Chapter 50
Physiology of Prokaryotic Cells
Chapter Outline
I. The Diversity of Prokaryotic Organisms
III. Energetics of Bacterial Cells
VI. Responding to the Environment
VIC. Phosphate Metabolism and “Two-Component” Regulatory Systems
VII. The Physiology of Pathogenesis
I The Diversity of Prokaryotic Organisms
Prokaryotic cells represent the smallest and simplest form of life that can metabolize, grow and reproduce. They are presumed to resemble the earliest forms of life and they reproduce much more quickly than multicellular organisms do. Taken together, these two properties imply that prokaryotic organisms have had more opportunity to evolve (by orders of magnitude) than plants or animals have had; accordingly, this predicts that prokaryotes should have the most functionally efficient, diverse and specialized of cells, despite their structural simplicity. The key to this paradox is the recognition that genetic variation and natural selection should allow a unicellular organism to improve its performance and acquire new functions without becoming structurally complex. Furthermore, as environments, survival strategies and ecological niches change over time, the criteria of optimal cellular function change which, in turn, sets the stage for new rounds of optimization in various directions. The resulting diversification and specialization can also be expected to make certain features superfluous in certain lineages, leading to cells that may be even simpler than their predecessor.
Functional specialization and optimization of a structurally simple cell seems to account for the observed diversity of modern prokaryotes. Molecular measures of divergence, such as small-subunit ribosomal RNA sequence, indicate that two prokaryotic lineages separated very early and that each encompasses more molecular diversity than multicellular organisms (Fig. 50.1). The two groups distinguished by this early split, Bacteria and Archaea, each have phylogenetic status equivalent to that of all eukaryotic organisms and the three resulting taxonomic units have been termed Domains (Woese et al., 1990). This extensive divergence is also evident in terms of cellular function. Certain bacteria and archaea have metabolic properties not represented among eukaryotes, including N2 fixation, anoxic photosynthesis, additional routes of CO2 assimilation and adaptation to extreme environmental conditions. Similarly, archaea (singular: archaeon, or less commonly, archaeum) have cellular features and metabolic pathways not found in bacteria. These uniquely archaeal features include isoprenoid membrane lipids (found in all archaea) and the ability to make methane (found in a number of genera).
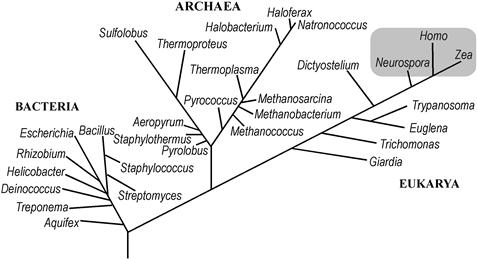
FIGURE 50.1 Phylogenetic relatedness of cellular organisms. Major groups of cellular organisms are indicated by genus names. The lengths of branches connecting two genera indicate the molecular divergence between them, as defined by the sequences of small-subunit ribosomal RNAs (16S rRNAs of bacteria and archaea, 18S rRNAs of eukarya [eukaryotes]). The gray box approximates the molecular diversity of multicellular organisms as measured by small-subunit rRNA.
One practical consequence of the deep diversity of the bacterial and archaeal lineages is that it precludes any one organism, such as the bacterium Escherichia coli, from modeling all aspects of prokaryotic physiology, even though this and several other species can be analyzed in great detail. It should also be noted that some components of eukaryotic cells have bacterial origins. In particular, at least two eukaryotic organelles, the mitochondrion and the chloroplast, resulted from endosymbiotic acquisitions of bacteria by progenitors of modern eukaryotic cells (Scwartz and Dayhoff, 1978). This relationship provides a context for understanding molecular structure and function of both the eukaryotic organelle and the bacterial cell.
II Prokaryotic Cytology
IIA Major Structural Types
Cells of bacteria and archaea generally measure about 1 μm in diameter and thus have about 0.1% or less of the volume and mass of a typical eukaryotic cell. This extremely small size limits the ability of light microscopy to visualize the structural features of these cells (Fig. 50.2). Prokaryotic cells generally have no obvious cytoskeleton, mitotic apparatus or intracellular organelles and span a limited range of structural complexity.
