Figure 5.1 Bacterial (A) and plant (B) cells. The plant cell has many membrane-enclosed compartments while the bacterial cell does not.

Chapter 5
Membrane transport and intracellular protein trafficking
In this chapter we will discuss how solutes move into and out of plant cells and organelles. We will also examine how macromolecules are transported within cells. Minerals, sugars, metabolites and other small molecules move across the limiting membranes of cells and organelles, whereas larger macromolecules, such as proteins and glycoproteins, are packaged within vesicles for transport to their specific destinations. Membranes form barriers that insulate the contents of cells from the harsh external environment and allow the partitioning of the cytoplasm into compartments that carry out specific metabolic functions. As evolution progressed, the partitioning of the cytoplasm into membranous organelles allowed eukaryotic cells to increase in size and adopt highly specialized functions. In contrast to prokaryotes, eukaryotic cells contain many specialized organelles responsible for diverse biosynthetic, catabolic and storage functions (Figure 5.1; see Chapter 4). Compartmentation of solutes and macromolecules within membrane-bound organelles not only concentrates reactants and catalysts, but also segregates incompatible processes.
Figure 5.1 Bacterial (A) and plant (B) cells. The plant cell has many membrane-enclosed compartments while the bacterial cell does not.

Movements of water and solutes are among the first changes that occur when dormant or quiescent seeds are activated. Mature seeds generally contain less than 10% water and the hydration of cells and tissues is one of the first events that occurs when seeds are imbibed. The movement of solutes across membranes is facilitated by transport proteins embedded within membranes (Figure 5.2). These transporters are selective for the compound being transported, and their activities are often highly regulated. Vesicular transport of macromolecules is also selective, distributing cargo to various destinations both inside and outside the cell. The selectivity of vesicular traffic can lie in information contained on the surface of the vesicle as well as in the cargo contained in the vesicle.
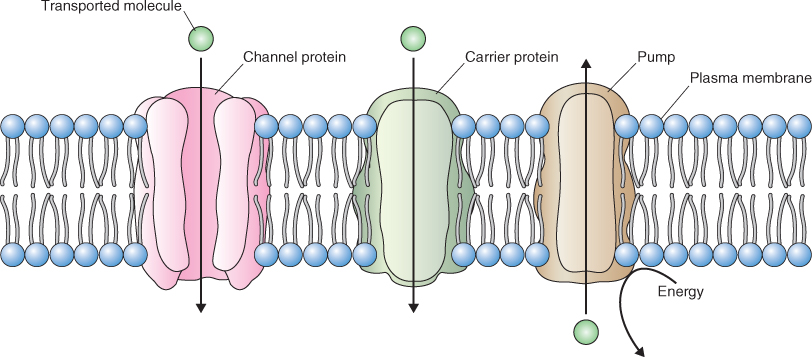
Figure 5.2 Channels, carriers and pumps are the principal classes of membrane transporters. Channels and carriers transport solutes down a favorable electrochemical gradient whereas pumps can move solutes against an electrochemical gradient by expending energy, usually in the form of ATP.
The field of membrane transport has benefited enormously from advances in molecular cloning and genome biology. The nucleotide and amino acid sequences of many transport proteins have been determined and, as you will see in this chapter, detailed information is now available on the molecular structure of many of these proteins. Another important advance in the field of solute transport came about with the development of the patch clamp technique more than 30 years ago. This method allows the behavior of single transport proteins to be studied in the membranes in which they are located, and for the cell physiologist it represents the ultimate molecular approach as the function of the molecule can be analyzed in near in vivo conditions.
Membrane and vesicular transport must be sufficiently precise to ensure that solutes are properly distributed within the cell and that macromolecules reach their final destinations. Differences in the concentrations of ions and small molecules across membranes can vary by several orders of magnitude and these differences in concentration are achieved in part by the activities of membrane transporters. Differences are also found in the distribution of macromolecules in cells. Soluble enzymes are present in all subcellular compartments, whereas membrane-bound proteins occur in the more than 14 different lipid bilayers that delimit these compartments (see Table 4.2). It is estimated that as many as 30% of all cellular proteins are found in membranes. Some proteins are unique to a particular structure, compartment or membrane; other very similar proteins with comparable amino acid sequences, structures and functions occur in more than one compartment. Clearly, the specificity of cellular function requires that the transport machinery in cells be highly specific and carefully regulated.
In this chapter we will first discuss the principles that govern the movement of solutes across cellular membranes. We will then look at the mechanisms by which this transport occurs. Finally, we will discuss intracellular sorting and the transport of macromolecules.
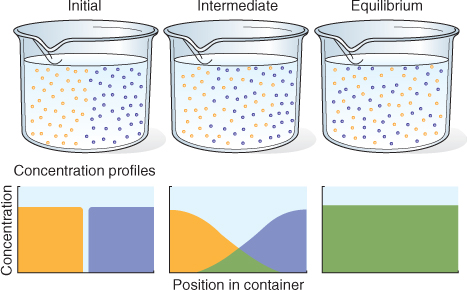
The driving force for the movement of a molecule either in solution or in the gas phase is a gradient in its potential energy. Except at absolute zero, molecules are in constant motion and will distribute themselves uniformly in the space that they occupy, as illustrated in Figure 5.3. This movement is called diffusion.
Figure 5.3 Random thermal motion of molecules dissipates concentration gradients, eventually leading to complete mixing. Initially the blue and yellow molecules are entirely separate; at equilibrium they are randomly distributed and therefore completely mixed. The diffusion of the molecules of each species is driven by its own concentration gradient. Diffusion is most rapid in the gas phase, slower in the liquid phase and much less rapid in the solid phase.
Diffusion occurs spontaneously; no additional energy must be provided. Diffusion occurs in biological systems, for example from one point in a cell to another or across a membrane. The rate at which a molecule, s, diffuses is related to the size of the molecule, the magnitude of its concentration gradient, the viscosity of the medium and temperature. This relationship is shown in Equation 5.1, known as Fick's law.
The rate of diffusion (Js) of species s is determined by its diffusion coefficient (Ds) and concentration gradient (ΔCs/Δx), i.e. the concentration difference (ΔCs) between two points (Δx). Js is expressed as moles per area per time. Small molecules in the gas phase diffuse rapidly, whereas large molecules, such as proteins or polysaccharides, in solution diffuse slowly. The negative sign in Equation 5.1 shows that substances move by diffusion down a concentration gradient.
The importance of Fick's law is that one can calculate the time that it takes a molecule to diffuse a given distance. The time that it takes substance s to move distance L is given by the expression L2/Ds. Thus the time it takes for s to diffuse increases with the square of the distance. The value of Ds for ions is around 10−9 m2 s−1 and for larger biological molecules Ds is around 10−11 to 10−10 m2 s−1. A molecule with a Ds of 10−9 m2 s−1 will take 2.5 s to move 50 μm, the diameter of an average plant cell, but to move a distance of 1 m would take 32 years. Thus diffusion may be an effective way for molecules and ions to move cellular distances, but it is ineffective for long-distance movement. As we shall see in Chapter 14, other mechanisms operate in plants to move solutes over long distances, e.g. from organ to organ.
Fick's law describes the flux of an uncharged solute; it only considers its concentration. When discussing transport of solutes in biological systems, it is important to consider the sum of the physical forces that act upon solute s. The chemical potential (μs) of a solute is its free energy per mole. It is affected by the concentration of the solute, its electrical charge and hydrostatic pressure (Equation 5.2).
In this equation 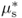 is the chemical potential of s under standard conditions; it provides the reference point for the measurement of the chemical potential of s. The term RTlnCs, the concentration component, includes the concentration C of s in moles per liter at standard pressure, R, and absolute temperature T. The term zsFE represents the electrical component; z is the charge on s (zero for an uncharged molecule and + 1, + 2 or − 1, − 2, etc. depending on the valence of the cation or anion), F is Faraday's constant and is equal to the amount of charge in one mole of electrons, and E is the electrical potential of the solution with respect to ground. The hydrostatic pressure component, VsP, includes the partial molal volume (V) of s and pressure (P).
is the chemical potential of s under standard conditions; it provides the reference point for the measurement of the chemical potential of s. The term RTlnCs, the concentration component, includes the concentration C of s in moles per liter at standard pressure, R, and absolute temperature T. The term zsFE represents the electrical component; z is the charge on s (zero for an uncharged molecule and + 1, + 2 or − 1, − 2, etc. depending on the valence of the cation or anion), F is Faraday's constant and is equal to the amount of charge in one mole of electrons, and E is the electrical potential of the solution with respect to ground. The hydrostatic pressure component, VsP, includes the partial molal volume (V) of s and pressure (P).
When considering the chemical potential of biological molecules at the cellular level, the contributions of V and P are generally small compared with the contributions of the chemical concentration and electrical properties of s. Therefore, for an ion or a charged molecule, Equation 5.2 can be simplified to Equation 5.3, which contains only the concentration and charge components.
The chemical potential of a charged species is often called its electrochemical potential. For an uncharged solute the electrical component of the equation equals zero and the equation for its chemical potential becomes:
Although the hydrostatic component of the chemical potential of solutes in cells is very small, in Chapter 14 we will see that pressure is an important factor when considering the chemical potential of water.
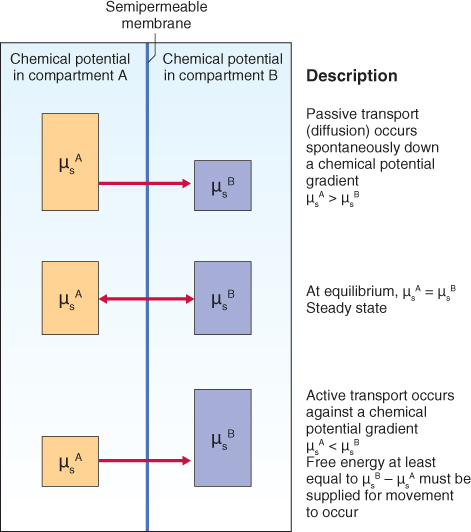
The driving force for diffusion of a solute into or out of a cell is the difference between its chemical potential (μs) inside the cell and that in the external solution (Figure 5.4). Therefore it is important to be able to describe this difference quantitatively. For uncharged solutes the difference in chemical potential (Δμs) between the inside (i) and the outside (o) of a cell can be described by an expanded expression of Equation 5.4 (Equation 5.5).
Figure 5.4 The magnitude of the chemical potential gradient drives the movement of solutes across membranes. The height of the boxes represents the chemical potential of solute s in each compartment; the membrane is permeable to solute s. When a difference in the concentration of s occurs across the membrane, s will flow passively down its chemical potential gradient (top) until equilibrium is reach (middle). To move s against its chemical potential gradient, free energy must be supplied (bottom), a process called active transport.
This equation reduces to:
The conclusion that can be drawn from Equation 5.6 is that the driving force for the diffusion of uncharged solute s is the magnitude of its concentration difference across the membrane. The sign of the product of this equation indicates the net direction of movement of s; a negative sign shows that s will tend to diffuse into the cell down its chemical potential gradient. The other factor that will determine the movement of s across a membrane is the membrane's permeability to s, a topic that we will explore in more detail below.
For charged molecules and ions, the electrical component as well as the concentration component of Equation 5.2 must be taken into account when calculating the gradient in electrochemical potential across the membrane, as shown in Equation 5.7.
This equation reduces to:
The term Ei − Eo in Equation 5.8 is often called the membrane potential or membrane voltage and is abbreviated Vm. We can conclude from Equation 5.8 that movement of a charged solute responds to two independent forces, namely the difference in the concentration of s across the membrane and the membrane potential. While only solute s can contribute to changes in its own concentration, any charged solute can change the membrane potential. This means that charged solutes can diffuse into or out of cells against their own concentration gradient if the membrane potential is favorable. We will discuss this idea in a little more detail below.
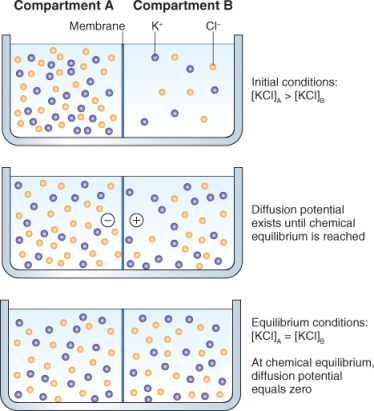
The rate at which different charged molecules and ions cross membranes is often not equal and even small differences in their distribution across the membrane can give rise to a membrane potential. The development of a membrane potential can be illustrated by the example shown in Figure 5.5; here two KCl solutions of different concentration are separated by a membrane that is more permeable to K+ than Cl−. In this illustration, both K+ and Cl− diffuse down their concentration gradients from compartment A to compartment B, but K+ diffuses through the membrane at a slightly higher rate than Cl−, giving rise to a difference in charge across the membrane. If electrodes were placed in compartments A and B, the difference in concentration of K+ and Cl− could be measured as a difference in voltage. Very small differences in the distribution of charge can give rise to significant differences in membrane potential. For example, it takes only a concentration difference of 0.001% in anions over cations across the membrane to produce a membrane potential of −100 mV. The membrane potential of most plant cells falls in the range − 100 to − 250 mV.
