UNSUPERVISED LEARNING includes a host of exploratory techniques that can be used to gain insight into data, often when there is no outcome variable or clear target. Although around for over a century, principal components analysis remains a data science staple and it is introduced along with MultivariateStats.jl. Probabilistic principal components analysis (PPCA) is considered next and an expectation-maximization algorithm for PPCA is implemented in Julia this is the first of two examples in this chapter that illustrate Julia as a base language for data science. Clustering, or unsupervised classification, is introduced along with the famous k-means clustering technique and Clustering.jl. A mixture of probabilistic principal components analyzers is then discussed and implemented, thereby providing the second example of Julia as a base language for data science.
Unsupervised learning differs from supervised learning (Chapter 5) in that there are no labelled observations. It may be that there are labels but none are known a priori, it may be that some labels are known but the data are being treated as if unlabelled, or it may be the case that there are no labels per se. Because there are no labelled observations, it might be advantageous to lean towards statistical approaches and models — as opposed to machine learning — when tackling unsupervised learning problems. In a statistical framework, we can express our uncertainty in mathematical terms, and use clearly defined models and procedures with well-understood assumptions. Statistical approaches have many practical advantages, e.g., we may be able to write down likelihoods and compare models using likelihood-based criteria.
Suppose n realizations x1, …, xn of p-dimensional random variables X1, …, Xn are observed, where Xi = (Xi1, Xi2, …, Xip)′ for i = 1, …, n. As mentioned in Section 3.1, the quantity = (X1, X2, …, Xn)’ can be considered a random matrix and its realization is called a data matrix. Recall also that a matrix A with all entries constant is called a constant matrix. Now, each Xi is a p × 1 random vector and, for our present purposes, it will be helpful to consider some results for random vectors. Let X and Y be p × 1 random vectors. Then,
and, if A, B, and C are m × n, p × q, and m × q constant matrices, respectively, then
Suppose X has mean μ. Then, the covariance matrix of X is
Suppose p-dimensional X has mean μ and q-dimensional Y has mean θ. Then,
The covariance matrix Σ of p-dimensional X can be written
The elements σii are variances and σij, where i ≠ j, are the co-variances. The matrix Σ is symmetric because σij = σji. Also, Σ is positive semi-definite, i.e., for any p × 1 constant vector a = (a1, …, ap)’,
Suppose C is a q × p constant matrix and c is a q × 1 constant vector. Then, the covariance matrix of Y = CX + c is
In general, the covariance matrix Σ is positive semi-definite. Therefore, the eigenvalues of Σ are non-negative, and are denoted
Eigenvalues are also known as characteristic roots. Write vi to denote the eigenvector corresponding to the eigenvalue κi, for i = 1, …, p. Eigenvectors are also known as characteristic vectors. Without loss of generality, it can be assumed that the eigenvectors are orthonormal, i.e.,
and
for i ≠ j. The eigenvalues and eigenvectors satisfy
(6.1) |
for i = 1, …, p.
Write V = v1, …, vp) and
Then, (6.1) can be written
Note that V′V = Ip, and
where
The determinant of the covariance matrix Σ can be written
The total variance of the covariance matrix Σ can be written
The determinant and total variance are sometimes used as summaries of the total scatter amongst the p variables. Note that some background on eigenvalues is provided in Appendix C.1.
6.2 PRINCIPAL COMPONENTS ANALYSIS
In general, observed data will contain correlated variables. The objective of principal components analysis is to replace the observed variables by a number of uncorrelated variables that explain a sufficiently large amount of the variance in the data. Before going into mathematical details, it will be helpful to understand the practical meaning of a principal component. The first principal component is the direction of most variation in the data. The second principal component is the direction of most variation in the data conditional on it being orthogonal to the first principal component. In general, for r ∈ (1, p], the rth principal component is the direction of most variation in the data conditional on it being orthogonal to the first r − 1 principal components. There are several mathematical approaches to motivating principal components analysis and the approach followed here is based on that used by Fujikoshi et al. (2010).
Let X be a p-dimensional random vector with mean μ and co-variance matrix Σ. Let κ1 ≥ κ2 ≥ … ≥ κρ ≥ 0 be the (ordered) eigenvalues of Σ, and let v1, v2, …, vp be the corresponding eigenvectors. The ith principal component of X is
(6.2) |
for i = 1, …, p. Which we can write as
(6.3) |
where W = (W1, …, Wp) and V = (v1, … vp).
Now, it is helpful to consider some properties of W. First of all, because ,
Recalling that = Σ = VKV′, we have
(6.4) |
Rearranging (6.3),
and left-multiplying by V gives
From (6.4), we can develop the result that
Now, the proportion of the total variation explained by the jth principal component is
Therefore, the proportion of the total variation explained by the first k principal components is
To understand how the theoretical explanation of a principal component relates to the intuition given at the beginning of this section, consider
such that v′v = 1. Now,
and we can write
for a′a = 1. Therefore,
(6.5) |
Now, maximizing (6.5) subject to the constraint a′a = 1 gives = κ1, i.e., a1 = 1 and aj = 0 for j > 1. In other words, is maximized at W = W1, i.e., the first principal component.
