Evolution and Conservation
H. Bradley Shaffer
OUTLINE
1. Evolution, genetics, and conservation
2. Process versus pattern and why both matter
3. The enemies to watch out for
4. What genomics brings to the table
5. Concluding thoughts and prospectus
Traditionally, evolutionary biology has had a distant relationship to conservation compared with ecology and field-based natural history. However, this situation has changed dramatically in the last two decades, particularly as abundant molecular data have become available for at-risk species of conservation concern. As the availability of genome-level data for these species increases, the role of evolutionary biology in conservation management continues to grow to a far greater extent. The combination of these new data from microevolutionary analyses with more traditional input from phylogenetics and systematics has elevated evolutionary biology to a position of primary importance in conservation science.
GLOSSARY
Ecotone. A transition area where two distinct ecological communities meet and integrate.
Endangered Species Act (ESA). The US law that protects critically at-risk species from extinction due to human activities. It was passed into law in 1973 under President Richard Nixon and remains one of the most powerful conservation laws in existence.
Landscape Genetics/Genomics. The fields that integrate population genetics (or genomics) data with features of specific landscapes to study how those features influence the movements of genes and individual. This is a computationally intensive discipline that has become a major part of many conservation programs.
Nongovernmental Organization (NGO). An organization that is independent of any government, and generally has an important advocacy role. Several leading NGOs play a critical role, both locally and globally, in biological conservation.
Phylogenetic Diversity (PD). The amount of character change that evolves along a branch of a phylogenetic tree. PD may evolve along internal or tip branches and may be nonsymmetrical along two branches derived from a common ancestor.
Phylogenetics. The discipline that reconstructs the genealogical relationships of species and lineages.
Systematics. The discipline that names, describes, and infers the evolutionary history of species and lineages.
1. EVOLUTION, GENETICS, AND CONSERVATION
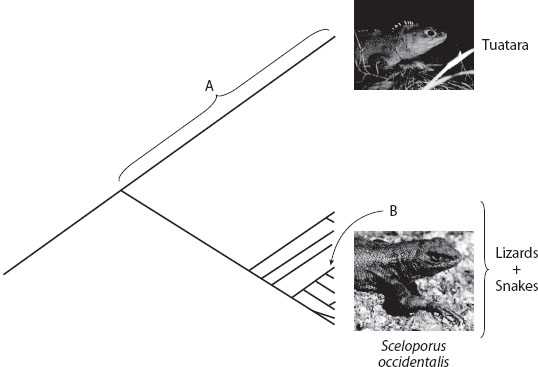
Suppose that you control environmental policy, and you have a choice: you can save either the New Zealand tuatara (Sphenodon punctatus) or the western fence lizard (Sceloporus occidentalis) from extinction. Whichever you choose, the other will go extinct. The tuatara is a lizard-like animal that is the sole surviving member of a once-diverse but now nearly extinct lineage of vertebrate life. Although that lineage was widespread and globally common 200 million years ago, it is currently down to one (or possibly two, virtually identical) species that occupies a handful of islands off the coasts of both main islands of New Zealand. If any lineage deserves the name “living fossil,” it may be the tuatara. The western fence lizard is probably the most common lizard in North America. It is a widespread, abundant, and somewhat-unremarkable member of one of the most diverse and adaptable genera of lizards on earth. As of this writing, 92 species in the fence lizard genus Sceloporus are recognized, and new ones are constantly being described and characterized. So, how do you decide?
This kind of “conservation triage” is one of the arenas where evolutionary biology plays a critical role in conservation decision making. Evolutionary biology cannot tell a manager which species is more important, but it can frame the question and provide quantitative insights that can help guide the decision-making process. In this particular case, virtually all policy-makers would choose the tuatara. The question is, Why? Conserving evolutionary history—that is, long branches of the tree of life that provide a record of the changes that have occurred during the history of life on earth—is a universally recognized component of conservation biology. The logic is that any lineage that took 200 million years to evolve is, in some real sense, more precious than another lineage that has many close relatives with which it shares most aspects of its morphology, ecology, and natural history. The phylogenetic uniqueness of the tuatara, its lack of close relatives, and the incredible length of its branch on the tree of life (figure 1) are all insights that come directly from understanding its evolutionary history and are the primary reasons why it is a global conservation icon. The same is true for many other important conservation targets, including the duck-billed platypus, the remaining rhinoceros species, and the California and Chinese redwoods.