Figure 5.5 A membrane potential can develop when a membrane is differentially permeable to ions such as K+ and Cl−. Upper panel: K+ and Cl− will diffuse across the membrane from compartment A to compartment B because of the electrochemical potential difference for each ion. Initially, because the membrane is more permeable to K+ than to Cl−, a difference in charge—a membrane potential—will develop (middle panel). At equilibrium (bottom panel), the chemical concentrations of K+ and Cl− are equal on both sides of the membrane and the membrane potential is zero.
Solutes moving into and out of cells by diffusion should reach equilibrium. When equilibrium between the inside and outside of a cell is reached for a given solute, Δμs is zero. If we rearrange Equation 5.8:
Equation 5.9 can then be further rearranged to show the difference in electrical potential, ΔE, between the inside and outside of the cell as shown in Equation 5.10:
For a monovalent cation (s) at 25° C, this equation can be simplified to Equation 5.11 (note that the logarithm term is now given as log(base 10) rather than natural logarithm (ln).
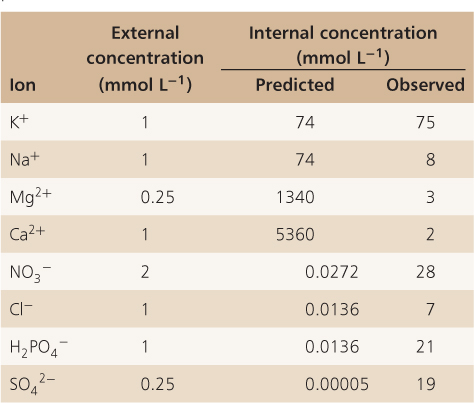
Equation 5.11 is known as the Nernst equation and the expression ΔE for a specific ion is known as the Nernst potential. The Nernst equation has great predictive value for cell physiologists. It can be used to determine whether or not ions are accumulated against their electrochemical potential gradient. If the membrane potential of a cell can be measured and if the concentration of solutes outside (Cso) and inside (Csi) the cell is known, it becomes easy to establish whether a solute is taken up by active transport or whether it moves down its electrochemical gradient. Table 5.1 shows data obtained from pea roots where the concentration of ions inside root cells and in the soil solution were determined at a membrane potential of −110 mV. What this table clearly demonstrates is that while K+ is at equilibrium in pea roots relative to soil solution, ions such as Cl−, H2PO4− and NO3− are present inside pea root cells at concentrations much higher than those predicted by the Nernst equation. Nitrate, for example, is accumulated to a concentration 1000-fold greater than predicted by the Nernst equation. From this it can be concluded that energy must be expended for nitrate to accumulate inside cells. Ions such as Ca2+ and Mg2+, on the other hand, are present inside pea root cells at far lower concentrations than those predicted by the Nernst equation, indicating that there are mechanisms that maintain these ions at a lower concentration, either by preventing their entry into cells or by pumping them out.
Table 5.1 Comparison of predicted and observed ion concentrations in pea root tissue, where the membrane potential was measured as −110 mV.
Biological membranes are selectively permeable. Lipophilic/hydrophobic molecules such as O2, CO2, N2, NH3 and H2O2 move fairly freely through the lipid bilayer (Figure 5.6). The lipid bilayer is even permeable to water, as well as to somewhat larger polar molecules such as urea. As long as favorable chemical potential gradients exist for these molecules they will diffuse through the membrane until there is no difference in their chemical potentials on either side of the membrane. Charged solutes and larger polar molecules such as nucleotides and sugars do not diffuse easily across membranes. Specific transport proteins are therefore required to facilitate their movement into and out of cells. Transport proteins are embedded in the membrane (see Figure 5.2). They generally consist of alternating blocks of hydrophobic and hydrophilic amino acids that allow them to span the phospholipid bilayer of the membrane and, at the same time, have hydrophilic domains exposed to the cytosol, the lumen of an organelle or the cell exterior (Figure 5.7).
Figure 5.6 A phospholipid bilayer is differentially permeable. It is freely permeable to gases such as CO2, O2 and N2 and to small, uncharged polar molecules such as ethanol. Other small, uncharged polar molecules such as urea and water have limited permeability. Membranes are impermeable to larger polar molecules, ions and macromolecules.
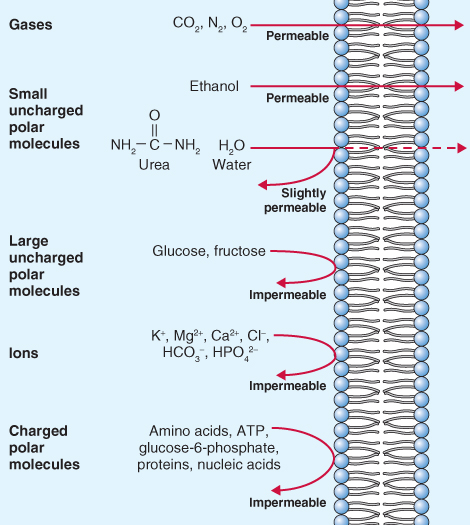
Figure 5.7 Configuration of membrane-associated ATPases. (A) Membrane disposition of the plasma membrane ATPase showing ten transmembrane domains and a long autoinhibitory C-terminal domain. The ATP-binding aspartate residue D is shown between loops 4 and 5. (B) Ribbon diagram of the H+-ATPase based on X-ray crystallographic data of the active form of the pump lacking the long C-terminal tail. The ten transmembrane domains are shown in orange, green and brown, the ATP-binding domain is labeled N, and bound Mg is labeled AMPPCP.
Solutes may cross membranes by diffusion, moving down their electrochemical potential gradients (Figure 5.8A). Solutes that freely cross the lipid bilayer are said to move by simple diffusion. Those that require the presence of a membrane transporter to diffuse across a membrane move by facilitated diffusion. There are two types of transporters that facilitate diffusion across membranes: channels and carriers.
Figure 5.8 Molecules can move across membranes by passive (A) or active (B) transport. Passive transport occurs down an electrochemical gradient by simple diffusion through the lipid bilayer or through proteinaceous channels or carriers. Active transport occurs against an electrochemical gradient and can occur through pumps, symporters or antiporters, all of which are transmembrane proteins.
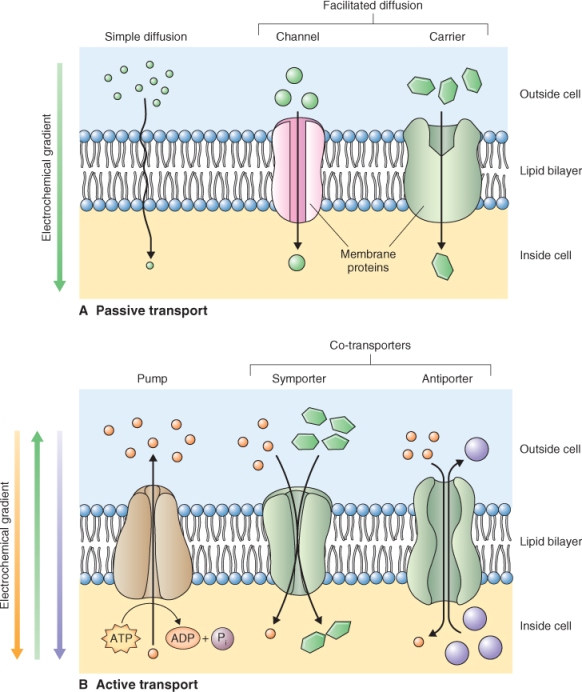
Protein transporters may also move solutes against their electrochemical gradients; this process consumes cellular energy and is called active transport (Figure 5.8B). Active transport can be subdivided into two categories: primary active transport and secondary active transport. During primary active transport, ATP or pyrophosphate is hydrolyzed to provide the energy required to establish ion gradients. Transporters that perform primary active transport are called pumps.
Secondary active transporters, called co-transporters, exploit the ion gradients established by primary active transport to move a second solute against its electrochemical gradient. They transport two solutes simultaneously; one solute moves down its electrochemical gradient while the second moves up its electrochemical gradient. Co-transporters can be divided into two groups: symporters and antiporters. Symporters move two solutes across a membrane in the same direction, while antiporters move one solute in one direction and the second solute in the opposite direction. In plant cells, secondary active transport usually exploits H+ gradients established by proton pumps. Let us examine the characteristics of each of these types of transport protein.
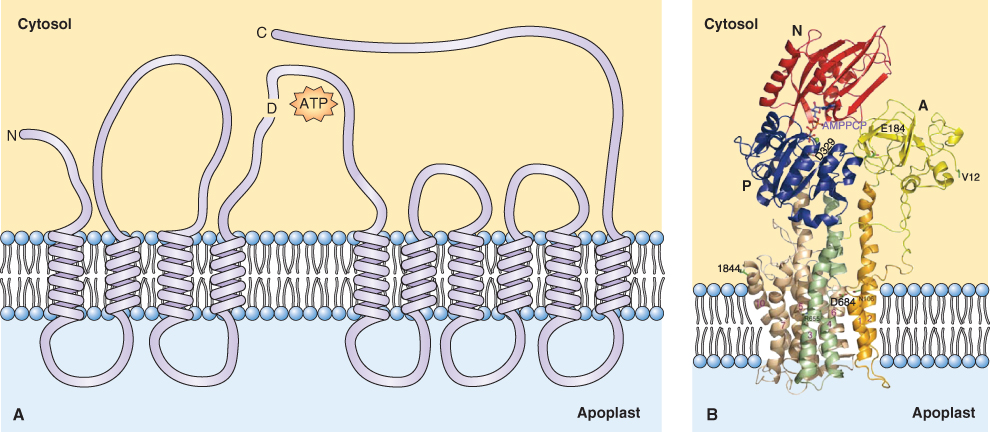
Pumps move solutes against their electrochemical gradients. They bind to the solute to be transported and hydrolyze ATP or pyrophosphate as a source of energy. Pumps transport solutes at a rate of tens to hundreds of molecules or ions per second. They are located in most cellular membranes (Figure 5.9). Pumps that hydrolyze ATP can be divided into three categories based on their structure, inhibitor sensitivity and mechanism of action (Figure 5.10). F-type ATPases (ATP synthases), such as those found in chloroplasts and mitochondria, have multiple subunits and are insensitive to the inhibitor vanadate. The V-ATPase found in vacuole and other cellular membranes is related to F-type ATPases, having multiple subunits, and is also vanadate-insensitive (see Section 5.4.4). P-type ATPases have a simpler structure than the other types of ATPase. They form phosphorylated intermediates after hydrolyzing ATP (Figure 5.11) and are inhibited by vanadate. Members of the ABC superfamily of transporters are P-type ATPases. These transporters consist of four core domains, two that span the membrane and two that are cytosolic. Their cytosolic domains bind and hydrolyze ATP and the energy released drives solute movement across the membrane.
Figure 5.9 Subcellular location of H+ pumps in plant cells. H+ pumps establish proton gradients across cellular membranes that are utilized in the synthesis of ATP by the F-type enzyme, or in powering the transport of solutes in the case of the P-type and V-type pumps and the proton-pumping pyrophosphatase. The pyrophosphatase and V-ATPase are found on membranes other than the tonoplast, including the plasma membrane, Golgi apparatus, trans-Golgi network and prevacuolar compartment.

Figure 5.10 Three types of ATP-hydrolyzing pumps are found in membranes. P-type and V-type pumps transport H+, and ABC-type pumps transport a variety of solutes. Note that in plants some P-type pumps such as the plasma membrane H+-ATPase and the Ca2+-ATPase, lack a β-subunit. A, ATP-binding site; T, transmembrane domain.