Now suppose that W is uncorrelated with the first r − 1 < p principal components W1, …, Wr−1. We have
and so ci = 0 for i = 1, …, r − 1. Maximizing (6.5) subject to the constraint ci = 0, for i = 1, …, r − 1, gives = κr. In other words, is maximized at W = Wr, i.e., the rth principal component.
Illustrative Example: Crabs Data
The following code block performs principal components analyses on the crabs data.
using LinearAlgebra
## function to do pca via SVD
function pca_svd(X)
n,p = size(X)
k = min(n,p)
S = svd(X)
D = S.S[1:k]
V = transpose(S.Vt)[:,1:k]
sD = /(D, sqrt(n-1))
rotation = V
projection = *(X, V)
return(Dict(
"sd" => sD,
"rotation" => rotation,
"projection" => projection
))
end
## crab_mat_c is a Float array with 5 continuous mearures
## each variable is mean centered
crab_mat = convert(Array{Float64}, df_crabs[[:FL, :RW, :CL, :CW, : BD]])
mean_crab = mean(crab_mat, dims = 1)
crab_mat_c = crab_mat .- mean_crab
pca1 = pca_svd(crab_mat)
## df for plotting
## label is the combination of sp and sex
pca_df2 = DataFrame(
pc1 = pca1["projection"][:,1],
pc2 = pca1["projection"][:,2],
pc3 = pca1["projection"][:,3],
pc4 = pca1["projection"][:,4],
pc5 = pca1["projection"][:,5],
label = map(string, repeat(1:4, inner = 50))
)
More than 98% of the variation in the data is explained by the first principal component, with subsequent principal components accounting for relatively little variation (Table 6.1).
Table 6.1 The proportion, and cumulative proportion, of variation in the crabs data that is explained by the principal components (PCs).
PC |
Prop. of Variation Explained |
Cumulative Prop. |
1 |
0.9825 |
0.9825 |
2 |
0.0091 |
0.9915 |
3 |
0.0070 |
0.9985 |
4 |
0.0009 |
0.9995 |
5 |
0.0005 |
1.0000 |
Interestingly, even though the second and third principal components explain a total of 1.61% of the variation in the data, they give very good separation of the four classes in the crabs dataset; in fact, they give much better separation of the classes than the first two principal components (Figures 6.1 and 6.2). This example illustrates that principal components that explain the vast majority of the variation within data still might be missing important information about subpopulations or, perhaps unknown, classes.
6.3 PROBABILISTIC PRINCIPAL COMPONENTS ANALYSIS
Probabilistic principal components analysis (PPCA; Tipping and Bishop, 1999b) is a data reduction technique that replaces p observed variables by q < p latent components. The method works well when the q latent components explain a satisfactory amount of the variability in the p observed variables. In some situations, we might even have substantial dimension reduction, i.e., q ≪ p. There are interesting relationships between PPCA and principal components analysis and, indeed, between PPCA and factor analysis (Spearman, 1904, 1927) — these relationships will be discussed later. In what follows, the notation and approach are similar to those used by McNicholas (2016a).
Consider independent p-dimensional random variables X1, …, Xn. The PPCA model can be written
(6.6) |
Figure 6.1 Scallerplot depicting the first two principal components for the crabs data, coloured by class.
Figure 6.2 Scallerplot depicting the second and third principal components for the crabs data, coloured by class.
for i = 1, …, n, where Λ is a p × q matrix of component (or factor) loadings, the latent component , and , where ψ ∈ ℝ+. Note that the Ui are independently distributed and independent of the ϵi, which are also independently distributed. From (6.6), it follows that the marginal distribution of Xi under the PPCA model is There are
free parameters in the covariance matrix ΛΛ′ + ψΙp (Lawley and Maxwell, 1962). Therefore, the reduction in free covariance parameters under the PPCA model is
(6.7) |
and there is a reduction in the number of free parameters provided that (6.7) is positive, i.e., provided that
The log-likelihood for p-dimensional x1, x2, …, xn from the PPCA model is
(6.8) |
where
(6.9) |
The maximum likelihood estimate for μ is easily obtained by differentiating (6.8) with respect to μ and setting the resulting score function equal to zero to get . An EM algorithm can be used to obtain maximum likelihood estimates for Λ and Ψ.
6.4.1 Background: EM Algorithm
The expectation-maximization (EM) algorithm (Dempster et al., 1977) is an iterative procedure for finding maximum likelihood estimates when data are incomplete or treated as such. The EM algorithm iterates between an E-step and an M-step until some stopping rule is reached. On the E-step, the expected value of the complete-data log-likelihood is computed conditional on the current parameter estimates. On the M-step, this quantity is maximized with respect to the parameters to obtain (new) updates. Extensive details on the EM algorithm are provided by McLachlan and Krishnan (2008).