Figure 1. A phylogeny showing the relationships between the tuatara and its closest relatives, the snakes plus lizards. If the tuatara goes extinct, that species plus all the evolutionary history that occurred along the branch leading to it (labeled A in the figure) will be lost forever. If the western fence lizard (Sceloporus occidentalis) goes extinct, only that species plus the unique evolution on the much shorter branch B will be lost.
At least three different components of evolutionary biology speak directly and forcefully to problems in conservation biology. The first is systematics and the related discipline of phylogenetics, and the tuatara is one of the classic examples (see Section II: Phylogenetics and the History of Life for additional detail). Both disciplines now rely heavily on molecular—usually DNA-level—data to make inferences about organisms, and both seek to describe the diversity and interrelationships of life on earth. Given that probably the single most important tenet of conservation biology is that “You cannot protect what you don’t recognize,” and that one goal of systematics is the delimitation of species and lineages, it seems clear that we require a catalog of life on earth before we can realistically plan for protecting it. For example, until fairly recently it was widely considered that the living tuatara consisted of a single species. However, in 1990, Daugherty and colleagues evaluated the variation found among remnant tuatara populations and hypothesized that the animals on Brothers Island actually constituted a different species, for which they used the name Sphenodon guntheri. In so doing, they simultaneously presented the world with one of the rarest species of vertebrates on earth and removed one population from the small catalog of known breeding populations of the critically endangered northern tuatara, S. punctatus. However, more recent work, based on additional data and sampling, reversed that decision, instead concluding that the tuatara “is best described as a single species that contains distinctive and important geographic variants.” By studying one endangered species in ever-greater detail, this research team has continued to refine our understanding of the evolutionary history of tuataras and thus the populations and potential species in need of conservation actions.
A second, related area in which modern evolutionary biology informs conservation and management is phylogeography. Originally introduced by evolutionary geneticist John Avise in 1987, phylogeography uses genetic data to understand lineage formation and evolutionary diversification within, rather than among, species of organisms (see chapter II.5). As the name implies, a key goal of phylogeographic research is determining the relationship between the geographic location of populations and genetic differentiation among those same populations. In many cases, the recognition of deeply separated lineages within species has led to their independent protection and conservation. For example, recent phylogeographic work from our laboratory on the California tiger salamander (Ambystoma californiense) demonstrated that the species consists of at least three genetically independent lineages; two of these are geographic isolates in the south (Santa Barbara County) and the north (Sonoma County), while the third is the larger central group from the Great Central Valley. When the species was protected under the US Endangered Species Act, the combination of different levels and types of threats and the phylogeographic recognition of three lineages led to the independent protection of salamanders from Santa Barbara and Sonoma counties as endangered, while the rest of the species’ range was separately listed as threatened under the ESA. These different listing levels (threatened versus endangered) actually do matter and could not have been proposed or implemented without this phylogeographic research.
Finally, population genetics has traditionally been the cornerstone of evolutionary biology’s contribution to conservation, and this tradition has grown in the last few years. Three decades ago Frankel and Soulé (1981) emphasized the close connections between population genetics and conservation in conceptual areas ranging from minimum viable population sizes to the relationship between inbreeding depression and genetic drift. Frankel and Soulé’s book, the first to use the words “conservation” and “evolution” in a single title, was also among the first to explicitly point out the expected relationship between small effective population size (Ne) and population health that is predicted from population genetics theory. Because inbreeding is generally detrimental to most outbreeding populations (see chapter IV.6), Frankel and Soulé argued that small populations would be particularly vulnerable to inbreeding depression and coined their “Basic rule of conservation genetics,” which relates the change in inbreeding coefficient, ΔF, to the likelihood that a population will survive into future generations. In particular, they suggested that ΔF greater than about 1 percent constitutes “a threshold rate of inbreeding, above which fitness relentlessly declines” and populations go extinct. A related concept is Frankel’s 50/500 rule, which states that on average, populations with a persistent Ne less than 50 may be in immediate danger of extinction, whereas over long time periods, populations with Ne less than 500 may not contain enough genetic variation to adapt to changing conditions (Braude and Low 2010). Although controversial, these “rules” emphasize a key point—when populations become too small and isolated, genetic drift can overcome natural selection, and low-fitness genotypes can rise in frequency by chance alone (see chapter IV.1). If this happens too often, or for too long, extinction may follow.