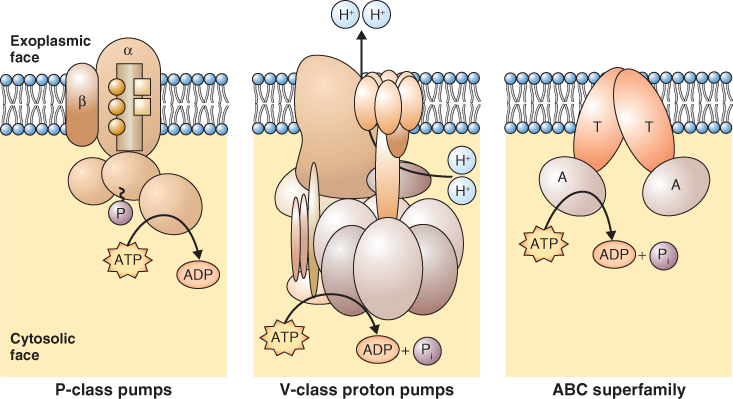
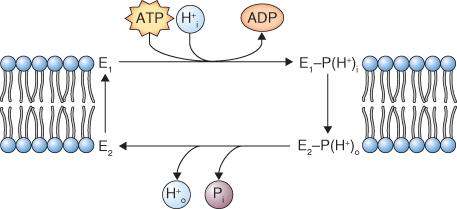
Figure 5.11 Reaction cycle of P-type ATPase in the plane of the plasma membrane. Phosphorylation of the ATPase (E1) leads to binding of H+ on one side of the membrane, leading to formation of E2. E2 has a lower affinity for H+ leading to the release of protons and inorganic phosphate on the other side of the membrane. Hydrolysis of inorganic phosphate from E2 returns it to the E1 state.
Proton (H+ )ATPases play a very important role because protons constitute one of the major energy currencies of the plant cell. At the inner mitochondrial membrane and at the thylakoid membrane, a transmembrane H+ gradient is generated using light or chemical energy; the gradient is then used to energize the synthesis of ATP by H+ pumps (ATP synthases). At all other membranes in the cell, H+ pumps hydrolyze ATP or pyrophosphate to power the transport of protons out of the cytosol. The electrochemical gradient in protons is called a proton motive force (pmf). The pmf can be used in secondary active transport to power the transport of other ions and solutes across the membranes (see Figure 5.8B).
The transport of protons by H+ pumps is not balanced by the movement of anions. Therefore the movement of protons establishes a charge gradient that changes the membrane potential. Pumps that create a charge gradient are said to be electrogenic. A very small difference in the actual number of positive and negative charges across the membrane is responsible for the change in the membrane potential Vm. Activity of H+ pumps in the plasma membrane makes its Vm more negative, i.e. hyperpolarizes the Vm; if the activity of these pumps decreases, the Vm depolarizes, becoming less negative. As we shall see these changes have an important role in the uptake of ions.
The plasma membrane (PM) H+-ATPase is a Mg-dependent, P-type ATPase; it is a single subunit, 100 kDa protein with ten membrane-spanning domains (see Figure 5.7). One H+ is pumped out of the cytosol for each ATP hydrolyzed; the reaction cycle of this enzyme is illustrated in Figure 5.11. The activity of this pump is key to the transport activities of the plant cell and it is a major consumer of cellular ATP. It is estimated that 30–50% of ATP produced by a cell is hydrolyzed by the PM H+-ATPase. This pump establishes the cell's pmf, powering the activity of symporters and antiporters (see Figure 5.8B). It also influences the Vm, thus affecting the movement of ions through channels. Acidification of the cell wall by the H+ pump also influences growth and development. Auxin activates the PM H+ pump within minutes, and the resulting acidification of the cell wall increases its extensibility, allowing turgor pressure to bring about cell expansion (see Chapter 12).
Another important function of the PM H+ pump is the maintenance of cytosolic pH. The pH of the cytosol is maintained in the range 7.3–7.5 despite the fact that many of the reactions of intermediary metabolism generate an excess of H+. An interesting aspect of the activity of the PM H+-ATPase is that its pH optimum is 6.6. Therefore, an accumulation of H+ in the cytosol results in the activation of the pump.
Given the central role of the plasma membrane H+ pump in plant cell physiology, the regulation of its activity has been intensively studied. This pump is encoded by a large multigene family; 11 genes have been identified in Arabidopsis. Two of these genes are expressed in all Arabidopsis cells and there is evidence that individual cells can express multiple isoforms of the gene.
Despite the physiological importance of the PM proton pump, its expression is not strongly regulated at the level of transcription or translation. Instead, evidence points to the regulation of the pump at the level of the enzyme's activity. One way in which the activity of this pump might be regulated is through the action of a 14-3-3 protein. 14-3-3 proteins are a family of conserved proteins found in all eukaryotic cells that bind a range of target proteins and modify their activity. The name 14-3-3 comes from the protein separation pattern after ion exchange chromatography and gel electrophoresis. The PM H+ pump has a C-terminal regulatory, autoinhibitory domain (Figure 5.12) that contains several phosphorylation sites. When a conserved threonine residue in the autoinhibitory domain of the pump is phosphorylated, a 14-3-3 protein binds to the C-terminal domain and forms an active pump −14-3-3 complex (Figure 5.12). Dephosphorylation of the conserved threonine releases the14-3-3 protein and deactivates the pump.
Figure 5.12 Activation of the plasma membrane H+-ATPase by 14-3-3 proteins and the fungal toxin fusicoccin. The autoinhibitory C terminus of P-type ATPase can bind two 14-3-3 molecules at a phosphorylated threonine residue (Thr), leading to relief from autoinhibition. Fusicoccin can also activate the enzyme by binding with a single 14-3-3 molecule at the C terminus.
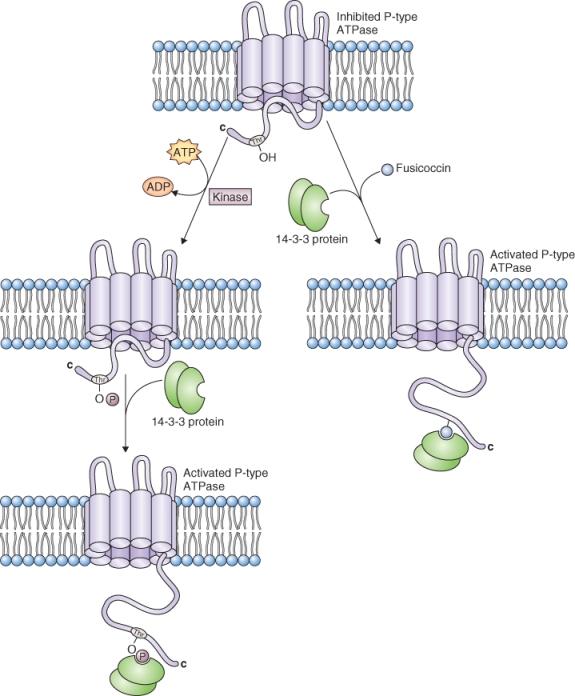
The identification of 14-3-3 proteins has helped to explain the mechanism of action of the fungal toxin, fusicoccin. Work in the 1970s showed that fusicoccin promotes the acidification of the cell walls of cells in various tissues. It was proposed that fusicoccin caused wall acidification by increasing the activity of the plasma membrane H+ pump. We now know that fusicoccin stimulates H+ pumping activity by binding to a site that is formed by the interaction of 14-3-3 proteins and the plasma membrane H+ pump (Figure 5.12).
Calcium is an important regulator of cellular activities; because it forms insoluble salts with phosphate, its concentration in the cytosol is maintained in the low nanomolar range. The concentration of Ca2+ in the cytosol is regulated by a number of transporters, among them a Ca2+-ATPase that is found on almost all cellular membranes including the membranes of chloroplasts and mitochondria. Like the plasma membrane H+-ATPase, the Ca2+ pump belongs to the P-type family of ATPases. It is a single polypeptide of about 110 kDa with an extended N-terminal domain (Figure 5.13A). ATP hydrolysis by this enzyme is accompanied by the transport of two Ca2+ ions across the membrane (Figure 5.13B).
Figure 5.13 Configuration of membrane-associated Ca2+-ATPase. (A) Membrane disposition of the vacuolar Ca2+-ATPase showing ten transmembrane domains and an extended N-terminal calmodulin-binding domain. D, ATP-binding domain. (B) Ribbon diagram of the Ca2+-ATPase based on X-ray crystallographic data, showing the ATP-hydrolyzing domain of a Ca2+ pump from an animal sarcoplasmic reticulum (SR; equivalent to the endoplasmic reticulum (ER)) and probable route of Ca2+ transport.
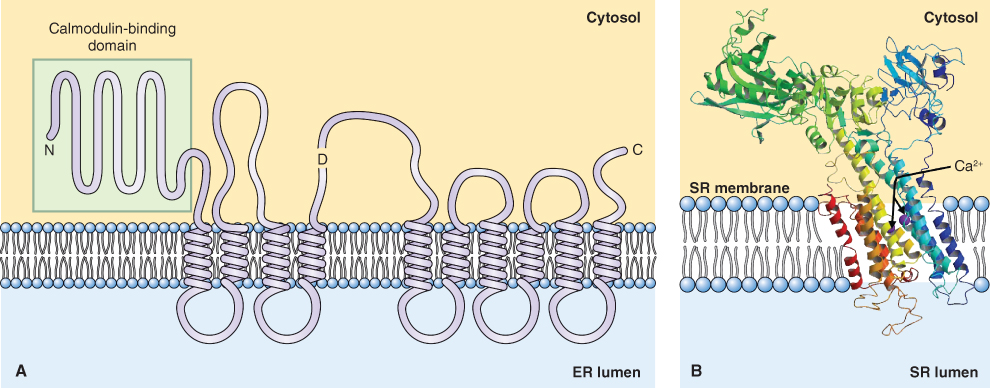
The role of Ca2+ pumps is to maintain cytosolic Ca2+ in the range of 50–200 nM by pumping Ca2+ out of the cell and into the vacuole and other organelles. Two broad classes of Ca2+-ATPases have been identified in plant cells based on their ability to bind calmodulin, a cellular Ca2+ sensor. The calmodulin-binding class of Ca2+-ATPases has an N-terminal, autoinhibitory domain that binds calmodulin in a Ca2+-dependent manner. When calmodulin binds to the N terminus of the Ca2+-ATPase, it inhibits the pump and thus raises the cytosolic Ca2+ concentration. This is a good example of a negative feedback loop that maintains cytosolic Ca2+ homeostasis.
Vacuolar H+-ATPases (V-ATPases) are found in almost all living cells; they share a common origin with F-type ATPases. V-ATPases have a complex multisubunit structure that is reminiscent of F-type ATPases (Figure 5.14). In Arabidopsis V-ATPases are made up of 13 subunits, each of which can be encoded by any one of 27 genes. If subunit isoforms can combine randomly, it is clear that an enormous number of different V-ATPase complexes could be assembled. V-ATPases pump about three H+ per ATP hydrolyzed. They are electrogenic and contribute to the pmf and membrane potential (Vm) of the vacuole membrane, the tonoplast.
Figure 5.14 Model of V-ATPase from the yeast Saccharomyces cerevisiae. The multiple subunits of this complex enzyme are numbered 1 to 13 with an additional subunit labeled Vph1p.
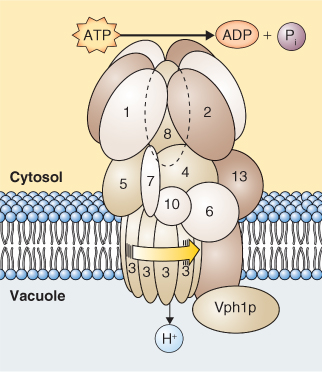
Although described as vacuolar enzymes, V-ATPases are ubiquitously distributed among the endomembranes of plant cells, but are absent from the membranes of chloroplasts and mitochondria. The V-ATPases on the membranes of the Golgi apparatus are thought to be involved in exocytosis and vesicle trafficking in the secretory pathway (see Section 5.8). The H+ pumping activity of the V-ATPase at the tonoplast causes vacuolar acidification. The net positive charge of the vacuole lumen is balanced by the accumulation of organic anions such as malate and oxalate and inorganic anions such as chloride. The pH of the vacuole in most plant cells is about 5.5 but more specialized vacuoles, such as those of lime fruits, have a pH around 1.7, and those of Oxalis spp. are between 1.9 and 2.6.
Plants, some protozoa and many species of archaebacteria and eubacteria possess an H+ pumping pyrophosphatase (H+-PPase), a protein of about 80 kDa. This pump is abundant on the tonoplast membrane but it is also located in the Golgi, trans-Golgi network (TGN), multivesicular body (MVB) and plasma membrane. The functional H+-PPase is a homodimer. Two types of H+-PPase are found in plants. Type I pumps are activated by cytosolic K+ and inhibited by Ca2+, whereas Type II H+-PPases are strongly activated by Ca 2+ and are insensitive to K+. H+-PPases have a remarkable degree of homology across phyla; all H+-PPases from plants and bacteria share >85% sequence homology at the amino acid level.