The complete-data comprise the observed x1, …, xn together with the latent components u1, …, un, where . Now, noting that , we have
Now, the complete-data log-likelihood can be written
where C is constant with respect to μ, Λ, and ψ.
Consider the joint distribution
It follows that
(6.10) |
where β = Λ′(ΛΛ′ + ψΙp)−1, and
(6.11) |
Therefore, noting that and that we are conditioning on the current parameter estimates, the expected value of the complete-data log-likelihood can be written
where is a symmetric q × q matrix, , and
(6.12) |
can be thought of as the sample, or observed, covariance matrix.
Differentiating Q with respect to Λ and ψ, respectively, gives the score functions
and
Solving the equations and gives
The matrix results used to compute these score functions, and used elsewhere in this section, are listed in Appendix C.
On each iteration of the EM algorithm for PPCA, the p × p matrix needs to be computed. Computing this matrix inverse can be computationally expensive, especially for larger values of p. The Woodbury identity (Woodbury, 1950) can be used in such situations to avoid inversion of non-diagonal p × p matrices. For an m × m matrix A, an m × k matrix U, a k × k matrix C, and a k × m matrix V, the Woodbury identity is
Setting , ,, and C = Iq gives
(6.13) |
which can be used to speed up the EM algorithm for the PPCA model. The left-hand side of (6.13) requires inversion of a p × p matrix but the right-hand side leaves only a q × q matrix and some diagonal matrices to be inverted. A related identity for the determinant of the covariance matrix,
(6.14) |
is also helpful in computation of the component densities. Identities (6.13) and (6.14) give an especially significant computational advantage when p is large and q ≪ p.
Similar to McNicholas and Murphy (2008), the parameters and can be initialized in several ways, including via eigendecomposition of . Specifically, is computed and then it is eigen-decomposed to give
and is initialized using
where d is the element-wise square root of the diagonal of D. The initial value of ψ can be taken to be
There are several options for stopping an EM algorithm. One popular approach is to stop an EM algorithm based on lack of progress in the log-likelihood, i.e., stopping the algorithm when
(6.15) |
for ϵ small, where l(k) is the (observed) log-likelihood value from iteration k. This stopping rule can work very well when the log-likelihood increases and then plateaus at the maximum likelihood estimate. However, likelihood values do not necessarily behave this way and so it can be worth considering alternatives — see Chatper 2 of McNicholas (2016a) for an illustrated discussion.
As McNicholas (2016a) points out, Böhning et al. (1994), Lindsay (1995), and McNicholas et al. (2010) consider convergence criteria based on Aitken’s acceleration Aitken (1926). Aitken’s acceleration at iteration k is
(6.16) |
and an asymptotic estimate of the log-likelihood at iteration k + 1 can be computed via
(6.17) |
Note that “asymptotic” here refers to the iteration number so that (6.17) can be interpreted as an estimate, at iteration k + 1, of the ultimate value of the log-likelihood. Following McNicholas et al. (2010), the algorithm can be considered to have converged when
(6.18) |
6.4.7 Implementing the EM Algorithm for PPCA
The EM algorithm for PPCA is implemented in two steps. First, we define some helper functions in the following code block. They are used by the EM algorithm function. Specifically, we have implemented a function for each of the identities (6.13) and (6.14) along with functions for each of the equations (6.8) and (6.16). Doing this helps keep the code modular, which facilitates testing some of the more complex mathematics in the algorithm.