2. PROCESS VERSUS PATTERN AND WHY BOTH MATTER
A key question in conservation biology is deciding what to conserve and why. The resolution of this question depends on many factors. The country where the action is taking place may have strong conservation laws like the US ESA, or it may have virtually no history or capacity for even the weakest protection of taxa or landscapes. Nongovernmental organizations (NGOs) may be prominent partners that have their own opinions and agendas, local jurisdictions may interact in a positive or negative way with national governments and NGOs, and international organizations like the United Nations or World Bank may enter into the conversation.
Regardless of the organization, its politics, or its agenda, anyone who considers human-mediated extinction to be an outcome that should be avoided is really trying to conserve an aspect of evolutionary biology. At the broadest level, one can think about this problem in two ways. First, one can focus on conserving evolutionary history that has already occurred. Alternatively, or in addition, one can attempt to conserve the potential for future evolutionary change. Interestingly, these two approaches sometimes lead to very similar actions, and sometimes to radically different conservation priorities.
Conserving Evolutionary History
The US ESA is one of the most powerful pieces of conservation law in the world. Essentially, it simply states that species (including subspecies) should not be allowed to go extinct and that actions that lead to the further decline of listed species require special permission from the federal government. For evolutionary biologists, this means that the ESA seeks to protect one of the key products of the evolutionary process—species, including incipient species and subspecies. This theme of conserving the outcome of the evolutionary process is at the core of most conservation efforts. It is a very retrospective view of what to conserve—it requires that evolutionary biologists provide a clear picture of how many species, subspecies, and distinct population segments exist in a region, an indication of whether those populations are increasing or decreasing, and a measure of how distinct they are from one another. It is then up to the conservation community to take those results and use them to prioritize species and landscapes and try to preserve as much evolutionary history as possible.
Several specific approaches to conserving evolutionary history above and beyond the basic tenet that species extinction should be avoided are worth discussing in a bit more detail. First is the issue of retaining as much of a phylogenetic tree as possible—the tuatara problem that opened this essay. Two basic approaches have dominated the thinking on this topic. First, one can attempt to conserve the phylogenetic branch length (i.e., the sum of the branches of a phylogeny)—in units of time—that might be lost if an extinction event occurs. In the case of the tuatara, different opinions exist as to when it last shared a common ancestor with its closest relatives, but 271.5 million years seems to be a reasonable estimate. That is, if you lose the tuatara, you lose not only that species but also the 271.5 million years of evolutionary history that it uniquely represents among living organisms.
An alternative approach proposed by Faith (1992) is to conserve phylogenetic diversity, or PD. Faith proposed PD as an explicitly character-based approach to identifying taxa to conserve—those that have evolved lots of unique features (characters) contain more important evolutionary history than those that have changed relatively little and therefore remain relatively similar to other taxa. An example might be the human-chimp-gorilla trio of species. Although they all shared a common ancestor about 6 to 8 million years ago, the human lineage has changed considerably more, in a wide variety of biologically important ways, than has the chimp or gorilla from its most recent common ancestor. Thus, in this case, humans would have a far greater PD than the chimpanzee and would be a higher conservation priority if the two species were equally threatened. To the extent that unique features accumulate over time, branch lengths and PD will prioritize species for conservation in the same order. However, evolution does not always proceed in a tidy, time-dependent manner, as the human example points out. In those cases, one must decide what matters most—time or evolutionary novelties—as a target for conservation.
