Evolution and Development: Molecules
Antónia Monteiro
OUTLINE
1. The goals of molecular studies in evolutionary developmental biology
2. Mapping genotype to phenotype during development
3. Mapping genotype to phenotype during evolution
4. The evolution of novel traits and their underlying gene regulatory networks
5. Future areas of research in evolutionary developmental biology
In this chapter, major research themes and approaches in evolutionary developmental biology, commonly referred to as evo-devo, are presented from a molecular perspective. The field is concerned primarily with connecting changes at the DNA level to changes in developmental pathways and gene regulatory networks that lead to the evolution of morphology, physiology, and behavior. Researchers in the field are interested in identifying whether mutations in DNA are altering the regulation or the function of proteins, and describing how these changes alter the output of larger gene regulatory networks and ultimately the adult phenotype. In addition, interest is mounting in understanding how novel gene regulatory networks originate and evolve, and how the environment interacts with these gene regulatory networks to promote either robustness or adaptive phenotypic plasticity in organismal form.
GLOSSARY
Candidate Gene. A gene that is suspected of playing a role in the evolution of a trait typically because its expression domain or temporal pattern of expression is associated with the development of that trait.
Enhancer or Cis-Regulatory Element. A sequence of DNA that regulates the temporal and spatial expression of flanking protein-coding genes when bound by specific transcription factors.
Homologous Trait. A trait found in two lineages is homologous if it derives from the same trait present in the common ancestor. For example, pectoral fins in fish and arms in humans are homologous traits.
Modular Gene Regulatory Network. An interacting group of genes that are activated together in response to simple inputs, and in a largely context-independent manner during development. For example, a gene regulatory network specific to the fruit fly eye can be activated in multiple places in the body (e.g., wings and antennae) in response to the expression of specific transcription factors.
Phenotypic Plasticity. The ability of some organisms to modify their phenotype in response to their rearing environment.
Quantitative Trait Loci (QTL) Mapping. A method that involves discovering the genomic position and relative effect of loci responsible for producing phenotypic differences between two individuals that can be crossed.
Selector Gene. A regulatory gene that specifies cell, tissue, organ, or regional identity in animals.
Serial Homologous Traits. Repeated traits within the same body that use a similar gene regulatory network during their development. Examples of serial homologous traits include arthropod segments, teeth, vertebrae, and pelvic and pectoral fins (or arms and legs).
Transcription Factor (TF). A protein that binds to DNA to effect changes in the transcription of flanking protein-coding genes.
Transgenic Organism. An organism that has been genetically modified to carry additional genes. Typically, transgenic organisms are used to test the function of particular protein-coding sequences during development. Transgenics can also be used to test the function of candidate enhancer sequences by attaching them to reporter genes such as green fluorescent protein (GFP), and monitoring GFP expression during development.
1. THE GOALS OF MOLECULAR STUDIES IN EVOLUTIONARY DEVELOPMENTAL BIOLOGY
Over the course of life, organisms have evolved into myriad sizes, shapes, and forms. They also evolved different physiologies, life histories, and behaviors. Most of this diversity is encoded in the molecule that unites all of life, DNA, and a challenge for biologists lies in understanding how variation in this molecule actually produces organismal diversity.
Connecting variation at the DNA level with organismal diversity can be broken into two separate challenges. One involves understanding how DNA sequences in any one organism lead to the development of the traits of that organism. This endeavor is also dubbed “identifying the genotype-phenotype map” for each species. The other challenge involves identifying the relevant changes in the genotype-phenotype map that cause different organisms to evolve different traits. The first challenge falls in the domain of developmental biology, and the second challenge in the domain of evolutionary developmental biology, or evo-devo.
In molecular studies of evo-devo (compared with organismal level studies; see chapter V.10), the goal is to understand how the process of genomic evolution, including changes in gene number, gene structure, and gene regulation, is translated via developmental mechanisms into the evolution of morphology, behavior, or physiology. Because phenotypes such as behavior are only just beginning to be studied, this chapter will mostly highlight the various ways that biologists are probing the developmental mechanisms that underlie the evolution of morphology. There have been two main routes to this type of work; one involves investigating the entire genome, whereas the other investigates candidate genes to identify DNA sequences or loci responsible for morphological evolution. The chapter also addresses the role of developmental modules and of modular genetic architecture in body plan evolution and the evolution of novel complex traits, and concludes with some of the unexplored aspects of molecular developmental evolution, including the molecular basis of plasticity.