Physiologists have puzzled over the existence of two types of H+ pumps on the tonoplast of plant cells. One explanation proposes that the H+-PPases evolved to exploit the abundant supply of pyrophosphate in plant cells, where its concentration can reach millimolar levels. Pyrophosphate is produced during several reactions of carbohydrate metabolism, e.g. during the synthesis of ADP-glucose and uridine diphosphate (UDP) glucose. The concerted actions of the V-ATPase and H+-PPase acidify the vacuole and maintain a tonoplast Vm of about 20–30 mV, with the vacuole being positive relative to the cytosol.
ABC (ATP-binding cassette) transporters make up a large family of transporters that hydrolyze ATP and transport a variety of organic molecules. They belong to the P-type class of ATPases. More than 120 ABC transporter genes have been identified in Arabidopsis and rice. All ABC transporters are made up of two basic structural elements: integral, membrane-spanning domains and cytoplasmically orientated, nucleotide-binding folds that are involved in ATP hydrolysis (Figure 5.15). ABC transporters transport uncharged solutes out of the cytosol and are therefore not electrogenic.
Figure 5.15 Model of a vacuolar ATP-binding cassette (ABC) transporter from Arabidopsis. Two nucleotide-binding folds, NBF1 and NBF2, are separated by transmembrane domains containing multiple, integral-membrane helices.
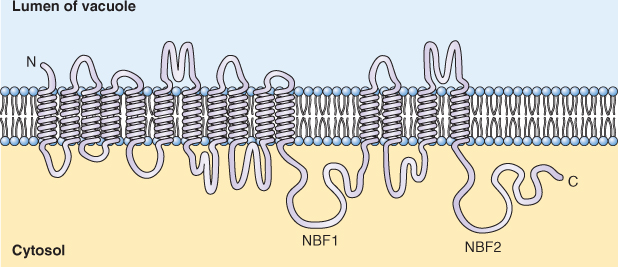
Although they were first discovered on vacuolar membranes, ABC transporters are also found on the plasma membrane. Among the compounds transported by ABC transporters are anthocyanins, chlorophyll catabolites, antifungal compounds and the plant hormone auxin. The BR2 gene of maize, which is known to be involved in basipetal auxin transport, has now been shown to encode an ABC transporter. ABC transporters also have been implicated in the transport of waxes to the surfaces of leaf cells.
ABC transporters are thought to be important in the sequestration of potentially harmful metabolites and xenobiotic (from the Greek: literally, xenos = foreign, bios = to life) molecules in the vacuole. Many xenobiotic molecules are first modified by the addition of the tripeptide glutathione, catalyzed by a family of enzymes called glutathione transferases (GSTs). The addition of glutathione acts as an address label that targets the solute to an ABC transporter. Among the xenobiotics conjugated to glutathione are synthetic herbicides.
Two types of channels have been well characterized in plants, ion channels and aquaporins that allow water movement. An ion channel forms a selective, water-filled pore in the membrane (Figure 5.16). Unlike carrier-mediated transport (see below), there is little interaction between the channel and the ion. Fluxes through ion channels are therefore extremely rapid with up to 108 ions moving per second, compared to carriers and co-transporters that move between 102 and 104 ions per second. Ion channels are therefore ideally suited to permit the transport of large numbers of ions or molecules very rapidly.
Figure 5.16 A plant K+ channel. (A) Top view, based on ribbon structures showing four channel subunits that come together to form the central K+ translocating pore. (B) Side view, showing disposition of the six transmembrane domains of one subunit of Arabidopsis AKT1. The P-domain between transmembrane loops 5 and 6 contributes to pore formation, whereas the charged amino acid residues in domain S4 contribute to the voltage sensitivity of the channel.
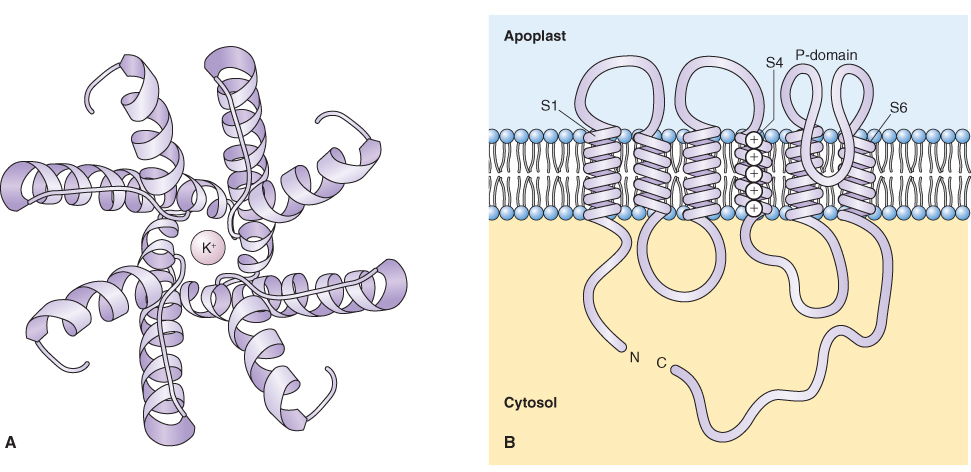
Ion channels are ubiquitously distributed in the membranes of plant cells and they play important regulatory roles. For example, they regulate osmotic concentration by allowing the flux of potassium into and out of cells, and set the concentration of cytosolic Ca2+, an ion that is important in cellular signaling. Movement through ion channels is passive and is dictated by the electrochemical potential for the particular ion. The diffusion of ions through channels is strongly affected by the membrane potential (see Equation 5.8). Because the Vm of the plasma membrane is usually negative, cations tend to diffuse into the cytosol and anions tend to diffuse out.
Most ion channels show strong selectivity for either anions or cations. The selectivity filter of channels operates on the basis of size exclusion. Cation channels can be divided into those that are selective for K+ over other monovalent cations, others that are relatively non-selective among monovalent cations, and those that are selective for Ca2+. Most plasma membrane anion channels allow the passage of a wide range of anions, including Cl−, 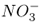 and organic acids. There are anion channels in the vacuolar membrane that select specifically for malate.
and organic acids. There are anion channels in the vacuolar membrane that select specifically for malate.
The patch clamp technique can resolve the activity of a single ion channel molecule as it catalyzes ion translocation. The technique allows the detection of small electric currents carried by ions as they move through channels, made possible by advances in solid-state electronics that allowed accurate measurement of picoampere (10−12 A) currents. A high-resistance seal is produced when a blunt-tipped glass micropipette is pressed against a biological membrane with simultaneous application of suction. The high resistance of this seal forces currents flowing into or out of the pipette to go through the membrane patch covering the pipette tip (Figure 5.17). This configuration is called the cell-attached mode; it can record the activities of individual ionic channels in the membrane patch at the tip of the micropipette. Pulling the pipette away from the rest of the membrane generates an inside-out patch, in which the cytosolic face of the membrane is exposed to the bathing medium (Figure 5.17). As with cell-attached recording, the activities of single ionic channels can be assessed, but the solution composition on both sides of the membrane is defined. Alternatively, the membrane patch that covers the pipette tip can be disrupted with a high-voltage pulse or suction, thereby giving electrical access to the inside of the cell, a configuration called whole-cell mode of recording. The relatively large volume of medium in the pipette exchanges rapidly with the cell contents, and becomes the defined intracellular solution. Finally, if the pipette is pulled away from the cell after attainment of the whole-cell mode, a membrane bleb is also pulled away and reseals itself across the pipette tip as an outside-out patch. This recording mode is particularly useful for testing the effects of putative cytosolic regulators on channel activity.
Figure 5.17 Recording modes used in the patch clamp technique. The initial, electrically-tight gigaohm seal yields a cell-attached mode of recording. When the pipette is pulled away from the rest of the membrane, an inside-out patch is formed in which the cytosolic face is exposed to the bathing medium. Alternatively, if the membrane patch that covers the pipette tip in the attached cell mode is disrupted by a high-voltage pulse or suction, a whole-cell mode of recording is produced. If the pipette is pulled away from the cell after attainment of the whole-cell mode, an outside-out membrane patch is formed.
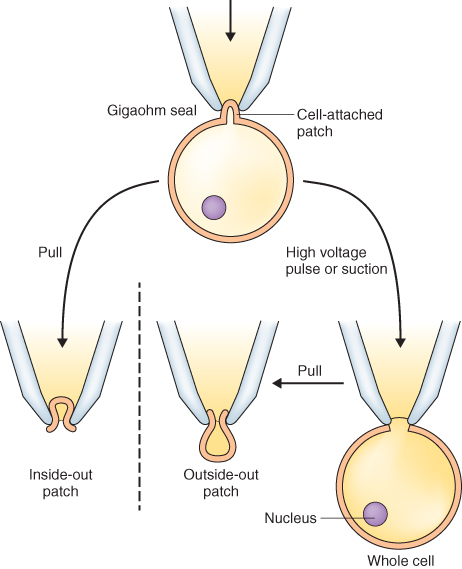
A major advantage of the patch clamp technique is that transport activity can be analyzed in relatively small cells. Thus most if not all types of plant cells are amenable to this approach. In contrast, classic microelectrode impalement techniques can irreparably damage small cells and thus are mostly limited to very large cells.
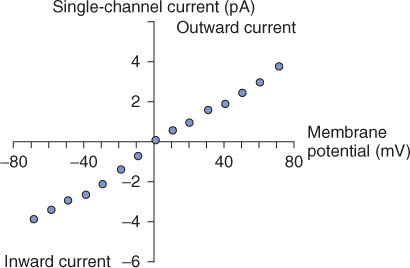
The movement of ions through channels results in the flow of current. This current can be used as a measure of the activity of a channel. Current flow is a function of membrane potential and the resistance that the membrane offers. Ohm's law (I = V/R, where current, I, is the membrane potential, V, divided by the resistance of the membrane, R) can therefore be used to model the movement of ions across the membrane (Figure 5.18). The resistance of a membrane to a particular ion depends on its selectivity and the number of channels. No current will cross a completely impermeable membrane, whereas membranes that are freely permeable will behave as a resistance-free circuit where I = V.
Figure 5.18 Current–voltage relationship of a single ion channel. This graph depicts the current that flows through the channel at different membrane potentials. Current–voltage plots allow determination of reversal potential, Erev, and can help define the permeability of the membrane to specific ions.
An example of the currents that can be measured using the whole-cell patch clamp technique is shown in Figure 5.19. Figure 5.19A shows current flow through the plasma membrane when different voltages ranging from − 100 to + 100 mV are applied to the membrane. What is apparent is that the membrane of this cell has channels that respond to voltage and open at either a positive or a negative Vm. Thus, with an increase in either negative or positive voltage, there is a concomitant increase in current flowing through channels in the membrane. An I–V plot of the data from the experiment illustrated in Figure 5.19A is presented in Figure 5.19B; the curve shows that activation of these channels occurs only after a given threshold voltage has been achieved. This is illustrated most clearly when positive voltage is applied, as channel activity is only activated when a considerable threshold is reached.
Figure 5.19 Whole-cell ion currents in a plant cell as measured by the patch clamp technique. (A) A pipette is sealed to the plasma membrane in the whole-cell configuration. In response to a series of 1-second voltage pulses, current flows through channels in the plasma membrane. The 13 current traces, corresponding to 13 different voltages between − 100 and + 100 mV, are superimposed in this plot. (B) Current (I)–voltage (V) relationship for the time-dependent component of currents in (A). Note that this whole-cell I–V curve is decidedly non-linear.