A very different approach is to recognize that the primary reason for human-mediated extinction is habitat alteration and destruction and that certain landscapes or regions tend to accumulate a great number of unique organisms. Certain regions of the world, like New Zealand or the Appalachian Mountains of eastern North America, are rich in endemic taxa found nowhere else on earth. The same can be said, of course, for many parts of the world—the Amazon basin is also very species rich, and most of the species found there are restricted to the Amazon. However, regions like New Zealand contain a disproportionately large number of old, unique lineages, like tuatara, flightless kiwis, and southern beech forests, while the Amazon abounds in species that are often members of widespread tropical genera. Many factors can contribute to these patterns, including the geological age and stability of a landscape, its isolation from other parts of the world, and the extent to which humans have disrupted ecological processes that naturally occur in the area. As phylogenies, and particularly as time-calibrated evolutionary trees, accumulate for the world’s fauna and flora, conservation biologists can prioritize those regions that harbor the greatest depth and breadth of the tree of life and try to protect them into the future. In so doing they are saving species, but they are also saving the longest branches in the tree of life.
Conserving the Potential for Future Evolutionary Change
In 1997 Tom Smith and colleagues promoted a very different approach to using insights from evolutionary biology to conserve biodiversity. Smith argued, based on an analysis of a dozen populations of an African bird species, that there is a tremendous level of morphological differentiation between birds in the forest and the same species in the ecotone between forest and savanna habitats, and that this variation persists in the face of ongoing movement of individuals and gene flow. While the argument is fascinating in its own right, Smith and colleagues took it one step further, arguing that the ecotone habitat selects for morphologically very different birds from those in the rain forest. Their conclusion was quite radical: if you want to preserve evolutionary processes that generate diversity, you should preserve ecotones in addition to pure rain forest.
The importance of preserving habitats critical for the functioning of normal evolutionary processes within species has gained considerable traction in the last decade. The entire discipline of landscape genetics, including the emerging subdiscipline of landscape genomics, focuses on exactly this issue, and it represents one of the major growth areas in research on the genetics of natural populations. Here, the goal is to use standard population genetics data, in combination with geographic information system (GIS) data layers, to quantify the ways that organisms move across the landscapes they occupy. The approach is directly relevant to landscape management and conservation planning because it takes genetic data from organisms on landscapes and asks whether potential migration corridors and barriers to gene flow function to promote or to disrupt population connectivity. The results can provide unexpected insights into the ways organisms use their environment and can identify those habitat patches and corridors that are most important for maintaining normal evolutionary processes. Such data can take years to collect with traditional mark-recapture methods, but only weeks or months using the insights gained from landscape genetics. Particularly for threatened species, for which decision makers must act quickly, landscape genetics is a powerful conservation tool.
Finally, a very different kind of evolutionary process with enormous conservation consequences has recently been recognized. Given the impact of people on natural landscapes, human activities have the potential to exert strong selection pressures on populations in the wild, and recent studies have demonstrated that organisms are responding to this selection, sometimes in surprising ways. Although the study of rapid evolutionary change due to human activities is still in its infancy, this phenomenon has the potential to profoundly affect conservation outcomes. To take one example, human fisheries generally remove the largest individual fish, which are often both old and female (most fish species have indeterminate growth and continue growing throughout their life). This intense selection on the largest, most fecund females leads to an evolutionary decrease in body size, an earlier age at first reproduction (since having a few babies at a small body size is better than waiting and being caught by a fisherman), and a lower reproductive output for individuals and the species overall. Population models show that this shift from a life history in which individuals wait many years to reproduce, grow to a large size, and have many young, to the alternative strategy of early reproduction at a small size with few offspring leads to lower total biomass, decreased population sustainability, and a greater likelihood for population collapse or extinction. Recent models on the effects of Marine Protected Areas (MPAs) in preventing these conservation disasters have found that some MPAs ameliorate such evolutionary responses to human fishing, but others do not. For example, if there is extensive gene flow between fishing grounds and MPAs, then smaller females will migrate to, and breed in the MPA, and larger, protected females will migrate outside MPAs, where they will be caught and killed; in this case, little is gained from the MPA. Alternatively, if the MPA is very large, or if the fish tend not to migrate extensively, then the larger, protected females will remain in the MPA and provide a constant source of young, genetically unmodified individuals to the outside fishing grounds.