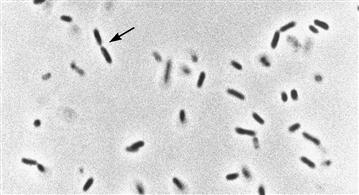
FIGURE 50.2 Live bacteria under the microscope. This is a light micrograph (phase contrast) of Escherichia coli cells suspended in growth medium. The average width of the cells is about 0.5 μm; arrowheads show site of constriction (septation) in a cell undergoing division. The micrograph illustrates the limited structural information obtainable by optical methods due to the small size of prokaryotic cells.
This range is illustrated in Fig. 50.3 as four structural types. Cells of the simplest structural type (Fig. 50.3, panel I) have only a nucleoid, a cytoplasm and a cytoplasmic membrane, each of which is described in more detail below. Examples of this truly minimal cell are relatively rare in nature and include members of the genera Mycoplasma and Thermoplasma. The former are bacteria, some of which are opportunistic pathogens, while the latter are archaea found in heated acidic soils (Kletzin, 2007). Both genera are osmotically fragile, due to an absence of cell walls (see below).
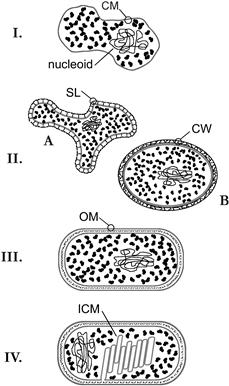
FIGURE 50.3 Examples of prokaryotic cellular structure. The limited range of cellular complexity found among prokaryotic cells has been represented in this diagram by four structural types. I: The minimal cell includes only a nucleoid, cytoplasm and cytyoplasmic membrane (CM). II. Most archaea (A) have an additional S-layer (SL), composed of glycoprotein subunits, providing external structural support for the cell membrane, whereas Gram-positive bacteria (B) have a thick cell wall (CW) composed of peptidoglycan. III. Gram-negative bacteria have a thin cell wall sandwiched between the CM and a second diffusional barrier, the outer membrane (OM). IV. Gram-negative photosynthetic bacteria produce a system of intracytoplasmic membranes (ICM). These contain specialized light-harvesting pigments and may be topologically continuous with each other and the CM.
IIB Nucleoid
Most prokaryotes have a single chromosome consisting of a single circular DNA of several million base-pairs which is neither confined within a nucleus nor evenly dispersed throughout the cytoplasm. The DNA instead occupies a convoluted region near the center of the cell, called the nuclear region or nucleoid, which bears a functional analogy to the eukaryotic nucleus (see Fig. 50.3, panel I). The nucleoid replicates and partitions itself into daughter cells during growth and cell division and maintains its compacted form despite ongoing replication, transcription and repair. Cations, including Mg2+, spermidine and other polyamines and small “histone-like” proteins contribute to this condensation. In E. coli, longer-range organization occurs in the form of 50 to 100 topologically constrained domains or “loops”. Lysis of cells under appropriate conditions (non-ionic detergent and high salt concentration) releases nucleoids in a compact, relatively intact form and allows the loops to be visualized by electron microscopy (EM) (Pettijohn, 1996).
IIC Cytoplasm
All other components of the cell interior are collectively termed the cytoplasm or cytosol. Although fluid, this mixture is very concentrated and probably reflects a complex and dynamic series of macromolecular associations. The cytosol of an E. coli cell adapted to growth in simple glucose medium at 37°C contains about 19 000 ribosomes which, together with transfer RNA and various translation factors, account for nearly half of the total cell mass. This large investment of cellular resources in the machinery of protein synthesis enables the E. coli cell to reproduce itself every 40 min under these conditions (Neidhardt et al., 1990). Enriching the simple glucose medium with other nutrients (such as peptides) increases the ribosome content and growth rate even more.