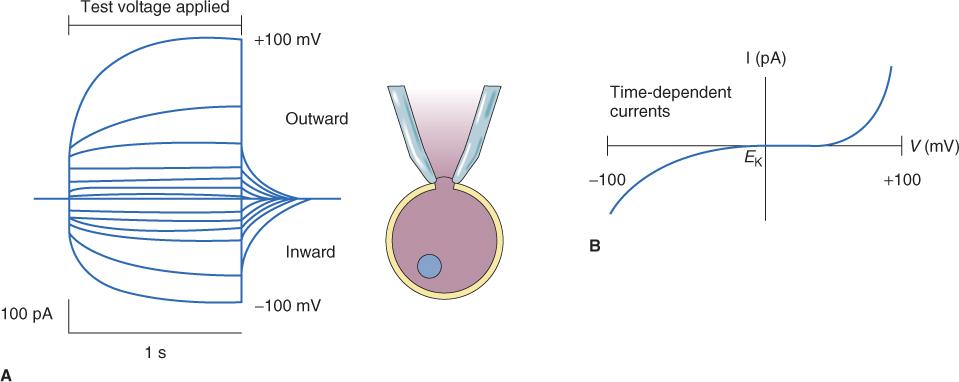
Because ion channels allow the movement of large numbers of ions per unit time, their activities must be tightly regulated. The regulation of channel opening and closing is often referred to as gating. Many ion channels function as part of a feedback loop where membrane potential is one of the key regulators of channel opening and closing. Voltage-gated K+ channels are important in maintaining the cell's membrane potential, because a shift in Vm can be compensated for by altering the opening and closing of channels. The plasma membrane has channels that allow either inward or outward movement of K+. These operate like valves and are said to rectify. Those channels that allow inward movement of K+ are referred to as inwardly rectifying channels whereas those that allow outward movement of K+ are called outwardly rectifying channels. By opening and closing in response to voltage these channels can keep Vm constant. On the other hand, changes in Vm can be used to regulate osmotic concentration by altering the electrochemical potential gradient of ions. Ion channels can also be gated by ligands (chemicals that bind to the channel), such as hormones, Ca2+, G-proteins and pH. Other channels are stretch-sensitive and are gated by changes in the turgor pressure of a cell.
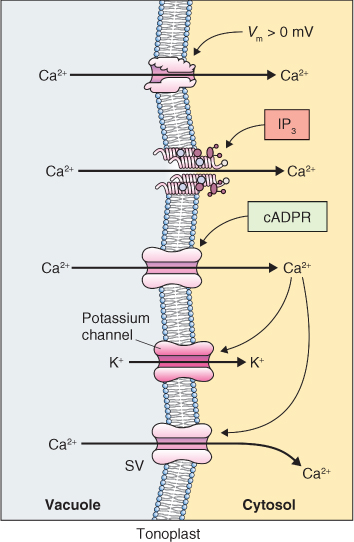
Complex regulatory circuits can be built up by the interaction of these various regulators of ion channel activity and the way in which these regulators interact is nicely illustrated by what happens at the tonoplast of guard cells when stomatal closure is induced by abscisic acid (ABA) (Figure 5.20). The tonoplast has at least four Ca2+ channels, one regulated by voltage, one by Ca2+ and the other two by other ligands. The presence of multiple Ca2+ channels gated by different signals allows for dynamic changes in cytosolic Ca2+ in response to a variety of stimuli.
Figure 5.20 Ca2+ and K+ fluxes through channels in the tonoplast of a stomatal guard cell during abscisic acid-induced stomatal closure. Shown are: a K+ channel that is regulated by Ca2+; several Ca2+ channels that are regulated by voltage (Vm), inositol trisphosphate (IP3) or cyclic-ADP-ribose (cADPR); and the slow vacuolar Ca2+ channel (SV).
It is somewhat counterintuitive that the lipid bilayer of biological membranes is highly permeable to water. It is perhaps even more surprising that membranes contain specialized water channels called aquaporins. Aquaporins in plant cells were initially found on the tonoplast but they are also localized to the plasma membrane as well as some other endomembranes including the endoplasmic reticulum. They belong to relatively large gene families in plants; for example Arabidopsis has 35 aquaporin genes, maize has 36 and rice 33. By definition, aquaporins act as water channels but there is growing evidence that they may facilitate the movement of CO2, NH3, H2O2, boron and silicon.
Aquaporins fall into four subgroups (Figure 5.21); there is high sequence homology ( > 97%) among members of each subgroup. The TIPs and PIPs are tonoplast and plasma membrane intrinsic proteins, respectively. Nodulin intrinsic proteins (NIPs) are found in the peribacteroid membranes of symbiotic nitrogen-fixing nodules and are present in non-leguminous plants as well. The fourth class of aquaporins, the SIPs (small basic intrinsic proteins), which are encoded by a small family of two to three genes, are found in the endoplasmic reticulum. The four subgroups of aquaporins are found in all land plants, from mosses to the angiosperms.
Figure 5.21 Diagram showing the subcellular localization of aquaporins. Plasma membrane intrinsic proteins, PIP1 and PIP2, and nodulin intrinsic protein, NIP, localize to the plasma membrane. Tonoplast intrinsic proteins, TIP1 and TIP2, localize to vacuoles, TIP3 to protein storage vacuoles, and NIP and small basic intrinsic protein (SIP) are found on the endoplasmic reticulum.

Aquaporins are relatively small proteins, around 30 kDa in size, and have six transmembrane domains (Figure 5.22). The C- and N-terminal sequences of all aquaporins localize to the cytosol as do connecting loops II and IV. The I, III and V connecting loops of PIPs face the apoplast, whereas those of TIPs and SIPs face the lumen of the vacuole and endoplasmic reticulum, respectively. Four aquaporin monomers make a functional complex but it is noteworthy that each subunit of the tetramer forms a water channel (Figure 5.23).
Figure 5.22 Membrane disposition of a PIP-type aquaporin. Six transmembrane helices are shown, forming five amino acid loops with I, III and V in the apoplast and II and IV facing the cytoplasm. Several phosphorylation sites are located in the long C terminus of the protein and in loop II, whereas regulatory sites that are methylated (Met) are shown at the N terminus.
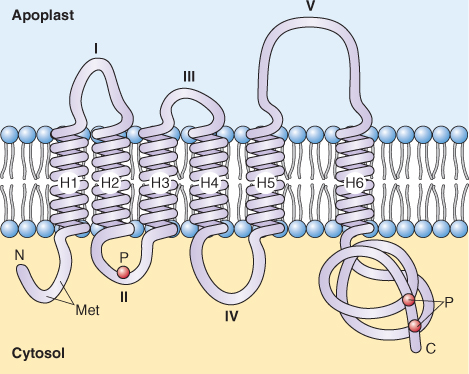
Figure 5.23 Ribbon diagrams of aquaporins based on crystallographic data. (A) A view from above the membrane of four aquaporin subunits; each subunit forms a water-transporting pore; one subunit is shown in a space-filling model to illustrate its pore. (B) A side view of one subunit showing the orientation of the amino acid chain in the plane of the membrane; water molecules (blue spheres) move through the pore.

Aquaporin activity is regulated at various levels. Many aquaporins are regulated by changes in the abundance of their mRNAs. Indeed, there is evidence for the coordinated downregulation of aquaporins in response to nutrient and water stress. They are also subject to regulation at the mRNA level by hormones and other regulatory molecules.
Aquaporins are extensively modified post-translationally by phosphorylation and methylation of amino acids at the N and C termini. Phosphorylation occurs at multiple amino acids at the C terminus of most aquaporins, and methylation of amino acids at the N terminus has been shown for PIPs (see Figure 5.22). Phosphorylation of aquaporins has been shown to regulate the opening of aquaporins, and the observation that Ca2+-dependent protein kinases may catalyze this phosphorylation suggests a link between Ca2+ signaling and the regulation of water movement between cells.
Another interesting aspect of aquaporin regulation comes from observations that changes in cytoplasmic pH alter water movement. When Arabidopsis roots are subjected to anoxia, the cytoplasm becomes highly acidic (see Chapters 7 and 15) and water transport is dramatically reduced. The effect of anoxia on water movement can be mimicked experimentally by acidifying the cytoplasm of root cells, suggesting that it is the pH of the cytoplasm that is affecting water flux.
Carriers and co-transporters belong to a large and diverse class of proteins that move a wide range of ions and metabolites across cellular membranes. They are ubiquitously distributed in cellular membranes. In addition to their association with the plasma membrane, they are found in the endomembrane system as well as the envelopes of chloroplasts and mitochondria, where they are especially abundant. The array of ions and molecules translocated by these transporters is vast. They play roles in the uptake of inorganic nutrients, including NH4+, NO3−, SO42− and H2PO4−, and are important in loading sugar into the phloem for long-distance transport. Co-transporters and carriers are involved in the exchange of metabolites across mitochondrial and chloroplast membranes. Co-transporters on the tonoplast have a role in the storage of ions and organic solutes in the vacuole.
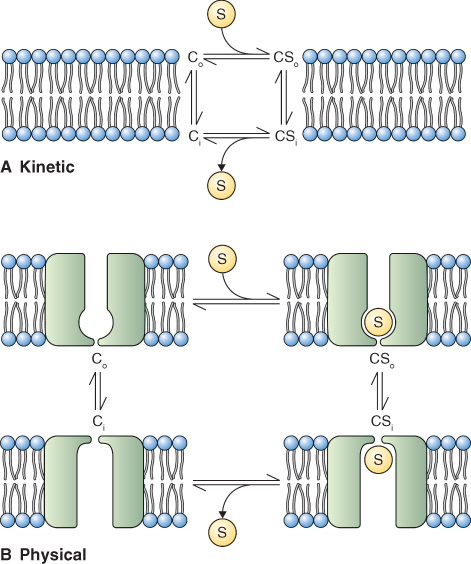
In transport mediated by carriers and co-transporters, the solute being transported binds to the transporter and causes a conformational change (Figure 5.24). This conformational change is thought to lead to the movement of the solute across the membrane. Because these transport proteins interact with solutes, their properties are reminiscent of those of enzymes. Carriers and co-transporters show saturable kinetics with increasing substrate concentration from which a Michaelis–Menten constant, Km, can be derived (see Figure 2.24A). The interaction between transporter and solute confers very high selectivity and these proteins can discriminate between stereoisomers of sugars or amino acids.
Figure 5.24 Models of the reaction cycle of a solute carrier, C. (A) Kinetic modeling of C as it interacts with solute S on each side of a membrane (Co and Ci). (B) Model of changes in the structure of C as it interacts with S. Differences in Co and Ci lead to binding and release of S.
Carriers facilitate the diffusion of organic solutes across membranes. For example, vacuolar uptake of glucose and some amino acids is driven by concentration gradients in these molecules. Co-transporters are involved in secondary active transport, coupling the downhill movement of ions, such as H+, to the uphill movement of inorganic ions and organic solutes. The H+-ATPases of the plasma membrane and tonoplast pump protons out of the cytosol, creating proton gradients across these membranes. Therefore protons tend to move energetically downhill across these membranes in the direction of the cytosol. Transporters that catalyze solute flux in the same direction as H+ flux are known as symporters (see Figure 5.8B). Symporters typically drive uptake of solutes into the cytosol, either from the external medium or from intracellular compartments. Proton-coupled symporters include the H+/sucrose symporter that is involved in loading sucrose into the phloem, several different H+/anion symporters and a number of H+/amino acid symporters. Conversely, secretion of solutes from the cytosol can be accomplished by antiporters, which exchange solutes for protons. Antiporters are present at the plasma membrane and at endomembranes. Proton-coupled antiporters include the H+/Ca2+ antiporter on the tonoplast and the Na+/H+ antiporter on the plasma membrane. Both symporters and antiporters tend to dissipate the proton gradient (pmf).
In addition to co-transporters that are powered by a proton gradient, there are those that utilize gradients of other ions to energize transport of their substrates. For example, the most abundant protein in the chloroplast envelope is a phosphate translocator that exchanges inorganic phosphate for triose phosphate (see Chapter 9). Because there are many types of carriers and co-transporters, it is difficult to provide a generalized model for their structure or regulation.
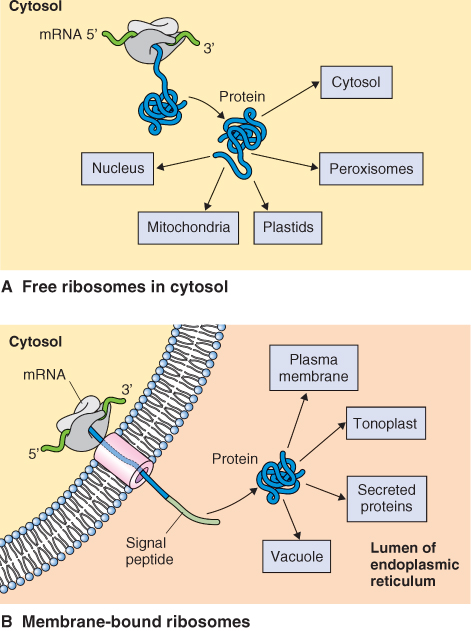
Plant genomes encode tens of thousands of proteins. Nearly all of a cell's proteins are encoded by nuclear DNA and synthesized by cytosolic ribosomes, which may or may not be attached to the endoplasmic reticulum. Proteins encoded by mitochondrial and chloroplast DNA (around 100 or so) are synthesized by ribosomes within these organelles and incorporated directly into the organelle compartments. However proteins synthesized in the cytoplasm or endoplasmic reticulum must be targeted to the compartment or membrane in which they will be active (Figure 5.25). To ensure that a protein reaches its proper location, it needs labels that designate that location. We will examine the nature of these address labels and the machinery that reads those labels. In addition, these proteins must often pass into or through various cellular membranes. We shall see that this also requires special cellular machinery. Finally, we will examine how proteins that are destined for the cell wall and several internal organelles are processed, packaged and targeted for transport.