## EM Algorithm for PPCA -- Helper functions
function wbiinv(q, p, psi, L)
## Woodbury Indentity eq 6.13
Lt = transpose(L)
Iq = Matrix{Float64}(I, q, q)
Ip = Matrix{Float64}(I, p, p)
psi2 = /(1, psi)
m1 = *(psi2, Ip)
m2c = +(*(psi, Iq), *(Lt, L))
m2 = *(psi2, L, inv(m2c), Lt)
return(-(m1, m2))
end
function wbidet(q, p, psi, L)
## Woodbury Indentity eq 6.14
Iq = Matrix{Float64}(I, q, q)
num = psi^p
denl = *(transpose(L), wbiinv(q, p, psi, L), L)
den = det(-(Iq, den1))
return(/(num, den))
end
function ppca_ll(n, q, p, psi, L, S)
## log likelihood eq 6.8
n2 = /(n, 2)
c = /(*(n, p, log(*(2, pi))), 2)
l1 = *(n2, log(wbidet(q, p, psi, L)))
l2 = *(n2, tr(*(S, wbiinv(q, p, psi, L))))
return(*(-1,+(c, l1, l2)))
end
function cov_mat(X)
n,p = size(X)
mux = mean(X, dims = 1)
X_c = .-(X, mux)
res = *(/(1,n), *(transpose(X_c), X_c))
return(res)
end
function aitkenaccel(l1, l2, l3, tol)
## l1 = l(k+1), l2 = l(k), l3 = l(k-1)
conv = false
num = -(l1,l2)
den = -(l2, l3)
## eq 6.16
ak = /(num, den)
if ak <= 1.0
c1 = -(l1, l2)
c2 = -(1.0, ak)
## eq 6.17
l_inf = +(l2, /(c2, c1))
c3 = -(l_inf, l2)
if 0.0 < c3 < tol
conv = true
end
end
return conv
end
function ppca_fparams(p, q)
## number of free parameters in the ppca model
c1 = *(p, q)
c2 = *(-1,q, -(q, 1), 0.5)
return(+(c1, c2, 1))
end
function BiC(ll, n, q, ρ)
## name does not conflict with StatsBase
## eq 6.19
c1 = *(2, ll)
c2 = *(ρ,log(n))
return(-(c1, c2))
end
The ppca() function, illustrated in the following code block, carries out the EM algorithm described previously. It uses the eigen-decomposition of the sample covariance matrix to initialize Λ, which is used in turn to initialize ψ. A while loop is used to iteratively perform the (post-) E-step updates for β and Θ as well as the M-step updates of Λ and ψ. After the M-step, the log-likelihood is calculated and its value is stored in an array. The values in the array are used to calculate the Aitken acceleration to check convergence. The Woodbury identity is used to speed up the beta updates and the log-likelihood computations. The function returns a Julia dictionary with the PPCA results. The results include the number of iterations the EM algorithm ran, the array of log-likelihoods, the model parameters, the number of latent components q, the standard deviations from the eigen-decomposition, the orthonormal coefficients, and the projections of the data onto the latent space.
using LinearAlgebra, Random
Random.seed!(429)
include("chp6_ppca_functions.jl")
## EM Function for PPCA
function ppca(X; q = 2, thresh = 1e-5, maxit = 1e5)
Iq = Matrix{Float64}(I, q, q)
qI = *(q, Iq)
n,p = size(X)
## eigfact has eval/evec smallest to largest
ind = p:-1:(p-q+1)
Sx = cov_mat(X)
D = eigen(Sx)
d = diagm(0 => map(sqrt, D.values[ind]))
P = D.vectors[:, ind]
## initialize parameters
L = *(P, d) ## pxq
psi = *(/(1,p), tr(-(Sx, *(L, transpose(L)))))
B = zeros(q,p)
T = zeros(q,q)
conv = true
iter = 1
ll = zeros(100)
ll[1] = 1e4
## while not converged
while(conv)
## above eq 6.12
B = *(transpose(L), wbiinv(q, p, psi, L)) ## qxp
T = -(qI, +(*(B,L), *(B, Sx, transpose(B)))) ## qxq
## sec 6.4.3 - update Lambda_new
L_new = *(Sx, transpose(B), inv(T)) ## pxq
## update psi_new
psi_new = *(/(1,p), tr(-(Sx, *(L_new, B, Sx)))) #num
iter += 1
if iter > maxit
conv = false
println("ppca() while loop went over maxit parameter. iter =
$iter")
else
## stores at most the 100 most recent ll values
if iter <= 100
ll[iter]=ppca_ll(n, q, p, psi_new, L_new, Sx)
else
ll = circshift(ll, -1)
ll[100] = ppca_ll(n, q, p, psi_new, L_new, Sx)
end
if 2 < iter < 101
## scales the threshold by the latest ll value
thresh2 = *(-1,ll[iter], thresh)
if aitkenaccel(ll[iter], ll[(iter-1)], ll[(iter-2)], thresh2)
conv = false
end
else
thresh2 = *(-1,ll[100], thresh)
if aitkenaccel(ll[100], ll[(99)], ll[(98)], thresh2)
conv = false
end
end
end ## if maxit
L = L_new
psi = psi_new
end ## while
## orthonormal coefficients
coef = svd(L).U
## projections
proj = *(X, coef)
if iter <= 100
resize!(ll, iter)
end
fp = ppca_fparams(p, q)
bic_res = BiC(ll[end], n, q, fp)
return(Dict(
"iter" => iter,
"ll" => ll,
"beta" => B,
"theta" => T,
"lambda" => L,
"psi" => psi,
"q" =>
"sd" => diag(d),
"coef" => coef,
"proj" => proj,
"bic" => bic_res
))
end
ppca1 = ppca(crab_mat_c, q=3, maxit = 1e3)
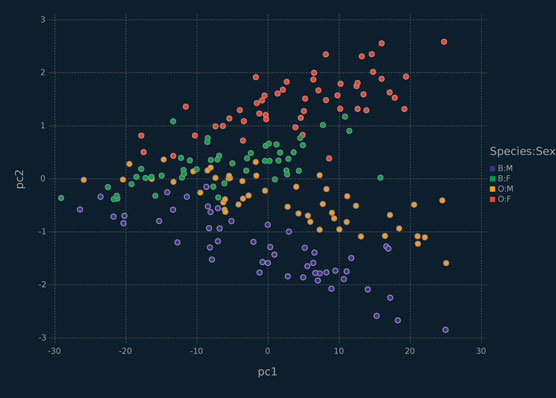
It should be noted that the Λ matrix is neither orthonormal nor composed of eigenvectors (Tipping and Bishop, 1999b). To get orthonormal principal components from it, a singular value decomposition is performed on Λ and the left singular vectors are used as principal component loadings. The projections are plotted in Figure 6.3. With three latent components, we see a clear separation between the crab species.