2. MAPPING GENOTYPE TO PHENOTYPE DURING DEVELOPMENT
While developmental biology aims to understand how genes are involved in building organismal traits via the developmental process, evolutionary developmental biology focuses on the subset of genes and developmental mechanisms responsible for the evolution of morphological variation within and between species. Variation at the DNA level impacts developmental mechanisms and ultimately phenotypes in multiple ways. This section briefly illustrates the fundamental steps connecting DNA to morphology via development, or the unfurling of the genotype-phenotype map.
Mapping genotype to phenotype involves the process of reading the DNA molecule through the course of development. All multicellular organisms start off as single cells that subsequently divide and differentiate into multiple cell types, tissues, and organs. Cell division and differentiation involve complex orchestrations of gene regulation. Genes are inactive in most cells of early embryos, but as development progresses, different genes are activated in different cells of the embryo, producing asymmetries in regulatory states (on/off). These asymmetries in gene expression across the body later translate into visible phenotypic differences. Certain cells will become differentiated to produce pigments that give rise to color patterns, other cells will become muscle cells, and yet others will secrete crystalline proteins that agglomerate to give rise to the lens of an eye.
Asymmetries in regulatory states of genes inside cells are produced by asymmetries in the distribution of important regulatory proteins, transcription factors (TFs). TFs induce (or repress) gene transcription by binding to specific regulatory sequences flanking a protein-coding sequence, also called enhancers or cis-regulatory elements (see chapter V.7). Binding of TFs to enhancers (usually more than one TF is involved) leads to the recruitment of the RNA polymerase enzyme to the promoters of those genes, and transcription is initiated. Enhancers contain information about when and where a gene will be expressed during development because they contain clusters of TF binding sites that will lead to gene activation (or repression) only when bound by the respective TFs. If a gene contains more than one enhancer, it can be expressed in very different developmental contexts, depending on the sequence of each of its enhancers. The earliest stages of development usually start with TFs that are asymmetrically distributed in the cytoplasm of the single-celled egg, and are responsible for beginning the process of cell differentiation. If a certain cocktail of TFs is present in the right combination and concentration in one part of the embryo but absent from another, then genes responsive to that exact complement and concentration of TFs (i.e., with binding sites for those TFs in their enhancer sequences) will be turned on only in those cells of the embryo. TFs can also induce the expression of signaling molecules that can diffuse some distance within the embryo. These molecules can, in turn, activate a novel set of TFs in the surrounding cells. The process of development is essentially a process of subdividing a growing and uniform field of cells into separate domains expressing unique combinations of TFs at unique concentrations. These TFs then control downstream target genes that build different traits in different parts of the body. Intermediate levels of gene regulation occur after a gene is transcribed, and before traits are built, for instance, by posttranscriptional modifications to proteins, but so far, not much work in evo-devo has explored evolution at this level.
This section has established, in broad brushstrokes, the mechanisms by which genomic information is translated into a phenotype during the course of development; the next section will focus on the methods used by researchers to identify the alterations to developmental programs that lead to distinct morphologies that are characteristic of different species.
3. MAPPING GENOTYPE TO PHENOTYPE DURING EVOLUTION
Two main approaches are used to investigate the genomic loci and/or developmental mechanisms that have been altered to produce morphological change across species: (1) the candidate gene approach and (2) the quantitative trait locus (QTL) mapping approach. These two approaches have different strengths and limitations. The candidate gene approach can be undertaken to investigate morphological change in any set of species, whereas the QTL mapping approach is limited to species that can be crossed in the lab or that cross naturally in the field. In addition, the candidate gene approach can highlight differences in developmental programs that are characteristic of different genera, family, or even phyla, and that have been established deep in the tree of life, whereas the QTL mapping approach usually addresses more recent divergence in developmental programs that result in species-level differences. The main distinction between these approaches is that while the candidate gene approach can identify how developmental programs have changed across species, it rarely can pinpoint the causative mutations that lead to these changes. The QTL approach, on the other hand, can zoom in on the exact genomic loci that have mutated and are responsible for alterations in developmental programs across closely related species (see chapter V.13). Examples of both these approaches are provided below.