The lack of internal compartmentalization in most prokaryotic cells contributes to their ability to grow quickly and to respond quickly to environmental change. In general, prokaryotic cells have no diffusional barriers segregating the sources of energy, raw material or sequence information needed for DNA, RNA and protein synthesis; this lack of compartmentalization and the small dimensions of the cell ensure that molecular diffusion is extremely rapid throughout the cell interior. Transcription and translation are temporally and spatially coupled, so that ribosomes begin “reading” the mRNA before the mRNA itself has been completed and released from the transcription complex. This coupling not only supports rapid growth, but also provides uniquely prokaryotic modes of genetic regulation, as demonstrated by the phenomenon called attenuation. In this regulatory mechanism, adequate levels of particular amino acids lead to disruption of the normally tight coupling between ribosome and RNA polymerase as it begins to transcribe the corresponding amino acid biosynthetic genes. This transient uncoupling, in turn, causes RNA polymerase to terminate transcription before it reaches the biosynthetic genes, thereby avoiding wasteful overproduction of the corresponding enzymes (Yanofsky and Crawford, 1987).
IID Cytoplasmic Membrane
The third component of the minimal prokaryotic cell structure (see Fig. 50.3 panel I) is a unit membrane called the cytoplasmic (cell) membrane (CM) which chemically separates the cytosol and nucleoid from the external environment. A bacterial CM generally conforms to the fluid mosaic model of biological membranes (see Chapter 3). Its matrix is a phospholipid bilayer impermeable to ionic or large polar molecules but intrinsically permeable to gases and water. The selective impermeability is essential for viability, because without it the cell cannot maintain ion potentials or retain metabolites (see below). However, cellular function also requires some chemical exchange with the surroundings, including nutrient uptake and environmental sensing. These functions are provided by a large number of different integral and peripheral proteins which breach the phospholipid bilayer with solute-specific pumps and gates. By means of these proteins, which comprise about 70% of the CM by weight, bacteria maintain transmembrane gradients and controls transmembrane fluxes of critical solutes. Archaeal membranes appear to have analogous structure and function, although the polar lipids that provide the lamellar barrier differ radically from the phospholipids of other cells. Archaea contain only sn-(2,3 di-O-alkyl)-glycerol membrane lipids, in which the hydrocarbon chains are C20 or C40 saturated isoprenoids attached to the glycerol backbone via ether linkages (Boucher, 2007).
IIE Cell Wall
Most archaea and bacteria have some form of cell wall immediately outside the cytoplasmic membrane (see Fig. 50.3, IIA, IIB). This structure solves a specific threat to the cell created by hypotonic environments, namely, that the CM cannot block the diffusion of water into the cell interior. Hypotonic environments thus promote cell swelling and rupture, i.e. lysis, of cells. Bacterial cell walls prevent lysis by enclosing the cell in a container that, like the cellulosic cell walls of plants, resists expansion. In contrast to plant cell walls, however, the bacterial cell wall is composed of a covalently cross-linked polymer, called peptidoglycan or murein. The repeating unit (two amino sugars linked via β-1,4 glycosidic bonds) is linked to form linear glycan chains, which are joined at frequent intervals by peptide bridges (Fig. 50.4). This results in one huge, bag-like macromolecule of high tensile strength that completely surrounds the cell membrane. This structural support allows the bacterial cell to maintain an osmotic (turgor) pressure (see Chapter 16), which can be as high as 20 atm. A morphological consequence of constitutive turgor pressure is that the bacterial cell wall determines cell shape and therefore plays a major role in cell division. This can be demonstrated in the effects on cell morphology of antibiotics that specifically disrupt the normal synthesis of peptidoglycan. In rod-shaped bacteria, low concentrations of β-lactam antibiotics, which block cell division but not cell growth, lead to elongated cells of normal diameter. At higher antibiotic concentrations, bulges (rather than constrictions) form at the normal site of bacterial cell division and, at very high antibiotic concentrations, the rod-like cells swell into spheres and lyse (Schwarz et al., 1969).
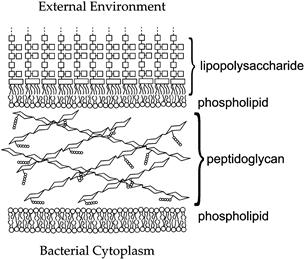
FIGURE 50.4 The cell envelope of Gram-negative bacteria. The bacterial cell wall (CW) is composed of peptidoglycan, whose repeating unit consists of a dimer of two modified amino sugars: N-acetyl muramic acid and N-acetyl glucosamine. Each repeating unit has a short peptide (circles) attached to the N-acetyl muramic acid; the peptides of adjacent glycan chains become joined by enzymes in the periplasm to form the periodic cross-links found in the mature peptidoglycan polymer. The outer membrane (OM) consists of one leaflet of lipopolysaccharide (LPS) and one of phospholipid. Each LPS molecule has a large polysaccharide chain (linked squares) whose precise structure depends on the bacterial species. The cytoplasmic membrane (CM) is composed of phospholipid. In addition to lipid, both the OM and CM contain protein species, which are not depicted in the figure.