Figure 6.3 Scallerplot depicting the second and third principal components from the PPCA model with three latent components for the crabs data, coloured by sex and species.
The choice of the number of latent components is an important consideration in PPCA. One approach is to choose the number of latent components that captures a certain proportion of the variation in the data. Lopes and West (2004) carry out simulation studies showing that the Bayesian information criterion (BIC; Schwarz, 1978) can be effective for selection of the number of latent factors in a factor analysis model, and a similar approach can be followed for PPCA. The BIC is given by
(6.19) |
where is the maximum likelihood estimate of , is the maximized (observed) log-likelihood, and ρ is the number of free parameters in the model. The BIC values for PPCA models with different numbers of latent components run on the crabs data are displayed in figure 6.4. The clear choice is the model with three latent components, which has the largest BIC value based on equation 6.19. Clearly this model is capturing the important information in the third principal component, visualized in figure 6.3.
Figure 6.4 BIC values for the PPCA models with different numbers of latent components for the crabs data.
The goal of PPCA is not to necessarily give better results than PCA, but to permit a range of extensions. The probabilistic formulation of the PPCA model allows these extensions, one of which will be illustrated in Section 6.6. Because the PPCA model has a likelihood formulation, it can easily be compared to other probabilistic models, it may be formulated to work with missing data, and adapted for use as a Bayesian method. Noted that, as ψ → 0, the PPCA solution converges to the PCA solution (see Tipping and Bishop, 1999b, for details).
The k-means clustering technique iteratively finds k cluster centres and assigns each observation to the nearest cluster centre. For k-means clustering, these cluster centres are simply the means. The result of k-means clustering is effectively to fit k circles of equal radius to data, where each circle centre corresponds to one of the k means. Of course, for p > 2 dimensions, we have p-spheres rather than circles. Consider the k-means clustering of the x2 data in the following code block. Recall that the x2 data is just a mixture of three bivariate Gaussian distributions (Figure 6.5).
Figure 6.5 Scallerplot depicting the x2 data, coloured by class.
The x2 data present an interesting example of simulated data where classes from the generative model, i.e., a three-component Gaussian mixture model, cannot quite be taken as the correct result. Specifically, an effective clustering procedure would surely put one of the green-coloured points in Figure 6.5 in the same cluster as the yellow points.
In the following code block, the choice of k is made using the elbow method, where one plots k against the total cost, i.e., the total within-cluster sum of squares, and chooses the value of k corresponding to the “elbow” in the plot. Note that the total within-cluster sum of squares is
where Cg is the gth cluster and is its mean.
using DataFrames, Clustering, Gadfly, Random
Random.seed!(429)
mean_x2 = mean(x2_mat, dims=1)
## mean center the cols
x2_mat_c = x2_mat .- mean_x2
N = size(x2_mat_c)[1]
## kmeans() - each column of X is a sample - requires reshaping x2
x2_mat_t = reshape(x2_mat_c, (2,N))
## Create data for elbow plot
k = 2:8
df_elbow = DataFrame(k = Vector{Int64}(), tot_cost = Vector{Float64}())
for i in k
tmp = [i, kmeans(x2_mat_t, i; maxiter=10, init=:kmpp).totalcost ]
push!(df_elbow, tmp)
end
## create elbow plot
p_km_elbow = plot(df_elbow, x = :k, y = :tot_cost, Geom.point, Geom.line,
Guide.xlabel("k"), Guide.ylabel("Total Within Cluster SS"),
Coord.Cartesian(xmin = 1.95), Guide.xticks(ticks = collect(2:8)))
From Figure 6.6, it is clear that the elbow occurs at k = 3. The performance of k-means clustering on this data is not particularly good (Figure 6.7), which is not surprising. The reason this result is not surprising is that, as mentioned before, k-means clustering will essentially fit k circles — for the x2 data, two of the three clusters are long ellipses.