Often researchers target candidate genes for their role in causing differences in development across species because of prior knowledge that these genes are expressed during the development of the trait of interest. Genes known to be involved in building a homologous trait in a different species also make good candidate genes. Candidate gene approaches were used to implicate two genes, Bone morphogenetic protein 4 (Bmp4) and Calmodulin, in the generation of differently shaped beaks in Galápagos finches. These two genes were differentially expressed in finches with deep and broad or long beaks, respectively. When chickens with artificially modified levels of these genes were produced in the lab, they also showed significant changes in the depth/width and length of their beaks. Taken together, these data suggest that changes in the expression of Bmp4 and Calmodulin during the course of evolution caused changes in beak shape in these finches; however, these data do not necessarily suggest that these genes were themselves modified during the course of evolution to alter the beaks of these finches. Alterations to a gene’s expression can occur via alterations in the cis-regulatory elements of the gene itself, or by alterations to the cocktail of TFs that bind to these elements (the trans regulators) and regulate gene expression. So while the candidate gene approach identifies changes in developmental mechanisms—changes to amounts of Bmp4 or Calmodulin mRNA and protein present in beaks at particular times in development—it cannot always identify the locus that mutated to produce these differences. To further dissect where these differences lie, a reciprocal locus transplantation experiment using transgenics is needed (discussed below).
QTL approaches have also been used to identify genes responsible for morphological evolution across species (see chapter V.12). For example, Drosophila melanogaster as well as several other closely related species are covered in small hairs, or trichomes, on the dorsal part of their bodies when they are larvae; however, D. sechellia has few trichomes on its body. By performing QTL mapping in laboratory crosses between D. melanogaster and D. sechellia, the position of the causative locus that explained most of the variation in larval trichome patterns was mapped to the shavenbaby-ovo (svb) locus. Modifications to the sequence of at least three different cis-regulatory elements of svb, each driving expression of svb in different sections of the larval body, were responsible for trichome loss in D. sechellia. This gene, when overexpressed in epidermal cells without trichomes, was shown to be necessary and sufficient to initiate the developmental program that builds trichomes, so shutting it down by deletion of its multiple epidermal enhancers, is an effective and direct way to eliminate trichome development in D. sechellia.
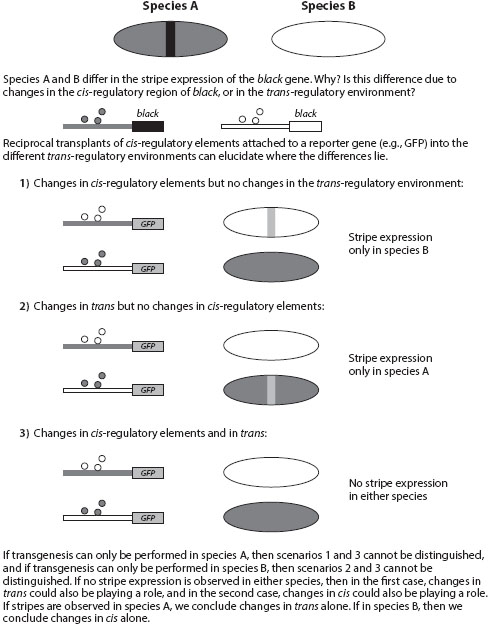
In the Drosophila case, unlike the case of the finches above, the ability to cross the two species with differing morphologies enabled the researchers to determine that cis-regulatory changes rather than changes to the trans-acting factors were responsible for the morphological changes that were observed between the species; however, when genetic crosses between species are not feasible because of reproductive incompatibilities, researchers can turn to transplantation experiments using transgenic tools. The rationale behind these experiments involves taking the candidate gene of one species and introducing it into the trans-regulatory environment of the other species, and then performing the reciprocal experiment with the orthologous gene from the second species (see figure 1). These transplantation experiments are commonly performed in only one direction, often because of limitations in transgenic technology in one of the two test species, but an example of a complete reciprocal transplantation experiment was performed with the lin-48 ovo gene in Caenorhabditis elegans and C. briggsae. Researchers hypothesized that differences in the expression pattern of this gene observed between species could be due either to changes in the cis-regulatory region of the gene or to changes in the trans-acting factors. By performing a complete set of transplantation experiments in which they took the regulatory regions of each gene attached to a reporter gene to monitor expression activity and transplanted them (transgenically) to the trans-regulatory environment of the other species, they were able to conclude that changes in both the cis-regulatory sequences and the trans-acting factors that meditate lin-48 expression contributed to the species-specific differences.