Some archaea synthesize a polymeric cell wall material similar to peptidoglycan, but most have only a layer of protein or glycoprotein that provides mechanical support for the CM. These archaeal surface- (S-) layers (Fig. 50.5) can be very strong structurally, despite the fact that they form by non-covalent association of individual subunits. For example, an experimental procedure used to purify bacterial cell walls as intact sacculi involves hot sodium dodecyl sulfate (SDS) extraction. Application of this method to Sulfolobus cells under mildly acidic conditions yields virtually intact S-layer sacculi. The non-covalent nature of the glycoprotein subunit interactions is seen, however, in the ease with which these structures dissociate into monomers at elevated pH (Grogan, 1996). Some archaeal S-layer cell walls appear to be relatively rigid and retain the cell shape, whereas others are flexible and thus not strictly equivalent to bacterial cell walls in all respects.
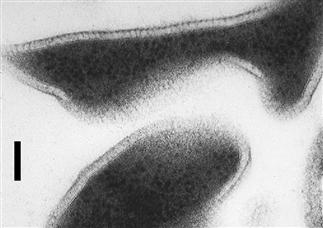
FIGURE 50.5 Cells of Sulfolobus acidocaldarius, an archaeon from geothermal environments (Section VII). The ultrathin sections illustrate the irregular cell shape and extracellular location of the glycoprotein S-layer. The S-layer is held at a fixed distance from the cytoplasmic membrane by a spacer (Jaenicke and Böhm, 1998); this results in a “picket fence” appearance for the boundary around cells or cell ghosts at certain points. Bar = 100 nm.
IIF Outer Membrane
A third, more complex cellular architecture seen in bacteria includes a second lipid bilayer, called the outer membrane, which surrounds the cell wall (see Fig. 50.3 panel III). This creates a protected compartment around the cell, bounded by the cytoplasmic and outer membranes, called the periplasm (Fig. 50.6). An important bacterial identification technique, the Gram stain, exploits the property of the peptidoglycan cell wall in these bacteria that it is relatively thin and does not retain a complex of crystal violet stain and iodine when rinsed with ethanol or acetone. This staining procedure therefore distinguishes bacteria which have the cell architecture shown in Fig. 50.3 panel IIB (Gram-positive), from those which have the more complex cell envelope illustrated in Fig. 50.3 panel III (Gram-negative). Table 50.1 summarizes the major differences between Gram-positive and Gram-negative bacteria.
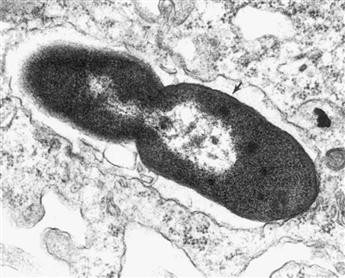
FIGURE 50.6 Electron micrograph (ultrathin section) of a Gram-negative bacterial cell. An E. coli cell is shown that has been engulfed by a macrophage and is in the late stages of cell division. The nucleoid is visible as a clear zone in each of the nascent daughter cells. The arrowhead indicates a region in which the three-layer structure of the Gram-negative cell envelope (CM-CW-OM) can be seen. (Photograph courtesy of A. Mukkada).
TABLE 50.1. Cellular Properties of Gram-Positive and Gram-Negative Bacteria
| Property | Gram-Positive Bacteria | Gram-Negative Bacteria |
| Cell wall | ||
| Chemical constituents | Peptidoglycan | Peptidoglycan |
| Teichoic, lipoteichoic and teichuronic acids | ||
| Typical thickness | 20–80 nm | 2–3 nm |
| Outer membrane | (Absent) | Composed of LPS and phospholipid |
| Intrinsic resistance to detergents, dyes, and certain antibiotics | Low | High |
| Dessication resistance | High | Low |
| Examples | Clostridium | Escherichia |
| Lactobacillus | Thiobacillus | |
The biochemical composition of the outer membrane (OM) differs from that of the bacterial CM with respect to both lipid and protein constituents. With respect to lipid, the OM is a hybrid membrane. Its inner leaflet, which faces the periplasm, incorporates the same phospholipids as the CM does. The outer leaflet, however, consists of much higher molecular weight class of lipid known as lipopolysaccharide, or LPS (see Fig. 50.4). With respect to protein, the OM has only a few protein species, but these are abundant (Nikaido and Vaara, 1985), which contrasts with the broad heterogeneity of CM proteins.