Regardless of how individual case studies play out, humans now constitute a potent force leading to rapid evolutionary change. We clearly need to add ourselves to the list of important processes at the intersection of evolution and conservation.
3. THE ENEMIES TO WATCH OUT FOR
Conservation biology is a complex business that involves equal parts of biology, politics, and economics if real progress is to be made and sustained. An essential element of conservation is to think clearly about what one wants to protect and what one wants to avoid in terms of conservation outcomes. Evolutionary genetics in particular has made very substantial contributions to the identification of these problems and their solutions. To take one of the highest-profile case studies yet conducted, consider the Florida panther, Puma concolor coryi. Designated the state animal of Florida in 1982, this endemic subspecies of panther (also known as puma, mountain lion, or cougar in other parts of the species’ vast range) was reduced to about 20 individuals in the 1970s, at which time it was showing clear signs of genetic inbreeding depression. As a result, the population was intentionally supplemented with panthers from the adjacent, much larger, and more outbred population in east Texas from the subspecies Puma concolor stanleyana. Hybridization occurred, the telltale signs of inbreeding depression disappeared, and Florida’s state animal appears to be in a strong phase of population growth and genetic recovery. Except—is it really recovering? There are definitely healthy panthers back in the Florida Everglades, but are they Puma concolor coryi? Or has that subspecies been driven extinct by an invasive hybrid panther? The example raises the critical question, What does one want to conserve, and why? Is one protecting native genes, naturally evolved lineages, or ecological roles? Is it better to keep some native Florida panther genes on the Florida landscape than none at all (the hybrid panthers definitely have a lot of native Florida genes), or is hybrid “impurity” worse than extinction? Evolutionary genetics can provide the data, but not the answers, to the moral dilemmas that these questions pose.
Hybridization
Hybridization happens all the time, both because of natural processes and because humans meddle with species and landscapes (see chapter IV.3). Evolutionary genetics can bring great clarity to the status of populations of plants and animals, including precise estimates of the fraction of the genome that is native versus derived from a different species. Most practitioners naturally assume that the primary goal of conservation biology is to preserve pure genetic lineages on the landscapes where they evolved. Thus, in a case that our lab has worked on for several years, human-transported, nonnative Barred tiger salamanders (Ambystoma tigrinum mavortium) from Texas and New Mexico have successfully hybridized with native, endangered California tiger salamanders (A. californiense) across much of central California, and at least 20 percent of the range of the California tiger salamander is now occupied by hybrids (Fitzpatrick et al. 2010). In this and many other cases, hybrids are viewed as a conservation threat, to be identified, eliminated, and replaced with pure natives if at all possible. Trout, salmon, escaped genes from agricultural plants, and domestic dog genes infiltrating wolf and coyote populations are a few of the better-studied examples of this phenomenon, and the evolutionary analysis of hybridization constitutes the key data on which conservation actions have been based. On the other side of the issue, “genetic rescue,” particularly for large mammal populations that have dipped below a genetically sustainable size, remains a viable and occasionally used strategy to augment populations that would otherwise go extinct without genetic intervention—the Florida panther is a classic example. Importantly, these are cases in which evolutionary biology can provide the key insights on expected and realized inbreeding depression, can track the fate of nonnative genes as they move through populations, and can measure the fitness consequences of hybridization. What it cannot do is tell us, as managers of the fate of populations, whether and when we should bring in foreign genes as a last-ditch conservation effort.
Population Bottlenecks, Population Isolation, and Effective Population Size
More books and papers discuss the interface of population genetics and conservation biology than any other aspect of evolutionary conservation biology. There are many reasons for this, but the primary one is that the connection between classical problems in population genetics and conservation biology is both direct and clear. Population geneticists tend to worry about the relationships between genetic drift caused by small effective population sizes, the efficacy of natural selection in shaping variation in the field, and the interplay among mutation, selection, and drift. Conservation biologists spend a great deal of time and energy trying to understand the health of populations in nature, including the effects of small population sizes. Both groups recognize that small populations have a higher chance of becoming inbred and that high levels of standing genetic variation are critical for the current and future health of populations and species. Any good field ecologist knows that populations fluctuate over time and that low numbers are sometimes unavoidable. However, when populations become completely isolated, then migrants from larger populations cannot help those reduced populations recover, either demographically or genetically. The result is small, isolated populations that lose genetic variation over time, become inbred, express deleterious mutations at a higher frequency, and have limited resilience to bounce back from unavoidable population crashes. And given their isolation, when they go locally extinct, they cannot be repopulated—they stay extinct.