Figure 1. Schematic of reciprocal genetic transplantation experiments that test whether changes in the cis-regulatory elements of a gene or the trans-regulatory factors that bind those elements are responsible for the expression differences observed between two species (A and B). In this case, expression differences correspond to the presence or absence of a stripe of black gene expression along the body (ellipse). Boxes correspond to protein-coding sequences. Black/white boxes: alleles of black candidate gene; gray box: reporter gene (GFP). Lines connected to boxes represent cis-regulatory sequences.
This type of transplantation experiment can also be done with the complete locus (cis-regulatory elements plus protein coding sequence) if alterations at the amino acid level are also suspected of contributing to particular phenotypic differences between species.
4. THE EVOLUTION OF NOVEL TRAITS AND THEIR UNDERLYING GENE REGULATORY NETWORKS
The examples discussed above monitor and dissect the evolution of developmental mechanisms from the perspective of individual genes. Mutations to single developmental genes, however, often modify the expression of many downstream targets and have a large impact on an organism’s final phenotype. The group of affected genes depends on the topology of the regulatory network, that is, how many targets are downstream of the mutated gene, including both direct and indirect targets (see chapter V.9).
Some gene-regulatory networks are modular in their effects and may be quite important in body plan evolution. For instance, the Distal-less and Pax6 TFs are important early regulators of limb and eye development, respectively, throughout the Metazoa. These genes, when ectopically expressed in several other parts of the body of a fly, are able to promote limb duplications and ectopic eyes; that is, they control the initiation of gene regulatory networks that lead to limb and eye differentiation. These networks have modular qualities in that they can be initiated in a context-independent manner at multiple locations in the body, somewhat independently of the cocktail of other TFs present at those locations.
The deployment and co-option of these modular networks into novel places in the body, and their recruitment to create repeated or serial homologous traits, and potentially also novel traits, is an active area of research in evo-devo. The idea is that the origination of novel traits may proceed by the co-option and the mixing and matching of modular networks, in novel combinations and at novel places in the body, rather than by the elaboration of preexisting networks one gene at a time (see figure 2). Evolution of novel traits would proceed via the genetic tinkering of modules of interacting genes by modification of the cis-regulatory regions of only a small set of individual genes regulating the initiation of each of these modules. The above-mentioned Distal-less (Dll) gene in the context of the evolution of appendages provides a nice example of the way these modular gene networks may originate.
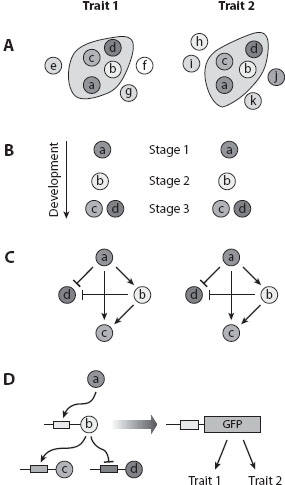
Figure 2. Different types of experiments aimed to test whether four genes (a, b, c, and d) expressed in two traits (1 and 2) are part of the same gene regulatory network that functions in the development of both traits. (A) A common set of genes (circled) is expressed during the development of the two traits. (B) The genes are expressed in a similar temporal order. (C) The genes display the same type of regulatory interactions (a represses d, b activates c, etc. Note that the regulatory interactions inferred may be direct or indirect). (D) Genes internal to the shared set (expressed at developmental stages 2 or 3, but not at stage 1) may contain unique cis-regulatory elements that drive gene expression in the two different developmental contexts. This is depicted by the isolation of the cis-regulatory element of the b gene, attaching it to a reporter gene (GFP), transforming the genome of the organism with this construct, and observing GFP expression in the tissue precursors of the two traits. (Modified from Monteiro 2012, Bioessays 34: 181–186.)
It is possible that in early metazoans, Dll became expressed in a novel cluster of cells as a result of evolution of novel positional information in its cis-regulatory region, “marking” these cells in a unique way. Other genes, by evolving binding sites for the Dll protein in their cis-regulatory regions, would be co-opted for expression in the same cluster of cells. Additional genes would have been gradually added to this basic gene network by developing binding sites either for Dll, or for any of the other gene products activated downstream of Dll. Perhaps, early in the process of building this network, a small outgrowth emerged from the body wall. If these outgrowths were useful in some way, the genomic information coding for the novel network would be retained. Later, with further network elaboration, the small outgrowths could become proper appendages. Such a network, scaffolded on Dll expression, is modular and context insensitive, i.e., when Dll is recruited to novel positions in the body, it is often able to direct the complete set of downstream targets and produce a novel outgrowth at these novel locations.