It is of interest to consider the solution for k = 2 (Figure 6.8) because it further illustrates the reliance of k-means clustering on clusters that are approximately circles. An example where k-means should, and does, work well is shown in Figure 6.9, where the clusters are well described by fitted circles of equal radius. It is also worth noting that k-means clustering works well for trivial clustering problems where there is significant spatial separation between each cluster, regardless of the cluster shapes.
Figure 6.6 Elbow plot for selecting k in the k-means clustering of the x2 data.
Figure 6.7 Scallerplot depicting the x2 data, coloured by the k-means clustering solution for k = 3, with red stars marking the cluster means.
Figure 6.8 Scallerplot depicting the x2 data, coloured by the k-means clustering solution for k = 2, with red stars marking the cluster means.
Figure 6.9 Scallerplot depicting a dataset where k-means works well, coloured by the k-means clustering solution for k = 3, with red stars marking the cluster means.
6.6 MIXTURE OF PROBABILISTIC PRINCIPAL COMPONENTS ANALYZERS
Building on the PPCA model, Tipping and Bishop (1999a) introduced the the mixture of probabilistic principal components analyzers (MPPCA) model. It can model complex structures in a dataset using a combination of local PPCA models. As PPCA provides a reduced dimensional representation of the data, MPPCA models work well in high-dimensional clustering, density estimation and classification problems.
Analogous to the PPCA model (Section 6.3), the MPPCA model assumes that
(6.20) |
with probability πg, for i = 1, …, n and g = 1, …, G, where Λg is a p × q matrix of loadings, the Uig are independently N(0, Iq) and are independent of the ϵig, which are independently N(0, ψ9Ip), where ψg ∈ ℝ+. It follows that the density of Xi from the MPPCA model is
(6.21) |
where ϑ denotes the model parameters.
Overview
Parameter estimation for the MPPCA model can be carried out using an alternating expectation-conditional maximization (AECM) algorithm (Meng and van Dyk, 1997). The expectation-conditional maximization (ECM) algorithm (Meng and Rubin, 1993) is a variant of the EM algorithm that replaces the M-step by a series of conditional maximization steps. The AECM algorithm allows a different specification of complete-data for each conditional maximization step. Accordingly, the AECM algorithm is suitable for the MPPCA model, where there are two sources of missing data: the unknown component membership labels and the latent components (or factors) uig, for i = 1, …,n and g = 1, …,G. Details of fitting the AECM algorithm for the more general mixture of factor analyzers model are given by McLachlan and Peel (2000), and parameter estimation for several related models is discussed by McNicholas and Murphy (2008, 2010) and McNicholas et al. (2010).
AECM Algorithm: First Stage
As usual, denote by z1, …, zn the unobserved component membership labels, where if observation i belongs to component g and otherwise. At the first stage of the AECM algorithm, the complete-data are taken to be the observed x1, …, xn together with the unobserved z1, …, zn, and the parameters πg and µg are estimated, for g = 1, …, G. The complete-data log-likelihood is
(6.22) |
and the (conditional) expected values of the component membership labels are given by
(6.23) |
for i = 1, …, n and g = 1, …, G.
Using the expected values given by (6.23) within (6.22), the expected value of the complete-data log-likelihood at the first stage is
where and
(6.24) |
Maximising Q1 with respect to πg and µg yields
(6.25) |
respectively.
AECM Algorithm: Second Stage
At the second stage of the AECM algorithm, the complete-data are taken to be the observed x1, …,xn together with the unobserved component membership labels z1, …,zn and the latent factors , for i = 1, …, n and g = 1, …,G, and the parameters Λg and ψg are estimated, for g = 1, …, G. Proceeding in an analogous fashion to the EM algorithm for the factor analysis model (Section 6.4), the complete-data log-likelihood is given by
where C is constant with respect to Λg and ψg. Bearing in mind that we are conditioning on the current parameter estimates, and using expected values analogous to those in (6.10) and (6.11), the expected value of the complete-data log-likelihood can be written
where
is a q × p matrix and
is a symmetric q × q matrix. Note that replaces µg in Sg, see (6.24).
Differentiating Q2(Λ, Ψ) with respect to Λg and , respectively, gives the score functions
and
Solving the equations and gives
The matrix results used to compute these score functions, and used elsewhere in this section, are listed in Appendix C.
AECM Algorithm for the MPPCA Model
An AECM algorithm for the MPPCA model can now be presented.
AECM Algorithm for MPPCA
initialize
initialize , , Sg, ,
while convergence criterion not met
update
if not iteration 1
update
end
compute Sg, , Θg
update ,
update
check convergence criterion
end
In the following code block, we define some helper functions for the MPPCA model.