Figure 5.25 Protein synthesis occurs on 80S ribosomes that are (A) free in the cytoplasm or (B) bound to the surface of the endoplasmic reticulum (ER). Proteins synthesized on free ribosomes are destined for the cytoplasm, nucleus, plastids, mitochondria and peroxisomes, whereas those synthesized on ER-attached ribosomes enter the secretory pathway.
Proteins that are synthesized on cytosolic ribosomes and destined for transport to other compartments or to cellular membranes have one or more targeting domains that act as address labels, specifying their final destination. Targeting domains are usually short peptides or amino acid motifs that can be located anywhere in the protein's amino acid sequence. Specific cellular machinery interacts with this information to translocate the protein into, or retain it within, the proper compartment. Each compartment and membrane system requires a different targeting domain and sorting machinery. Although the targeting domain is essential for protein transport, it may not be part of the active protein. Proteases in the target location often remove the targeting domain to create a functional, mature polypeptide.
For those proteins that remain in the cytoplasm or are targeted to chloroplasts, mitochondria, nuclei and peroxisomes, synthesis of the protein is generally completed on free ribosomes (Figure 5.25A). Proteins destined for the secretory pathway are synthesized on ribosomes that are attached to the endoplasmic reticulum (Figure 5.25B). Whether mRNAs are translated by free or membrane-bound ribosomes is determined by the presence of a signal peptide, a short amino acid sequence located at the N terminus of the protein. When the growing polypeptide chain contains a signal peptide, this targets the protein-synthesizing machinery to the surface of the endoplasmic reticulum.
Although there are major differences in the mechanisms that underlie targeting to simple organelles such as peroxisomes or to more complex organelles such as chloroplasts and mitochondria, the underlying principles are similar. Targeting domains are found on most proteins destined for cellular organelles. The targeting domain is recognized by a receptor at the surface of the organelle membrane. The specificity of both the targeting domain and the receptor ensures that proteins reach the correct destination.
As the polypeptide elongates, cytosolic chaperones keep the chain in the unfolded state in the cytosol. The movement of proteins through pores in the organelle membrane is facilitated by several polypeptides that make up a translocation motor, including those that hydrolyze nucleoside triphosphates (NTPs). As the translocated protein enters the lumen of the organelle, it interacts with another set of chaperones that catalyze protein folding (Figure 5.26).
Figure 5.26 Proteins that cross the lipid bilayer of membranes are targeted to receptors embedded in the membrane; they pass through a proteinaceous pore and require the activity of molecular chaperones on both sides of the membrane. Chaperones keep proteins in the unfolded state to facilitate transport through the pore and catalyze the folding of protein when transport is completed.
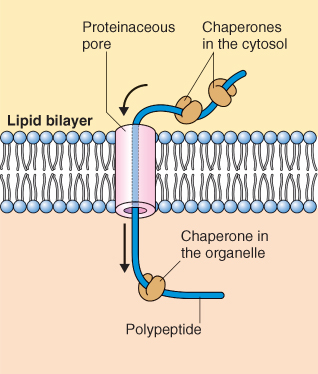
To enter target organelles, proteins must pass through at least one membrane. Most proteins have hydrophilic surfaces and therefore do not readily pass through the hydrophobic core of cellular membranes. Translocation through a membrane involves a proteinaceous pore or channel through which a protein must pass in an extended or unfolded configuration rather than in its final, folded form (Figures 5.26 and 5.27). As a polypeptide passes through a pore, it is assisted by molecular chaperones, proteins that bind the polypeptide during the folding and assembly process. Some cytosolic chaperones interact with newly synthesized proteins, keeping them unfolded so they can pass through a protein pore to an appropriate compartment or membrane. Other chaperones bind to the amino acid chain as it emerges from the membrane and facilitate folding. Still others function as repair stations to correct minor misfolding. The many members of three families of chaperones—the 60 kDa, 70 kDa and 90 kDa heat shock proteins (Hsp60, Hsp70 and Hsp90, respectively; see Chapter 15)—fulfill all these roles, interacting with a wide spectrum of proteins. Hsp70 chaperones are present in all cells. The synthesis of many heat shock proteins is upregulated in response to heat stress, perhaps to prevent misfolding and to repair misfolded proteins (see Chapter 15).
Figure 5.27 Proteins synthesized on cytosolic ribosomes are imported into the chloroplast and can be targeted to six distinct destinations: the two outer membranes of plastid, intermembrane space, stroma, thylakoid membrane and thylakoid lumen. Proteins synthesized in the plastid stroma on 70S ribosomes either remain in the stroma or are targeted to the thylakoid membrane, stroma or inner envelope of the plastid.
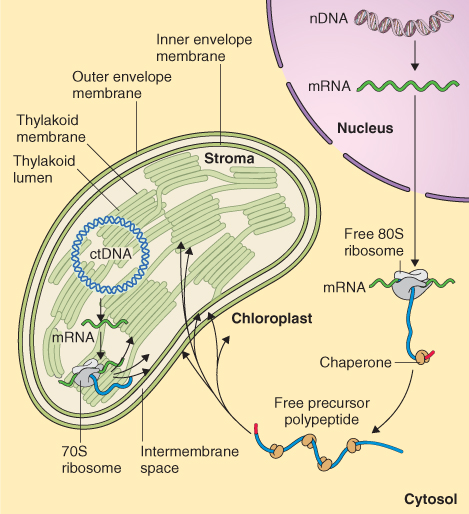
Chloroplasts and mitochondria are surrounded by two unit membranes (see Figures 4.25 and 4.27). In chloroplasts an additional set of membranes called the thylakoid membranes form from an infolding of the inner chloroplast membrane. These membrane barriers in turn enclose three aqueous compartments, the intermembrane space between the outer and inner chloroplast membrane, the stroma and the thylakoid lumen. Proteins must be targeted to each of the membranes and the compartments in these organelles (see Figure 5.27).
Although the principles involved in peptide targeting to chloroplasts and mitochondria are similar, the components of the molecular machinery involved are very different. Here we will describe the principles involved in targeting to chloroplasts. As indicated in Section 5.7.1, membrane receptors, peptide targeting domains, molecular chaperones and translocation proteins are required for proteins to enter the various compartments and membranes of chloroplasts. Figure 5.28 illustrates the processes involved in the targeting of proteins to the chloroplast stroma, thylakoid membrane and thylakoid lumen. Although the vast majority of chloroplast proteins are synthesized on free cytosolic ribosomes, some proteins are imported into the chloroplast after synthesis on endoplasmic reticulum-bound ribosomes and transported to the chloroplast via Golgi-derived vesicles.
Figure 5.28 Protein import into the plastid requires the participation of molecular chaperones, targeting sequences and energy. Stromal transit peptides target unfolded proteins to the outer plastid envelope and transport across the membrane is facilitated by transport complexes TOC and TIC. The transit peptide is cleaved by a protease and (1) stromal proteins interact with Hsp60/70 chaperones and assume their final conformation. Proteins destined for the thylakoid membrane are targeted there by one of two routes: (2) by a mechanism that requires neither targeting information nor energy, or (3) by using a signal peptide that interacts with a signal recognition particle (SRP) in the stroma and a receptor on thylakoid membrane. Those proteins that are inserted into the thylakoid membrane via route (3) require energy in the form of GTP to become incorporated into the membrane. (4) Transport of proteins destined for the thylakoid lumen have signal peptides to target them to transport machinery in the thylakoid membrane; the signal peptides are cleaved after transit through the membrane.
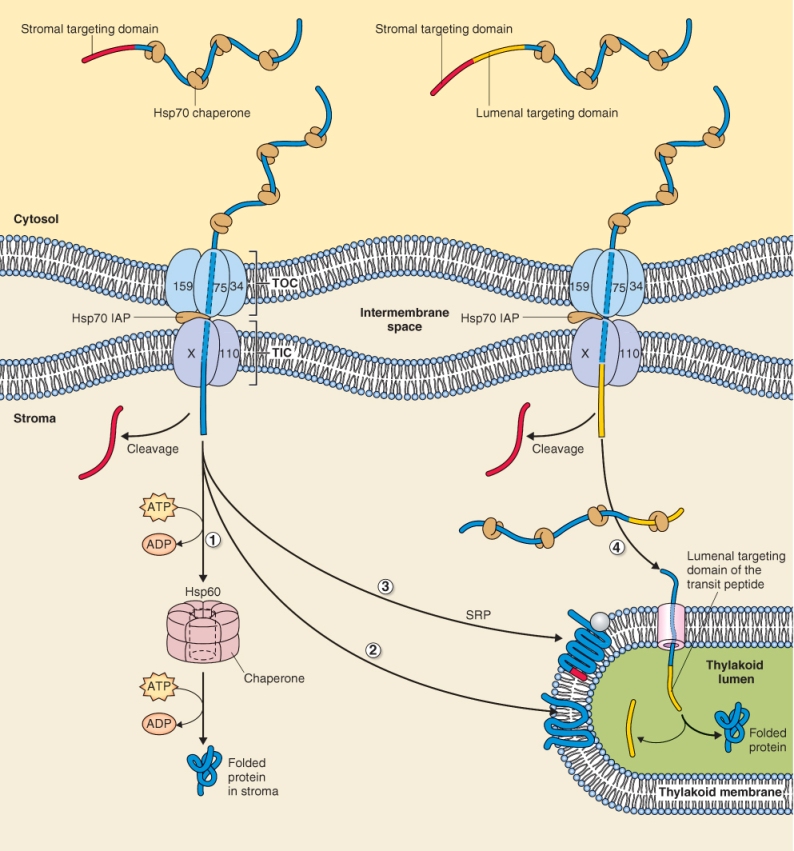
Most chloroplast proteins synthesized on free cytosolic ribosomes interact with molecular chaperones of the Hsp70 class before they are imported into the stroma. Two distinct transport complexes denoted TOC (translocon at the outer chloroplast envelope) and TIC (translocon at the inner chloroplast envelope) participate in protein import (Figure 5.28). Chloroplast-bound proteins have a short amino terminal transit peptide that targets them to the outer surface of the chloroplast envelope where they interact with a receptor in the TOC complex. Interaction of imported proteins with TOC and translocation through the outer membrane requires ATP and GTP hydrolysis whereas translocation through TIC requires only ATP. Unlike protein translocation into the mitochondrion, import into the stroma of the chloroplast does not involve transmembrane pmf. Although transport through TOC and TIC can be viewed as biochemically separate events they are thought to occur almost simultaneously at points where the outer and inner membranes of the plastid make close contact.
Once the protein enters the stroma, the transit peptide is cleaved by a protease. This is the final destination of a stromal protein. It interacts with chaperones of the Hsp60 and Hsp70 class and assumes its final conformation. The interaction of the protein with molecular chaperones and the formation of the mature folded protein require the expenditure of energy in the form of ATP (Figure 5.28).
Proteins targeted to the thylakoid membrane and lumen arrive at their destinations by different routes and use different mechanisms. Proteins destined for the thylakoid membrane are targeted there by one of two routes, either by a non-cleavable signal sequence similar to the cleavable peptide that targets proteins to the endoplasmic reticulum, or by a mechanism that requires neither targeting information nor energy. Those proteins that are inserted into the thylakoid membrane using a signal peptide require energy in the form of GTP in order to become incorporated into the membrane.
Transport of proteins destined for the thylakoid lumen can also occur via two separate mechanisms, both of which require signal peptides for targeting. In one case specific transporting machinery moves only unfolded proteins across the thylakoid membrane and requires ATP. The second transport machinery translocates folded and unfolded proteins and utilizes the transmembrane pmf to power transport. In both cases, once in the thylakoid lumen, cleavage of the signal peptide is catalyzed by proteases and folding of the protein requires molecular chaperones.
Peroxisomes differ from chloroplasts and mitochondria in several important respects. First, they are devoid of nucleic acids and the machinery for protein synthesis, and they are surrounded by a single unit membrane. These organelles, however, need to import proteins. First, they have the capacity to increase in size and to divide by binary fission (simply the pinching of the organelle into two daughter organelles). In addition, the function and enzyme content of peroxisomes are developmentally related. Following germination, cotyledons of lipid-storing seeds, such as those of the castor bean (Ricinus communis), convert lipid to sugar by the reactions of β-oxidation and gluconeogenesis in specialized peroxisomes often referred to as glyoxysomes (see Chapters 6 and 7). When the cotyledons are depleted of lipid they become green and photosynthesize. Photosynthesis requires the presence of peroxisomes to complete the reactions of photorespiration (see Chapter 9). During the transition from heterotrophic metabolism to autotrophy peroxisomes acquire a new suite of enzymes. This transformation occurs through the import of membrane and luminal proteins into the peroxisome. Redifferentiation of peroxisomes has also been observed to occur during senescence (see Chapter 18).