These two compositional features support the primary physiological function of the OM, which is shielding the Gram-negative cell from toxic, hydrophobic compounds in the environment. An intact LPS leaflet greatly impedes diffusion of soluble hydrophobic (i.e. lipophilic) molecules across the OM. This presumably reflects the permeability properties of the dense array of cooperatively interacting charged and polar sugar residues at the membrane surface, through which non-polar solutes must pass (Fig. 50.4). Experimental evidence of the importance of this screening function can be seen in the sensitivity of Gram-positive bacteria and LPS-depleted Gram-negative cells to toxic dyes, detergents and hydrophobic antibiotics. As in the case of the CM, the OM must nevertheless allow passage of nutrients, which is mediated by relatively non-specific solute channels, or pores. These structures form as trimers of corresponding proteins, called porins. In E. coli, OM pores exclude solutes with molecular weights of about 700 Da and above and tend further to discriminate against anionic solutes (Nikaido and Vaara, 1985).
IIG Intracellular Structures
Still greater complexity can be seen in a few bacterial cells which incorporate additional intracellular structures into the basic bacterial cell architecture described above. Examples of these specialized structures include photosynthetic membranes, internal cytoplasmic compartments and gas vesicles.
Although different groups of photosynthetic bacteria have biochemically diverse forms of photosynthesis, most involve specialized photosynthetic membranes located in the cytoplasm (see Fig. 50.3 panel IV). In the case of cyanobacteria, the intracellular photosynthetic membranes appear to be collapsed and lamellar in form and are called thylakoids, in conformity with the corresponding membranes of plant chloroplasts (see Chapter 51). For other types of photosynthetic bacteria, the membranes assume more rounded, vesicular shapes; these intracytoplasmic membranes (ICM) appear to be continuous with the CM (Dierstein et al., 1981).
Other membranes have been observed inside several genera of non-photosynthetic bacteria; most of the membranes have no confirmed metabolic function and they divide the cytoplasm into two or more compartments. These bacteria, collectively termed planctomycetes, all lack the peptidoglycan cell wall found in most bacteria and all have at least one large, internal membrane vesicle that divides the cytoplasm into two compartments of comparable volumes. One of these vesicles encloses the nucleoid, which tends to be highly condensed, forming a well-defined structure clearly visible in EM thin sections. In at least one species, a double-membrane structure surrounds the nucleoid, forming an analog of the eukaryotic nuclear membrane. Other species have multiple distinct membranes that create a nested series of cellular compartments (Fuerst, 2005).
Other bacteria and archaea produce small, regular vesicles in their cytoplasms which form from the self-assembly of protein subunits. The assembly process excludes water, so that the resulting vesicles contain only gas. The function of these gas vesicles (or gas vacuoles) appears to be to provide buoyancy, allowing the cell to migrate vertically in natural waters such as ponds. Consistent with this role, their synthesis is often regulated by environmental conditions, such a light intensity and oxygen concentration (Walsby, 1994).
IIH Extracellular Structures
Bacteria and archaea have several structurally distinct types of external filaments which are anchored in the CM. Flagella are helical protein filaments that support rapid swimming motility. Depending on the bacterial species, they may be single or numerous, clustered or dispersed over the cell, or absent altogether. Each flagellum extends several cell lengths (2–20 μm) and is rotated by a protein complex at its base; as a result, it acts like a propeller to push the cell through its liquid medium. Thus, bacterial flagella have a cellular function (i.e. motility) analogous to that of eukaryotic cilia and flagella, but no structural, molecular or mechanistic homology to them (see Chapter 47).