One of the holy grails of both population genetics and conservation biology has been to infer past and current demographic parameters using the standing genetic variation that exists in natural populations. At least for genetic markers that are unaffected by strong natural selection (so-called neutral genetic variation), there is a long, rich history of using patterns of variation within and among populations to estimate the amount of migration (or gene flow) among populations. Here, the idea is straightforward—if a mutation arises in one population, and no migrants successfully leave that population and reproduce in a new population, then the mutation will remain exclusively in its site of origin. Such “private” variants will build up over time, such that the longer a population remains in isolation, the greater will be its genetic distinctiveness from other populations. Sewall Wright, one of the pioneers in the field of population genetics, developed a series of statistical methods to quantify this kind of genetic differentiation, and his F-statistics remain the primary way in which such realized gene flow is measured in nature. High values imply little or no gene flow among populations, whereas low values suggest that successful migration and breeding occur regularly. Newer methods can measure the movement of individuals by recognizing that if an occasional migrant moves between somewhat-differentiated populations, that individual can be “assigned” to its population of origin based on its multigene identity. Conducting such assignment tests with confidence requires a great deal of genetic (or genomic) data, but it represents a powerful addition to the conservation biologist’s toolkit for measuring how organisms successfully traverse landscapes in nature.
Some of the most compelling and exciting new developments in population genetics allow conservationists to study, with far greater precision, the actual size of a breeding population in nature. Population biologists recognize two different ways to measure population size—Ne, or the effective population size, and Nc, the census population size. The difference is straightforward and absolutely critical: Nc is the number of individuals in a population, whereas Ne reflects the number of individuals who breed and contribute to the genetic variation in the species (note that this represents a simplification of a mathematically complicated concept). For example, if a population has 50 males and 50 females, its census size will be 100. However, if only one of those males and 10 of those females actually breed, its effective size will be close to 10; this latter population will suffer much greater genetic drift and potential inbreeding depression than one in which all 100 individuals breed. Both Nc, and Ne are important, and they measure different aspects of the health of a population. Recent advances in molecular population genetics have provided new tools to measure Ne in nature, sometimes from only a single individual. The math is complex and the requirements for both the number of genetic markers and the proportion of the population sampled may be large, but the results indicate that the effective population size can be estimated, often relatively easily and quickly.
4. WHAT GENOMICS BRINGS TO THE TABLE
Genomics means many things to many people, ranging from data on the full DNA sequence of an organism assembled into complete chromosomes to having “a lot of sequence data.” A truly complete genome, in which every base pair has been sequenced and assembled into contiguous chromosomes, has yet to be completed for any vertebrate, although several species, including humans, have essentially complete genomes. However, an increasingly large number of species have had many thousands of genes sequenced, sometimes for multiple individuals and populations. In either case, genomics always means having lots of data for each study organism—it may mean billions of nucleotides of sequence data (many vertebrate genomes are around 2–3 billion nucleotides in length), or it may mean thousands, but it is always a lot.
It seems clear that in the next few years, genomic data will dominate population genetics, phylogenetics, and conservation genetics research. Aside from the general truism that more data are always better than fewer data, this onslaught of new information should open several critical avenues of research at the interface of evolutionary and conservation biology (see chapters in Section V: Genes, Genomes, Phenotypes). First, genomic data allow one to study the genetics of functionally important genes as well as neutral ones not affected by natural selection. Presumably, conservation geneticists should focus on genetic variation in the functionally important genes, since they are most important to survival and the ability to adapt to future change. For neutral loci, genetic variation per se is not important to population health, but the standing levels of variation at those loci reflect the past and current effective population size and levels of gene flow or genetic isolation. Both are important to evolutionists and conservationists, but the two are very different. When a large part of the genome is subject to study, the neutral and selected loci can be neatly separated, leading to important insights from both genomic components.