Transport of macromolecules between the cytoplasm and the nucleus occurs through specialized pores in the nuclear envelope. Proteins enter the nucleus and ribosomal subunits, mRNAs, tRNAs and a host of other macromolecules leave the nucleus through nuclear pores. The nuclear envelope is two membranes thick and separates the cytoplasm from the matrix of the nucleus, which is called nucleoplasm. The outer membrane is often studded with ribosomes and it forms connections to sheets of rough endoplasmic reticulum (see Figures 4.14 and 4.15).
Nuclear pores are complex, radially symmetrical structures made up of more than 100 individual peptides (Figure 5.29). They facilitate the movement of materials into and out of the nucleus. The diameter of the nuclear pore is around 9 nm. This permits passive diffusion of molecules up to about 40 kDa in size. Active transport through the nuclear pore also occurs; proteins of up to 20 kDa can be transported by this mechanism. This process involves a targeting sequence called the nuclear localization signal (NLS), receptor proteins in the nuclear pore complex, soluble cytoplasmic factors and energy in the form of GTP. The process can be divided into two steps. First the NLS on the protein binds to receptors in the nuclear pore. This step requires both cytosolic factors and GTP. Then the protein is translocated through the pore; this step also requires GTP to supply additional energy.
Figure 5.29 The nuclear pore complex. (A) Model showing the organization of proteins in the nuclear pore complex. (B) Electron micrograph of a tangential section through several nuclear pore complexes. Part (A) shows a cross-section of membrane, whereas (B) shows a surface view of the nuclear membranes with embedded pore complexes (arrows).

Transport through the nuclear pore is regulated by a number of factors, including those that effectively mask the NLS on targeted proteins, and environmental cues, such as light. For example, the effect of light on photomorphogenesis is affected by a protein, COP1, which shuttles between the cytoplasm and nucleus in response to light. Note that this protein is different from the COPI coat protein that is involved in vesicle transport (see Section 5.8.3). The COP1 protein is a repressor of photomorphogenesis. COP1 normally resides in the nucleus in dark-grown plants, but it moves into the cytoplasm when plants are exposed to light. Movement of COP1 into the cytoplasm relieves repression of photomorphogenesis genes (see Chapter 8).
The secretory pathway forms an extensive membranous system, the endomembrane system, which branches throughout the cell. It consists of endoplasmic reticulum (ER), the Golgi apparatus and trans-Golgi network (TGN), multivesicular bodies (MVB) and various classes of vesicles and vacuoles (Figure 5.30). Although the synthesis of all proteins begins on free ribosomes, proteins destined for the endomembrane system or for export to the cell exterior have a signal peptide that targets the ribosome–protein complex to the surface of the ER where synthesis of the protein is completed (Figure 5.31). ER with attached ribosomes is called rough ER. It is particularly abundant in cells that specialize in protein secretion (e.g. cereal aleurone cells during seed germination; see Chapter 6) or in the storage of proteins in vacuoles (e.g. storage parenchyma cells in developing seeds).
Figure 5.30 The secretory pathway. Proteins synthesized on 80S ribosomes attached to ER are transported via the Golgi apparatus to the trans-Golgi network (TGN). The TGN plays a central role in directing vesicle traffic. (1) Proteins destined for the vacuole are transported from the TGN via multivesicular bodies (MVB) to protein storage vacuoles. (2) Proteins destined for the exterior are transported in vesicles to the plasma membrane for exocytosis. (3) Proteins imported into the cell by endocytosis move in clathrin-coated vesicles to the TGN (4) where they are sent via a MVB to lytic vacuoles. During this process, the TGN is functionally equivalent to an early endosome (EEnd) in animal cells and the MVB functions as a late endosome (LEnd).
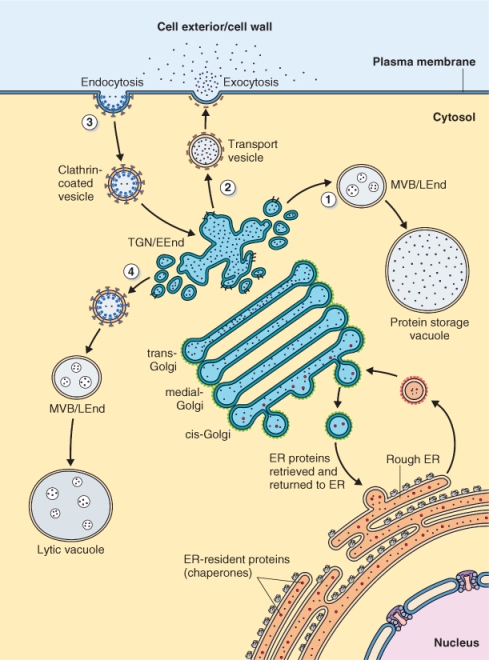
Figure 5.31 Targeting of proteins to the endoplasmic reticulum lumen involves (1) a signal sequence, (2) a signal recognition particle (SRP), (3) an SRP receptor, (4) a translocation complex, (5) signal peptidase and (6) molecular chaperones. These work in a coordinated fashion so that a folded protein synthesized in the cytosol resides in the lumen of the ER.
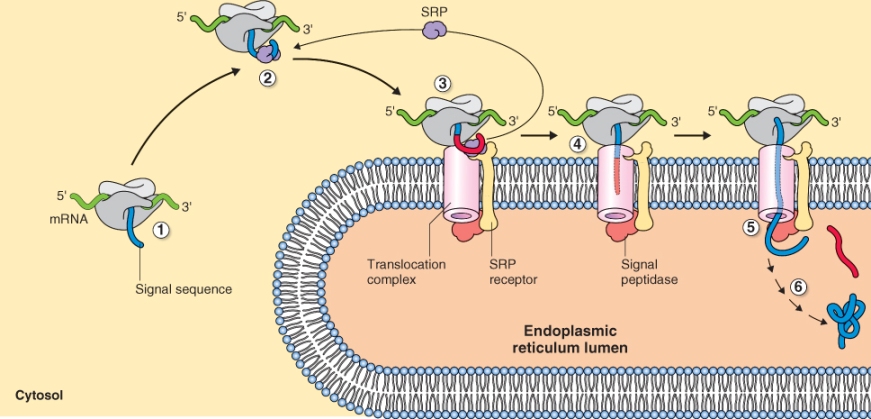
The mRNAs that encode proteins destined to function outside the cell, in the cell wall or inside the organelles of the endomembrane system, contain a signal sequence about 48–90 nucleotides in length that is located near the start codon where translation begins. Translation of this mRNA sequence results in the synthesis of a signal peptide of about 16–30 amino acids at the amino (N) terminus of the encoded protein (Figure 5.31). As the mRNA is translated and the N terminus of the resulting protein emerges from the ribosome, the signal peptide is recognized by the signal recognition particle (SRP), an RNA–protein complex in the cytosol that mediates the docking of the ribosome onto a protein complex within the ER membrane. Although the amino acid sequence of signal peptides vary enormously, almost all of them can function interchangeably to target proteins to the ER. For example, plant signal peptides can target animal proteins to the ER of animal cells and vice versa. Furthermore, addition of a signal peptide to the N terminus of a protein that is not normally secreted can target that protein to the secretory pathway.
When an SRP binds to a signal peptide, protein synthesis is temporarily arrested. The ribosome–nascent protein–SRP complex is recognized by an SRP receptor on the surface of the ER that allows docking of the complex on the membrane (Figure 5.31). When docking has occurred, the SRP and several other components of the SRP receptor dissociate using energy from the hydrolysis of GTP. Synthesis of the anchored protein is reinitiated, and the nascent protein is guided through a transmembrane channel in the ER. GTP hydrolysis also provides the energy required to move the protein through the membrane.
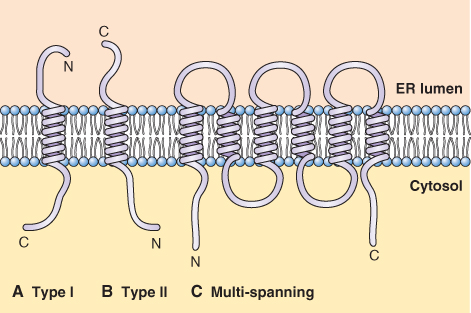
Several classes of proteins that are synthesized on rough ER do not completely cross the ER membrane; these include integral membrane proteins of the ER and other parts of endomembrane system. Their transport through the membrane is arrested by the presence of a hydrophobic sequence of amino acids called a stop transfer sequence. It is easy to imagine how a protein with an N-terminal signal peptide and a stop transfer sequence located half-way along its length can become stuck in the membrane, as illustrated in Figure 5.32A. These proteins, designated Type I, have their N terminus in the lumen of the ER and the C terminus in the cytosol. Type II proteins, whose C terminus lies in the lumen of the ER, do not use a signal peptide to target the ER; rather they use other hydrophobic amino acid sequences that allow their orientation (Figure 5.32B). Proteins that have multiple membrane-spanning domains, such as the membrane transporters described in earlier pages of this chapter, possess a series of alternating signal peptides and stop transfer sequences that work together to thread the protein into and out of the membrane (Figure 5.32C).
Figure 5.32 Orientation of N- and C-terminal domains of membrane-spanning proteins synthesized on ribosomes bound to the endoplasmic reticulum. (A) Type I proteins have cytosolic C-terminal domains, whereas (B) Type II proteins have their N terminus in the cytosol. (C) Proteins that have several membrane-spanning domains have both their N and C termini in the cytosol.
Soluble proteins enter the ER lumen in their entirety. In this case, the N-terminal signal peptide is removed by the action of a signal peptidase. The polypeptide is then folded with the aid of chaperones of the Hsp class as well as BiP (binding protein), calnexin and calreticulin, on the inner face of the ER membrane. Chaperone-catalyzed protein folding requires GTP. Metalloproteins bind metals as folding occurs. For example, the activity of cereal α-amylase is dependent on Ca2+ binding. That this binding occurs in the ER lumen is shown by the fact that α-amylase isolated from the ER is enzymatically active. Misfolded proteins are targeted for degradation in the lumen of the ER or else are transported out of the ER and degraded in the cytoplasm by the 26S proteasome (see Section 5.9).
The process of post-translational modification of proteins begins in the ER lumen. Informational amino acid sequences in these proteins determine whether and how they will be modified and whether they will progress further along the secretory pathway. Post-translational modifications that occur in the ER include the addition of glycan chains to glycoproteins, the modification of amino acids (e.g. conversion of proline to hydroxyproline), the formation of disulfide bonds catalyzed by protein disulfide isomerases, and the assembly of multimeric complexes, a process that requires chaperones.
Many soluble plant proteins, destined for the secretory pathway, are glycosylated by enzymes that add sugar chains. Glycans may be attached to the amide nitrogen of asparagine residues (N-linked) or to the OH groups of serine or hydroxyproline residues (O-linked). Glycosylation begins in the ER but may be completed in the Golgi. Proteins with N-linked glycans, including many legume storage proteins, have a characteristic amino acid sequence, Asn-X-Ser/Thr, where X can be any amino acid except Pro. Proteins with O-linked glycans also carry a specific amino acid motif. The hydroxyproline-rich cell wall protein extensin is a soluble protein with O-linked oligoarabinan chains attached to hydroxyprolines. There is no codon for hydroxyproline: it is produced post-transcriptionally by the hydroxylation of proline catalyzed by the enzyme prolyl hydroxylase. Thus the sequence of events in extensin modification in the ER is first the hydroxylation of prolines followed by the addition of arabinose. Specific amino acid signals dictate which proline residues are hydroxylated as well as which of these residues carry the arabinose chain.
Some soluble proteins are retained in the lumen of the ER for proper functioning. These include chaperones such as BiP, calnexin and calreticulin and enzymes such as protein disulfide isomerase and prolyl hydroxylase. Such proteins have a C-terminal ER retention signal, the amino acid sequence Lys-Asp-Glu-Leu, also referred to by the single letter amino acid code KDEL, or variants thereof such as HDEL (for amino acid abbreviations, see Figure 2.12). The ER retention signal is used to retrieve these proteins if they move out of the ER to the Golgi.