Many classic examples of the evolution of body plans involve changes to modular gene regulatory networks. These include examples where modular networks are modified by the action of region-specific TFs, named selector genes, or are duplicated, repressed, or co-opted into novel locations in the body to create serial homologous traits, or novel body plans. An example of network modification includes the evolution of arthropod appendages into a variety of different shapes and sizes by the action of Hox genes, selector genes that are differentially expressed along the anterior-posterior axis of the body and give each region of the body a unique identity. In crustaceans, limbs that develop in regions of the body where anterior Hox genes are expressed become feeding appendages, whereas limbs that develop in regions of the body where posterior Hox genes are expressed become walking legs. The Hox genes appear to bind to the cis-regulatory regions of many different genes within a limb network in order to modify their expression, and thus, the final limb phenotype. In addition, Hox genes expressed in the abdominal region directly bind to the early limb enhancer of Dll, thereby shutting down the limb network in the abdomen of flies, and perhaps most other insects.
Similar to the role of Hox genes in specifying the identity of modules along the anterior-posterior axis, modifications to other types of selector gene also underlie modifications to modular gene networks that are repeated in the body. For instance, the Pitx1 gene controls the identity and the development of the pelvic fins in stickleback fish, but Pitx1 has no role in pectoral fin development. Multiple independent deletions of a cis-regulatory element upstream of Pitx1 have occurred in different stickleback populations, resulting in the loss of Pitx1 expression in pelvic fins, and therefore, loss of the pelvic fin structure in these fish. Changes to Pitx1, because of its unique pelvic fin expression, are among the few places in the fin gene regulatory network that would allow a complete fin to be lost without impairing the development of the other serial homologue (the pectoral fin).
Modular gene regulatory networks, such as the limb or the eye network, may have also been co-opted into different regions in the body to give rise to novel body parts, or serial homologous traits. For instance, the appearance of horns in the heads of beetles may have originated via the co-option of the insect limb network to the head, as many of the genes found in limbs are also expressed in horns. The evolution of multiple eyes along the mantle of scallops is probably due to the co-option of an early expressed gene from the eye gene regulatory network to the mantel’s edge. And the evolution of the most posterior set of fins/limbs in vertebrates is due to the co-option of the vertebrate limb network, initially deployed only in the pectoral fin region of primitive fish, to a more posterior position along the anterior-posterior body axis, thus creating the vertebrate paired appendages.
Co-option of the modular gene regulatory networks mentioned above to the novel locations would involve the evolution of novel positional information for the expression of the network’s top regulatory gene. This positional information would be in the form of a novel enhancer sequence where binding sites for one or more TFs expressed at the novel body location would evolve and allow the top regulatory gene to be turned on at that location. It remains possible that completely novel and parallel gene networks were created de novo at these novel body locations; however, this is unlikely, as such networks would take a much longer period of time to evolve and would probably not be fully functional until complete. Many aspects of modular gene regulatory networks are still unclear, such as their frequency in developmental systems, their size distribution (e.g., how many genes are involved), and their evolution, but this information will likely become available as research progresses in this field.
Thinking of development as the temporal stringing together of modular gene regulatory networks also helps explain why there are sometimes dramatic differences between species at the early stages of development, while later stages of development are conserved. Early network modules can evolve as long as the connections to later modules are kept intact. An example involves the earliest steps in embryonic development in Drosophila: the determination of where the head is going to lie. This is achieved by a gradient of Bicoid protein that is set up by the mother before the egg is laid. She deposits and attaches Bicoid mRNA molecules to the anterior end of the egg. On translation, a gradient of protein is established, and high levels of protein activate a downstream target gene in the anterior half of the embryo, hunchback, which defines the head region of the fly. In the beetle Tribolium, Bicoid protein is not responsible for head patterning in the early embryo, but the function and expression of hunchback is still conserved. Another example of such modularity is the sex-determination pathway in animals where the upstream factors that determine the sex of the animal are very diverse, ranging from a sex chromosome to temperature induction (see chapter V.4), but the downstream effectors are very conserved and usually involve the gene doublesex and its homologues. So, gene regulatory networks can evolve in their very earliest steps while downstream components and the final phenotype remain unchanged.