## MPPCA helper functions
using LinearAlgebra
function sigmaG(mu, xmat, Z)
res = Dict{Int, Array}()
N,g = size(Z)
c1 = ./(1,sum(Z, dims = 1))
for i = 1:g
xmu = .-(xmat, transpose(mu[:,i]))
zxmu = .*(Z[:,i], xmu)
res_g = *(c1[i], *(transpose(zxmu), zxmu))
push!(res, i=>res_g)
end
return res
end
function muG(g, xmat, Z)
N, p = size(xmat)
mu = zeros(p, g)
for i in 1:g
num = sum(.*(Z[:,i], xmat), dims = 1)
den = sum(Z[:, i])
mu[:, i] = /(num, den)
end
return mu
end
## initializations
function init_LambdaG(q, g, Sx)
res = Dict{Int, Array}()
for i = 1:g
p = size(Sx[i], 1)
ind = p:-1:(p-q+1)
D = eigen(Sx[i])
d = diagm(0 => map(sqrt, D.values[ind]))
P = D.vectors[:, ind]
L = *(P,d)
push!(res, i => L)
end
return res
end
function init_psi(g, Sx, L)
res = Dict{Int, Float64}()
for i = 1:g
p = size(Sx[i], 1)
psi = *(/(1,p), tr(-(Sx[i], *(L[i], transpose(L[i])))))
push!(res, i=>psi)
end
return res
end
function init_dict0(g, r, c)
res = Dict{Int, Array}()
for i = 1:g
push!(res, i=> zeros(r, c))
end
return res
end
## Updates
function update_B(q, p, psig, Lg)
res = Dict{Int, Array}()
g = length(psig)
for i=1:g
B = *(transpose(Lg[i]), wbiinv(q, p, psig[i], Lg[i]))
push!(res, i=>B)
end
return res
end
function update_T(q, Bg, Lg, Sg)
res = Dict{Int, Array}()
Iq = Matrix{Float64}(I, q, q)
qI = *(q, Iq)
g = length(Bg)
for i =1:g
T = -(qI, +(*(Bg[i], Lg[i]), *(Bg[i], Sg[i], transpose(Bg[i]))))
push!(res, i=>T)
end
return res
end
function update_L(Sg, Bg, Tg)
res = Dict{Int, Array}()
g = length(Bg)
for i = 1:g
L = *(Sg[i], transpose(Bg[i]), inv(Tg[i]))
push!(res, i=>L)
end
return res
end
function update_psi(p, Sg, Lg, Bg)
res = Dict{Int, Float64}()
g = length(Bg)
for i = 1:g
psi = *(/(1,p), tr(-(Sg[i], *(Lg[i], Bg[i], Sg[i]))))
push!(res, i=>psi)
end
return res
end
function update_zmat(xmat, mug, Lg, psig, pig)
N,p = size(xmat)
g = length(Lg)
res = Matrix{Float64}(undef, N, g)
Ip = Matrix{Float64}(I, p, p)
for i = 1:g
pI = *(psig[i], Ip)
mu = mug[:, i]
cov = +(*(Lg[i], transpose(Lg[i])), pI)
pi_den = *(pig[i], pdf(MvNormal(mu, cov), transpose(xmat)))
res [:, i] = pi_den
end
return ./(res, sum(res, dims =2))
end
function mapz(Z)
N,g = size(Z)
res = Vector{Int}(undef, N)
for i = 1:N
res[i] = findmax(Z[i,:])[2]
end
return res
end
function mppca_ll(N, p, pig, q, psig, Lg, Sg)
g = length(Lg)
l1,l2,l3 = (0,0,0)
c1 = /(N,2)
c2 = *(-1,c1, p, g, log(*(2,pi)))
for i = 1:g
l1 += log(pig[i])
l2 += log(wbidet(q, p, psig[i], Lg[i]))
l3 += tr(*(Sg[i], wbiinv(q, p, psig[i], Lg[i])))
end
l1b = *(N, l1)
l2b = *(-1,c1, l2)
l3b = *(-1,c1, l3)
return(+(c2, l1b, l2b, l3b))
end
function mppca_fparams(p, q, g)
## number of free parameters in the ppca model
c1 = *(p, q)
c2 = *(-1,q, -(q, 1), 0.5)
return(+(*(+(c1, c2), g), g))
end
function mppca_proj(X, G, map, L)
res = Dict{Int, Array}()
for i in 1:G
coef = svd(L[i]) .U
sel = map .== i
proj = *(X[sel, :], coef)
push!(res, i=>proj)
end
return(res)
end
Having created helper functions in the previous code block, an AECM algorithm for the MPPCA model is implemented in the following code block. The values of can be initialized in a number of ways, e.g., randomly or using the results of k-means clustering.