The transfer of proteins from the ER to other destinations in the secretory pathway occurs by the movement of vesicles initially from the ER to the Golgi where proteins may undergo further modification and sorting. The movement of vesicles from the ER to the Golgi is referred to as anterograde transport (Figure 5.33). It occurs in vesicles that have a specific class of coat proteins called COPII on their surface. COPII provides the information that dictates the direction of transport.
Figure 5.33 Proteins can be recycled between the endoplasmic reticulum (ER) and Golgi apparatus. The movement of proteins from the ER to the Golgi is referred to as anterograde transport and occurs in vesicles having COPII coats, whereas recycling from the Golgi to the ER is known as retrograde transport and occurs in vesicles with COPI coats. Retrograde transport returns both soluble and membrane ER-resident proteins.
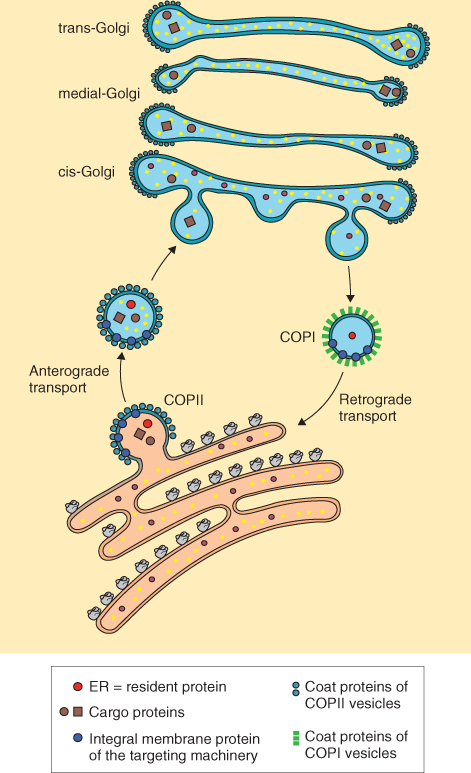
The retrieval of ER-localized, soluble proteins and membrane proteins from the Golgi occurs by a process called retrograde transport (Figure 5.33). For soluble proteins a KDEL receptor, ERD2, recognizes the KDEL motif on the protein. ERD2 is embedded in membranes of ER vesicles that shuttle proteins between the Golgi and ER. Membrane proteins are also recycled from the Golgi to ER but the amino acid motifs and receptors for these have yet to be identified in plants. In both cases receptors act as retrieval mechanisms for resident ER proteins. These proteins are returned to the ER in vesicles that have a second type of coat protein, COPI, on their surfaces.
From the Golgi, proteins are transported to other downstream destinations such as the TGN, MVB, vacuoles or cell exterior (Figure 5.33). Movement of vesicles from the TGN to downstream destinations such as vacuoles or the plasma membrane occurs in clathrin-coated vesicles (see Chapter 4). Soluble secretory proteins that lack sequence information, which acts as specific address labels, reach the cell surface via the so-called default pathway. They are packaged into vesicles at the trans-Golgi that then move to the TGN (Figure 5.34). From there vesicles containing these proteins move to and fuse with the plasma membrane, releasing their contents outside the cell. This process is called exocytosis. Cereal aleurone α-amylase is an example of this type of protein.
Figure 5.34 Electron micrograph of a Eucalyptus siebertii cell showing a Golgi apparatus (G) and an adjacent trans-Golgi network (TGN).
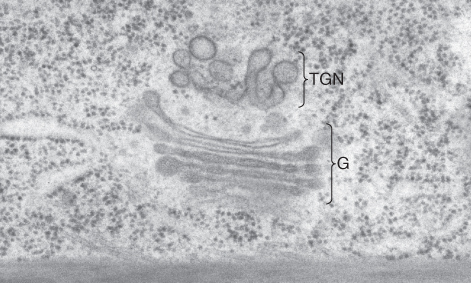
In contrast to the default pathway, protein trafficking to vacuoles requires specific sorting signals. In addition to the central vacuole that is present in all mature plant cells, cells may contain at least three other kinds of vacuoles: protein storage vacuoles, acidic lytic vacuoles and vacuoles with a neutral pH. Proteins move to these compartments along different routes that are determined by distinct vacuolar sorting sequences. Proteins, such as sporamin, a soluble storage protein from sweet potato tubers, that are destined for lytic vacuoles contain an N-terminal propeptide signal (NTPP). At the TGN, the NTPP is recognized by a vacuolar sorting receptor (VSR) that directs sporamin into clathrin-coated vesicles that move to the MVB from which the clathrin-coated vesicles move to lytic vacuoles. Proteins having C-terminal propeptide signals, on the other hand, are targeted to vacuoles that have a neutral pH along a route that has not been precisely defined. As we will discuss in Chapter 6, there are still more routes that vacuolar proteins can take after their synthesis on the ER. For example, pumpkin albumin, a vacuolar storage protein, is transported in precursor accumulating vesicles (PACs) from the ER to a protein storage vacuole bypassing the Golgi, TGN and MVB (see Figure 6.18).
In addition to the secretory pathway, proteins may be trafficked through the endocytotic pathway (see Figure 5.30). Vesicles produced by endocytosis, the invagination of the plasma membrane, have clathrin coats and are transported to the TGN. Here both membrane and soluble proteins are sorted and transported to vacuoles via the MVB. The TGN is thought to be the functional equivalent of the early endosome in animal cells and the MVB to be equivalent to the late endosome.
Protein degradation is required for normal cellular function, not only to break down mis-synthesized or misfolded proteins, but also to remove developmentally regulated proteins. Degradation of these proteins must be highly selective to prevent the loss of other, needed proteins. Proteins targeted for removal must be specifically tagged before they are broken down. Cells use the ubiquitin–proteasome system (UbPS) as one of the principal pathways of protein degradation. Proteins are first tagged using ubiquitin (Ub), a small 76-amino acid protein, and are then broken down by a complex proteolytic machine called the 26S proteasome; the designation 26S refers to the size of the proteasome in Svedberg units. The importance of protein removal by the UbPS in plants can be appreciated from genome analyses in Arabidopsis that show more than 1300 genes to be implicated in this pathway. In the next sections, we will first look at the process of ubiquitin addition to target proteins and then discuss the structure and function of the proteasome.
Ubiquitin addition to target proteins occurs in an ATP-dependent cascade of reactions that proceeds in three distinct steps (Figure 5.35) catalyzed sequentially by Ub-activating enzymes (E1), followed by Ub-conjugating enzymes (E2) and finally by Ub-ligating enzymes (E3). E1 activates Ub using ATP to catalyze the formation of an E1-Ub intermediate. Ub is then transferred to E2. Finally, a Ub ligase (E3) binds both E2-Ub and the target protein and then catalyzes transfer of ubiquitin to the target. After transfer of Ub to the target substrate, additional Ubs are added to form a polyubiquitin chain, often up to a total of seven Ub molecules. The process of ubiquitination is highly specific; specificity is determined largely by the E3 ligation process. There are more than 1200 E3 genes in Arabidopsis but only two genes encoding E1, and 37 encoding E2.
Figure 5.35 Protein degradation via the ubiquitin–proteasome system. Ub-activating enzyme E1 catalyzes the ATP-dependent addition of Ub. Ub is transferred from E1 to Ub-conjugating enzyme E2. The Ub ligase E3 binds both E2 and the target protein to be ubiquitinated facilitating specific transfer of up to seven Ub molecules to the target.
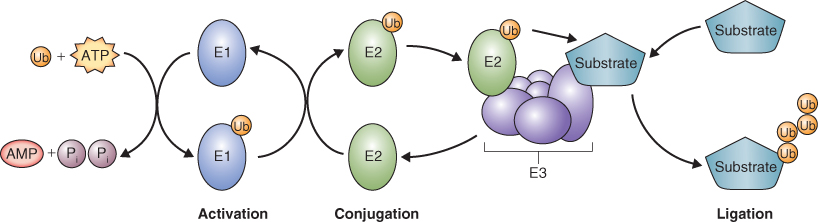
The E3 ligase complex has been classified into two groups based on the mechanism of Ub transfer to target proteins. HECT E3 ligases accept Ub from E2 and then transfer it to target proteins (Figure 5.36A). RING E3 ligases bind E2-Ub and the target protein and facilitate the direct transfer of Ub from E2 to the target. This latter group includes simple monomeric RING E3 ligases, which bind both E2-Ub and the target protein (Figure 5.36B), and complex multimeric RING E3 ligases such as the CULLIN/RING ligases. The CULLIN/RING ligases are complex assemblages of proteins, each of which includes: a RING finger domain protein that binds E2-Ub; a variable component, an adaptor that recognizes and binds the target protein; and a CULLIN-type protein that forms a scaffold for the rest of the complex (Figure 5.36C). Eleven different CULLIN proteins and hundreds of different adaptors have been identified in plants. The most complex of the CULLIN/RING E3 ligases are the APCs (anaphase-promoting complexes) that contain more than ten different subunits (see Chapter 11).
Figure 5.36 Two types of E3-Ub ligases are known. (A) In HECT-domain ligases Ub is transferred from the conjugating enzyme E2 to the HECT domain protein and it in turn transfers Ub to the target protein that is also bound to the HECT domain. (B, C) In the second type, Ub is transferred directly from E2 to the target protein. (B) RING/U-box domain E3 ligases are monomeric and directly bind both E2 and the target protein. (C) CULLIN/RING domain ligases are made up of a number of proteins. A CULLIN platform anchors the other proteins, which include a RING-domain protein that binds E2 and adaptor proteins that bind the target.
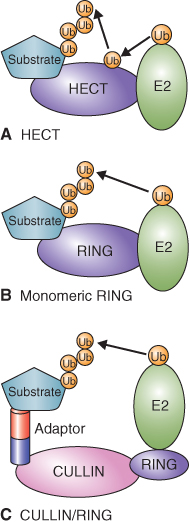
Ubiquitinated proteins are recognized by the 26S proteasome and undergo ATP-dependent proteolysis. The proteasome is a large complex made up of a 20S core protease (CP) and a 19S regulatory particle (RP) (Figure 5.37). The CP can be visualized as an open-ended barrel capped at both ends by the RP. The RP confers specificity to proteolysis by recognizing ubiquitinated proteins and allowing those that are to be degraded to enter the CP where proteolysis occurs. The RP also catalyzes removal of the ubiquitin tag and the unfolding of target proteins to allow for rapid proteolysis by the CP.
Figure 5.37 Model of the 26S proteasome showing key features of this protein-degrading machine. (A) The core protease consists of a 20S particle that is capped at each end by a 19S regulatory particle made up of a lid and base. The association of the regulatory particle with the core protease is ATP-dependent. (B) Ubiquitinated proteins are recognized by the regulatory particle and protein chaperones in the base unfold the target protein for proteolysis by the core protease.
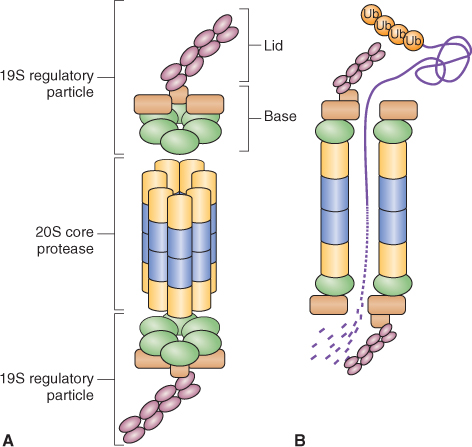
Proteolysis by the ubiquitin–proteasome system occurs in the cytosol and nucleus where it brings about regulated proteolysis of proteins synthesized on non-membrane-bound ribosomes. Surprisingly, defective proteins synthesized on the ER are also degraded by the cytosolic UbPS. As many as 30% of total cellular proteins are synthesized on the ER and it is estimated that a third of these newly synthesized proteins are degraded soon after synthesis. Because of the enclosed nature of the ER lumen there must be efficient mechanisms to degrade defective proteins synthesized in this compartment. The mechanism by which this is done is called ER-associated degradation (ERAD), a highly regulated process.
In ERAD, ER-localized chaperones recognize defective proteins in the ER lumen, unfold them and permit their transport to the cytosol to be degraded by the UbPS. These chaperones include the ER proteins, calnexin and calreticulin, and Hsp chaperones. Stresses that increase the proportion of defective proteins synthesized by the ER also increase the synthesis of ER-localized chaperones. The transport of aberrant proteins from the ER to the cytosol occurs via a translocation pore in the ER membrane; this process is called retrotranslocation. An interesting aspect of ERAD is that the 26S proteasome is enriched at the surface of the ER.