5. FUTURE AREAS OF RESEARCH IN EVOLUTIONARY DEVELOPMENTAL BIOLOGY
The Molecular Basis of Phenotypic Plasticity
While much is beginning to be known about the molecular details of morphological evolution, an area that is still lagging behind concerns investigating the molecular basis of the integration of environmental factors into regulatory gene networks to induce distinct phenotypes. Phenotypic plasticity, or the ability of the same genome to give rise to very different morphological, physiological, or behavioral traits depending on rearing environment, is still poorly understood at the molecular level. A variety of environmental factors such as temperature, light, pressure, food availability, and certain chemicals are known to induce alternative developmental pathways, but the molecular details of the mechanisms by which these factors influence gene regulatory networks are poorly understood.
The evolution of adaptive phenotypic plasticity usually involves changes to gene-regulatory networks that better adapt the organism to different and predictable environments. In many cases, hormones appear to play important roles in coordinating plastic development as they circulate among all the tissues in the body, and are thus able to coordinate changes in multiple modular gene regulatory networks underlying the development of various traits. But how these hormonal signaling systems evolve to interact with specific gene networks and how hormonal systems themselves become sensitive to the environment are still areas of active investigation.
Robustness
The flip side of plasticity is robustness, where developmental networks have evolved extreme insensitivity to environmental and/or genetic perturbations. At the molecular level, robustness is achieved by evolution of regulatory wiring that leads to gene expression homeostasis, by gene duplications, or even by cis-regulatory element duplications that lead to more robust patterns of gene expression in the face of perturbation. Robust gene networks can potentially accumulate many mutations that are buffered from affecting network output (creating cryptic genetic variation) by the architecture of the developmental gene network.
Understanding how these two types of gene networks, plastic and robust, bias or channel further evolutionary change is an important area of future research. In particular, the roles of natural and sexual selection are believed by many to be all-powerful in shaping the behavior and morphology of organisms, but these forces can exert change in systems only if these systems produce sufficient phenotypic variation for selection to act on (see chapter V.10). Plastic networks will readily produce variation in response to environmental variation, whereas robust networks will not. On the other hand, selection on phenotypes derived from plastic networks will not lead to evolutionary change, since the variation is not based in genetics but environmentally induced, whereas selection on phenotypes derived from robust networks will produce minimal change, because phenotypes will essentially be the same. Novel environments may favor evolutionary change, and this can lead to novel patterns of selection and changes to network topology in the case of plastic networks, and to the release of accumulated cryptic genetic variation in the case of robust networks, if these networks are altered beyond their natural buffering capacity.
Conclusion
In summary, molecular evo-devo has the ability to explain both micro- as well as more macro evolutionary changes in developmental programs and phenotypes, the evolution of novel traits, and the role played by the environment in modifying development to create plastic phenotypes. Future empirical work with additional species and traits, as well as modeling work, should eventually aim to produce a theory of morphological evolution based on gene networks, and gene interactions, that fully updates the modern synthesis.
FURTHER READING
Carroll, S. B., J. K. Grenier, and S. D. Weatherbee. 2005. From DNA to Diversity: Molecular Mechanisms and the Evolution of Animal Design. 2nd ed. Malden, MA: Blackwell. A comprehensive text in the field of evo-devo.
Erwin, D. H., and E. H. Davidson. 2009. The evolution of hierarchical gene regulatory networks. Nature Reviews Genetics 10: 140–148. A recent review of several of the major topics discussed in this chapter by leaders in the field.
Fielenback, N., and A. Antebi. 2008. C. elegans dauer formation and the molecular basis of plasticity. Genes and Development 22: 2149–2165. A nice review article that discusses the molecular basis of plasticity in a model organism.
Gilbert, S. F., and D. Epel. 2009. Ecological Developmental Biology: Integrating Epigenetics, Medicine, and Evolution. Sunderland, MA: Sinauer. An engaging and clear exposition of the ways in which the environment affects developmental programs, with many examples at the molecular level.
Nowick, K., and L. Stubbs. 2010. Lineage-specific transcription factors and the evolution of gene regulatory networks. Briefings in Functional Genomics 9: 65–78. A review of the ways in which evolution of TF sequence and TF duplications impact gene regulatory networks.
Stern, D. L. 2010. Evolution, Development, and the Predictable Genome. Greenwood, CO: Roberts & Company. A very readable account that discusses the reasons certain genes in networks become hot spots of morphological evolution.