In a similar vein, genomic data help the evolution and conservation community focus much more clearly on exactly what needs to be conserved. For example, the major histocompatibility complex (MHC) is a set of genes involved in the immune response to disease of many vertebrates. Certain diseases, including the fibropapilloma tumors in green sea turtles or the devil facial tumor disease in Tasmanian devils of Australia, may be involved in bringing these endangered taxa to the brink of extinction, and conserving and managing populations for MHC variation may be a way to increase their chances of survival. Genes that allow cold-adapted plants and animals to better cope with human-induced climate change in the next decade are another key class of functional genes that genomics may bring to the conservation table.
The impact of genomic data on evolutionary biology in general, and conservation in particular, is huge, multifaceted, and largely unexplored. It stands as perhaps the most important frontier at the intersection of evolution and conservation biology.
5. CONCLUDING THOUGHTS AND PROSPECTUS
Evolution is all about change—changes in allele frequencies over time, in population size and distributions, and in species composition due to extinction and speciation. Conservation is about managing for change—climate change, invasive species, hybridization, human habitat modifications, and a host of others. As large-scale genetic analyses become increasing available for nonmodel organisms, it seems inevitable that evolutionary genetic analyses will move to center stage in the conservation and management of declining species. Consider, for example, being able to track the reproductive output of captive-reared organisms that are repatriated into the wild, allowing resource managers to measure the impact of their conservation efforts, in the wild, in real time. Or imagine having the data to be able to determine, with very high accuracy, exactly how many individuals have moved between habitat patches historically, and using that information to mimic those patterns with human-assisted migration in fragmented habitats. Or being able to quantify, for any newly proposed protected park, exactly how much of the phylogenetic tree of life is contained in that park—not for specific taxa based on a few genes, but for all life. These are heady ideas, but as genomics, metagenomics, and phylogenomics become affordable and easier to accomplish, they are also very realistic. And they just might help conserve a bit more of our declining biosphere.
FURTHER READING
Allendorf, F. W., and G. Luikart. 2007. Conservation and the genetics of populations. Malden, MA: Blackwell. A wonderful reference, particularly for the more mathematical aspects of conservation genetics. This book is particularly strong on the interface of population genetics and conservation.
Braude, S., and B. S. Low, eds. 2010. An Introduction to Methods and Models in Ecology, Evolution, and Conservation Biology. Princeton, NJ: Princeton University Press.
DeWoody, J. A., J. W. Bickham, C. H. Michler, K. M. Nichols, O. E. Rhodes Jr., and K. E. Woeste, eds. 2010. Molecular Approaches in Natural Resource Conservation and Management. New York: Cambridge University Press. An edited volume, with some very specific chapters that may be of limited general interest, but others of quite broad appeal. A great source of recent examples and case studies using a wide range of molecular genetic tools to inform conservation.
Faith, D. P. 1992. Conservation evaluation and phylogenetic diversity. Biological Conservation 61: 1010. The paper that introduced the phylogeny-based concept of conserving character evolution into the mainstream of conservation thinking.
Fitzpatrick, B. M., J. R. Johnson, D. K. Kump, J. J. Smith, S. R. Voss, and H. B. Shaffer. 2010. Rapid spread of invasive genes into a threatened native species. Proceedings of the National Academy of Sciences USA 107: 3606–3610. Following a well-documented, human-mediated introduction, this paper shows that some invasive genes can sweep across landscapes at incredibly rates, while other genes are much slower.
Frankel, O. H., and M. E. Soulé. 1981. Conservation and Evolution. Cambridge: Cambridge University Press. The original book that brought together the fields of conservation biology and evolutionary genetics—a “must-read.”
Frankham, R., J. D. Ballou, and D. A. Briscoe. 2004. A Primer of Conservation Genetics. Cambridge: Cambridge University Press.
Höglund, J. 2009. Evolutionary Conservation Genetics. Oxford: Oxford University Press.
Schonewald, C. M., S. M. Chambers, B. MacBryde, and W. L. Thomas, eds. 2003. Genetics and Conservation. Caldwell, NJ: Blackburn.