Pili and fimbriae are structurally similar to each other and distinct from flagella. Both consist of straight protein fibers that protrude less than about 1 μm from the cell, giving it a bristled or hairy appearance in the electron microscope. Both types of appendages appear to mediate attachment of a bacterium to another cell. In many cases, pili attach a “donor” bacterium to a recipient cell for the subsequent transfer of DNA (conjugation), whereas fimbriae typically attach a pathogenic bacterium to specific cells of its host. In some bacteria, pili mediate a type of slow, surface-dependent motility (Strom and Lory, 1993).
Flagella, pili and fimbriae all form via self-assembly of small protein subunits. Bacterial flagella grow by adding protein subunits to the distal tip, which requires the subunits (monomers) to travel from the cytoplasm to the growing tip of the flagellum, several cell lengths away. This is accomplished by using the hollow core of the flagellum itself as a conduit. In contrast, pili and fimbriae appear to polymerize at the base of the fiber, where it attaches to the CM (Neidhardt et al., 1990).
In addition to these small, discrete structures, many bacteria secrete hydrophilic polymers (usually polysaccharides) that form a gelatinous matrix around the cell. If this matrix remains attached to the cell and forms a defined zone around it, it is called a capsule. Capsules inhibit the ingestion of bacteria by human phagocytes or by protozoa and may also help free-living bacteria survive temporary desiccation or starvation. Even if secreted polysaccharide does not form a defined capsule, it aids the non-specific adherence of bacteria to wetted solid surfaces, leading to the formation of “biofilms”. Bacteria in biofilms tend to survive much higher concentrations of antibiotics than bacteria suspended in fluids and, accordingly, pose serious health risks if they form on surfaces of heart valves or implanted devices (Costerton et al., 1987).
III Energetics of Bacterial Cells
With a few notable exceptions, the endergonic cellular processes of bacteria are driven via enzymatic coupling to the hydrolysis of ATP and thus require the cell continuously to re-supply this energy currency. An E. coli cell growing aerobically using glucose regenerates ATP by two fundamentally different strategies: substrate-level phosphorylation and chemiosmotic coupling. Other prokaryotes may use these two processes in ways that differ from E. coli and related bacteria, or may use only one of them.
IIIA Substrate-Level Phosphorylation
As also occurs in the eukaryotic cytosol, soluble enzymes in the E. coli cytoplasm convert one mole of glucose to two moles of pyruvic acid via the Embden–Meyerhof–Parnas pathway (“glycolysis”); this process results in phosphorylation of 2 moles of ADP to form ATP and the reduction of 2 moles NAD+ to NADH. If the cell has no exogenous electron acceptors available (see below), the two moles of ATP represents the cell’s sole energy harvest and the reduced co-factor (NADH) represents unusable electrons. If the electrons are not transferred to some other molecule, the oxidized cofactor (NAD+) will not be regenerated and additional glucose cannot be metabolized. Bacteria (and eukaryotic cells) solve the latter problem by transferring the electrons to pyruvate or its metabolites, thereby forming various organic compounds which the cell excretes; examples of these end-products include lactic, acetic, propionic or butyric acids and ethanol, butanol or acetone. This metabolic strategy of substrate-level phosphorylation, made possible by reduction of metabolites derived from the growth substrate, constitutes the biochemical definition of fermentation.
IIIB Chemiosmotic Coupling
The general features of chemiosmotic coupling in bacteria and archaea resemble those of mitochondria and chloroplasts, which are described elsewhere in this volume (see Chapters 5 and 51). Also called oxidative phosphorylation, the process has two distinct stages, each of which involves vectoral enzymatic processes at the CM (Fig. 50.7). The first (oxidative) stage of chemiosmotic coupling transfers electrons taken from carbon compounds during glycolysis and the tricarboxylic acid cycle to some electron acceptor, such as O2, via a series of enzymatically catalyzed, strongly exergonic, oxidation–reduction reactions. This respiratory electron transport is coupled at certain points to the extrusion of protons from the cytoplasm, creating a proton potential or protonmotive force (PMF) across the CM (Fig. 50.7). The second (phosphorylation) stage converts the PMF into ATP, using an F1F0 proton-translocating ATPase to couple the entry of three protons from outside the cell to phosphorylation of an ADP molecule (Harold and Maloney, 1996). The combination of these two stages yields many times more of ATP per glucose molecule than fermentation does.
