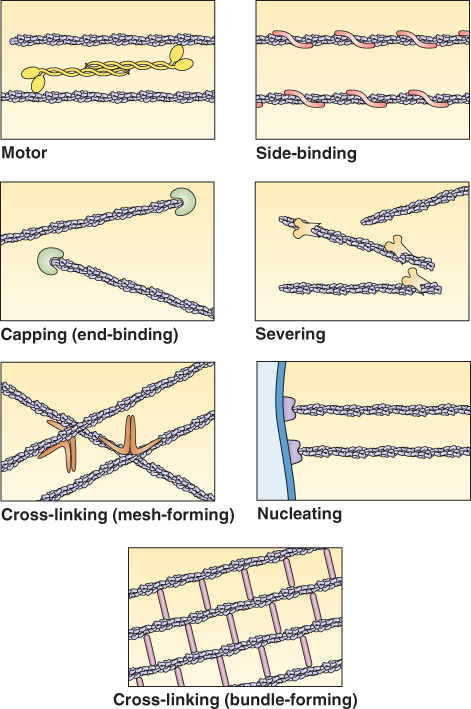
In addition to motor proteins, several other types of accessory proteins affect cytoskeleton assembly and function (Figure 4.38). Some, called ‘cross-linking’ or ‘bundling’ proteins, form bonds between cytoskeletal polymers of the same type. Some cytoskeletal accessory proteins stabilize polymers; others sever or weaken them. Some proteins bind specifically to one of the polymer ends. Others, including the pollen allergen profilin (see Chapter 16), bind soluble actin, lowering the concentration of subunits available for polymerization. Finally, some proteins cross-link the cytoskeleton to other components of the cell, such as membranes, biosynthetic enzymes or signal transduction components.
Figure 4.38 Various functions of accessory proteins. Proteins that associate with the cytoskeleton have various functions, as illustrated here with actin filaments. Although these proteins have been studied mainly in animal cells, some similar and possibly homologous proteins have been identified in plant cells.
In many plant cells, organelles such as mitochondria and plastids flow through the cytoplasm; this movement is called cytoplasmic streaming. Cytoplasmic streaming is thought to involve an interaction between myosin that is bound to the surfaces of organelles and actin filaments in the cytoplasm. The cytoskeleton also moves organelles to specific locations and anchors them at specific positions within the cell. For example in photosynthetic cells, actin filaments are involved in repositioning chloroplasts for optimal light absorption in response to changes in light intensity (Figure 4.39). Also actin filaments and microtubules can collaborate to reposition and anchor the nucleus.
Figure 4.39 Reorientation of chloroplasts in an Arabidopsis leaf. (A) Under dim light, chloroplasts move to walls parallel to the leaf surface, thus maximizing light absorption for photosynthesis. (B) In bright light, chloroplasts migrate to cell walls perpendicular to the leaf surface, thus minimizing light absorption and photodamage.

Actin filaments play a pivotal role in exocytosis in tubular cells that undergo ‘tip growth’ (also discussed in Chapter 12); in these cells exocytosis is localized to the extending apical end of the cell. In tip-growing cells (e.g. pollen tubes, root hairs), polarized actin filaments are arranged perpendicular to the tip and deliver secretory vesicles to the tip. The vesicles have myosin in their membranes and associate with actin filaments (Figure 4.40). The high rates of relative expansion sustained by tip-growing cells presumably requires the delivery of a large quantity of wall precursors without interruption. Whether other cell types use actin-dependent exocytosis remains to be determined.
Figure 4.40 Schematic, cross-sectional view of the movement of vesicles to the tip of a growing pollen tube. Cortical microtubules are shown in green, actin filaments and cables in orange (V, vacuole). The high cytoplasmic Ca2+ gradient in the tip region is represented by gray shading with black representing the highest Ca2+ concentration. Blue dots represent secretory vesicles.
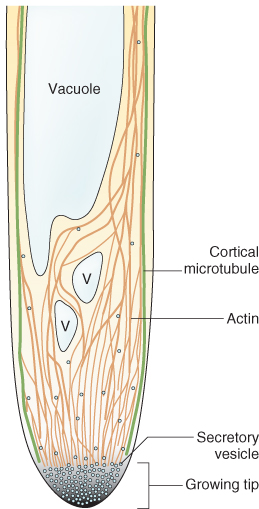
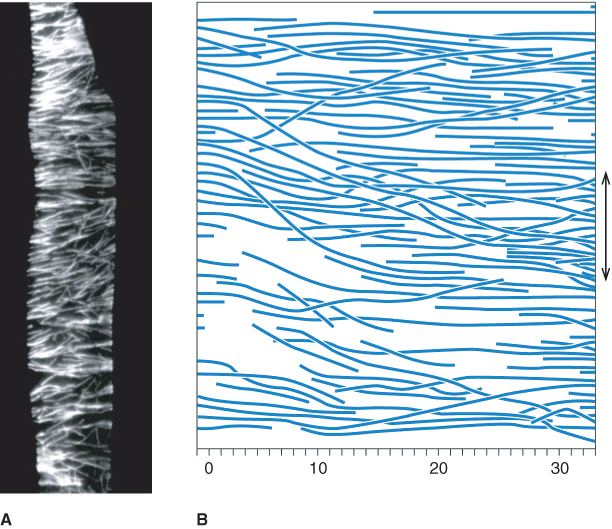
An array of microtubules usually lies just inside the plasma membrane of plant cells and perpendicular to the direction of most rapid expansion. In single cells, or cylindrical organs such as stems and roots, the direction of maximum expansion parallels the long axis of the organ. Hence, the cortical microtubules, orientated perpendicular to that axis, are commonly described as ‘transverse’ (Figure 4.41). The principal function of the cortical array is to influence the direction of cellulose microfibril deposition, which, in turn, specifies the direction of maximal cell expansion in growing cells.
Figure 4.41 Orientation of microtubules in an elongating Arabidopsis root cell. (A) Fluorescence micrograph of a cell from the root of Arabidopsis thaliana, showing the cortical array of microtubules. Microtubules are orientated mainly with their axes transverse to the long axis of the cell (and root). (B) Region of the cortical array from a cell in the root of Azolla pinnata. The double-headed arrow represents the direction of maximum cell expansion rate.