using Clustering
include("chp6_ppca_functions.jl")
include("chp6_mixppca_functions.jl")
## MPPCA function
function mixppca(X; q = 2, G = 2, thresh = 1e-5, maxit = 1e5, init = 1))
## initializations
N, p = size(X)
## z_ig
if init == 1
## random
zmat = rand(Uniform(0,1), N, G)
## row sum to 1
zmat = ./(zmat, sum(zmat, dims =2))
elseif init == 2
# k-means
kma = kmeans(permutedims(X), G; init=:rand).assignments
zmat = zeros(N,G)
for i in 1:N
zmat[i, kma[i] ] = 1
end
end
n_g = sum(zmat, dims = 1)
pi_g = /(n_g, N)
mu_g = muG(G, X, zmat)
S_g = sigmaG(mu_g, X, zmat)
L_g = init_LambdaG(q, G, S_g)
psi_g = init_psi(G, S_g, L_g)
B_g = init_dict0(G, q, p)
T_g = init_dict0(G, q, q)
conv = true
iter = 1
ll = zeros(100)
ll[1] = 1e4
# while not converged
while(conv)
## update pi_g and mu_g
n_g = sum(zmat, dims = 1)
pi_g = /(n_g, N)
mu_g = muG(G, X, zmat)
if iter > 1
## update z_ig
zmat = update_zmat(X, mu_g, L_g, psi_g, pi_g)
end
## compute S_g, Beta_g, Theta_g
S_g = sigmaG(mu_g, X, zmat)
B_g = update_B(q, p, psi_g, L_g)
T_g = update_T(q, B_g, L_g, S_g)
## update Lambda_g psi_g
L_g_new = update_L(S_g, B_g, T_g)
psi_g_new = update_psi(p, S_g, L_g, B_g)
## update z_ig
zmat = update_zmat(X, mu_g, L_g_new, psi_g_new, pi_g)
iter += 1
if iter > maxit
conv = false
println("mixppca() while loop went past maxit parameter. iter =
$iter")
else
## stores at most the 100 most recent ll values
if iter <= 100
ll[iter] = mppca_ll(N, p, n_g, pi_g, q, psi_g, L_g, S_g)
else
ll = circshift(ll, -1)
ll[100] = mppca_ll(N, p, n_g, pi_g, q, psi_g, L_g, S_g)
end
if 2 < iter < 101
## scales the threshold by the latest ll value
thresh2 = *(-1,ll[iter], thresh)
if aitkenaccel(ll[iter], ll[(iter-1)], ll[(iter-2)], thresh2)
conv = false
end
else
thresh2 = *(-1,ll[100], thresh)
if aitkenaccel(ll[100], ll[(99)], ll[(98)], thresh2)
conv = false
end
end
end ## if maxit
L_g = L_g_new
psi_g = psi_g_new
end ## while
map_res = mapz(zmat)
proj_res = mppca_proj(X, G, map_res, L_g)
if iter <= 100
resize!(ll, iter)
end
fp = mppca_fparams(p, q, G)
bic_res = BiC(ll[end], N, q, fp)
return Dict(
"iter" => iter,
"ll" => ll,
"beta" => B_g,
"theta" => T_g,
"lambda" => L_g,
"psi" => psi_g,
"q" => q,
"G" => G,
"map" => map_res,
"zmat" => zmat,
"proj" => proj_res,
"bic" => bic_res
)
end
mixppca_12k = mixppca(cof_mat_c, q=1, G=2, maxit = 1e6, thresh = 1e-3,
init=2)
Predicted Classifications
Predicted classifications are given by the values of (6.23) after the AECM algorithm has converged. These predicted classifications are inherently soft, i.e., [0,1]; however, in many practical applications, they are hardened by computing maximum a posteriori (MAP) classifications:
6.6.3 Illustrative Example: Coffee Data
The MPPCA model is applied to the coffee data, using the code in the previous block. Models are initialized with k-means clustering and run for G = 2, 3, 4 and q = 1, 2, 3. Using the BIC, we choose a model with G = 2 groups (i.e., mixture components) and q = 1 latent component (Figure 6.10). This model correctly classifies all of the coffees into their respective varieties, i.e., the MAP classifications exactly match the varieties.
Figure 6.10 BIC values for the MPPCA models with different numbers of groups and latent components for the coffee data.
Analogous to our comments in Section 5.8, it is worth emphasizing that only selected unsupervised learning techniques have been covered, along with Julia code needed for implementation. Just as Chapter 5 cannot be considered a thorough introduction to supervised learning, the present chapter cannot be considered a thorough introduction to unsupervised learning. However, the Julia code covered herein should prove most instructive. Furthermore, it is worth noting that two further unsupervised learning approaches — one of which can be considered an extension of MPPCA — are discussed in Chapter 7.
Two techniques for clustering, or unsupervised learning, have been considered herein. While there are a host of clustering techniques available (see, e.g., Everitt et al., 2011), the authors of the present monograph have a preference for mixture model-based approaches and a wide range of such approaches are available (see McNicholas, 2016a,b, for many examples). In the PPCA example, the BIC was used to select q and, when using mixtures thereof, the BIC was used to select both G and q. For completeness, it is worth noting that the BIC is a popular tool for mixture model selection in general (see McNicholas, 2016a, Section 2.4, for further details).