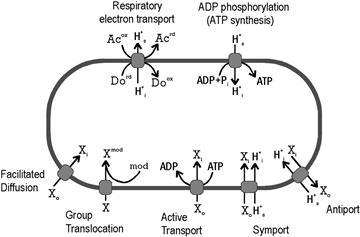
FIGURE 50.7 Energetics of solute transport across the CM. The diagram summarizes the salient features of major classes of energetic coupling to solute transport in prokaryotic cells. For example, the net result of respiratory electron transport is transfer of electrons from a donor (Do) to an acceptor (Ac) coupled to the extrusion of protons. Similarly, controlled entry of a proton through a membrane-bound ATPase drives the phosphorylation of ADP. The remaining processes are those which support growth by promoting the uptake of some necessary solute (X) from the environment. Cellular location is indicated by subscripts (o, outside; i, inside); “mod” indicates a chemical modification.
Oxygen represents only one of several terminal electron acceptors which prokaryotes can use to form a PMF. Accordingly, as summarized in Table 50.2, prokaryotes carry out various types of anaerobic respiration, but these may not be used under all conditions. E. coli, for example, can use four compounds in addition to oxygen as electron acceptors (Table 50.2), but it restricts the synthesis of the corresponding oxido-reductases according to a complex regulatory hierarchy. This hierarchy ensures that only the energetically most favorable electron acceptor is used, should more than one be available (Gunsalus, 1992).
TABLE 50.2. Respiratory Strategies of Prokaryotes
| Electron Acceptor | Reduced Product | Organism |
| Oxygen | Water | E. coli, other aerobes |
| Nitrate | Nitrite | E. coli, other bacteria |
| Fumarate | Succinate | E. coli, other bacteria |
| Dimethyl sulfoxide | Dimethyl sulfide | E. coli, other bacteria |
| Trimethylamine oxide | Trimethylamine | E. coli, other bacteria |
| Sulfate | Hydrogen sulfide | Desulfovibrio, Archaeoglobus∗ |
| Sulfur | Hydrogen sulfide | Thermoproteus∗ |
| Carbon dioxide | Methane | Methanobacterium∗ |
∗ Denotes archaea; all others are bacteria.
The PMF of a respiring bacterium represents an energetic intermediate in the regeneration of ATP by the oxidation of carbon compounds, yet even bacterial cells that are not respiring, as well as those that cannot respire, maintain a PMF. The lactic acid bacteria, for example, have no electron-transport chain and generate ATP only by substrate-level phosphorylation. They nevertheless have an F1F0 ATPase and use it in the reverse sense of respiring bacteria, i.e. to maintain a PMF at the expense of ATP hydrolysis. This may reflect the fact that certain basic prokaryotic processes use the PMF directly as their energy source. E. coli, for example, uses direct coupling to H+ influx, rather than ATP hydrolysis, to transport several nutrients (see below) and to drive flagellar rotation.
The PMF consists of two components: the electrical potential (Δψ) and the chemical potential (ΔpH). Although the small size of prokaryotic cells precludes direct electrical measurement of these potentials, they can be estimated using chemical probes. The Δψ can be measured by the fluorescence yield of triphenylmethyl- or tetraphenylphosphonium ions, or by the equilibrium distribution of radioactive K+ across the CM in the presence of valinomycin. The ΔpH can be estimated by the distribution of radioactive weak acids, such as acetic or benzoic acids, across the CM and the overall PMF can be independently estimated by the maximal accumulation of lactose by cells able to transport, but not metabolize, this sugar. According to these methods, which generally agree, a typical E. coli cell in medium at pH 6.5 has a Δψ of about 100 mV (inside negative) and a ΔpH that corresponds to an additional 100 mV (Harold and Maloney, 1996).
In those bacteria which live at pH values near 7, both components contribute significantly to the PMF. Extreme acidophiles, however, may maintain an electrical potential which is of opposite polarity as the normal Δψ, i.e. inside positive. This helps counteract the very large ΔpH across the cytoplasmic membrane, which can be more than 4 pH units in these organisms. In contrast, extreme alkaliphiles, which grow at external pH values of 10–12, have a negative ΔpH across the CM and, accordingly, a very large Δψ (White, 2000). A number of these organisms, and certain neutrophiles from marine environments, use the more abundant Na+ ion, rather than protons, to drive ATP synthesis via Na+-coupled ATP synthases (Skulachev, 1994).
IV Solute Transport
While certain membrane proteins generate the ion potentials and ATP needed to drive metabolism, other proteins use related processes to transport a wide range of organic compounds and inorganic ions into the cell as raw material for metabolism and growth. These systems employ different mechanisms and they differ with respect to functional properties, including energetic cost, solute specificity and ability to concentrate the solute inside the cell. These functional properties, in turn, affect biological properties of the organism, including its ability to scavenge critical nutrients from its environment. Fig. 50.7 summarizes the basic transport strategies used by bacterial cells, and the energetic consequences of these processes, as described below.
IVA Facilitated Diffusion
Although facilitated diffusion (see Fig. 50.7) is mechanistically simple and energetically cheap, it is rare among bacterial transport systems, presumably because it also relatively ineffective. One of the best-studied examples of a facilitated diffusion through a bacterial membrane is glycerol uptake in E. coli. In this case, extracellular glycerol diffuses passively through a polyol-specific membrane channel encoded by the glpF gene.
IVB Group Translocation
In an elaboration of facilitated diffusion, the solute becomes chemically modified upon entering the cytoplasm. The chemical modification, or “group translocation” (typically phosphorylation or phosphoribosylation), converts an uncharged solute molecule into an ion that cannot diffuse out through the same membrane-bound carrier. In this way, the uncharged species actually transported by the channel can remain at a lower steady-state concentration inside the cell than outside and the modified solute can be concentrated in the cytoplasm, due to the investment of chemical energy. In most cases, however, the “trapping reaction” doubles as the first step in metabolizing the solute, so the modified form does not necessarily accumulate to high steady-state concentrations.
IVC Active Transport
Active transport in bacteria is defined by the following features (Neidhardt et al., 1990):
1. specific steric recognition between the solute and a membrane-bound transport protein
2. release of the solute into the cytoplasm in its unmodified form
3. accumulation against a solute concentration gradient, and
Two types of active transport are distinguished from each other by the immediate source of energy used (see Fig. 50.7). The first type requires the hydrolysis of ATP or an equivalent high-energy phosphoryl bond. In these systems, the membrane-bound carriers typically consist of several subunits, one of which contains an ATP-binding site (Furlong, 1987). In Gram-negative bacteria, such a transporter often utilizes a non-membrane (i.e. soluble) protein located in the periplasm. Transport requires these periplasmic solute-binding proteins, as demonstrated by the fact that cold hypotonic shock treatment of bacterial cells, which releases only periplasmic proteins, also destroys transport capability. Studies of bacterial mutants and of cytoplasmic membrane vesicles confirm that the membrane-bound transporter interacts specifically not with external solute in its free form, but with the solute complexed to its cognate binding protein (Furlong, 1987).
The second type of active transport utilizes ionic potentials directly. In some cases, the solute enters via symport with a proton, in other cases, with an Na+ ion. Alternatively, certain ions, such as Ca2+, are pumped out of the cells by proton-coupled antiport systems (Harold and Maloney, 1996).
V Metabolic Strategies
Prokaryotes are metabolically diverse and mediate many different conversions of compounds in their environments as a way to harvest energy and other metabolic resources. These conversions contribute to the global cycling of many ecologically essential elements and several are unique to archaea and bacteria.
VA Autotrophy
A number of prokaryotes can derive all of their carbonaceous cell material from CO2. This capability, termed autotrophy, requires environmental sources of energy and reducing equivalents (i.e. electrons). For several diverse families of bacteria (collectively called photoautotrophs), light provides the energy, whereas various inorganic and organic compounds in the environment provide electrons. Photoautotrophic bacteria exhibit great diversity with regard to the biochemistry of photosynthesis. Among them, only the cyanobacteria carry out oxygenic photosynthesis like that of plant chloroplasts, which reflects the ability to use water as the electron donor. Cyanobacteria contain chlorophyll b and have two connected photosystems: photosystem II (PS2) oxidizes water to supply electrons to photosystem I (PS1; see Chapter 51). This is the scheme also used by plant chloroplasts, to which cyanobacteria are evolutionarily related (Schwartz and Dayhoff, 1978).
The remaining groups of photoautotrophic bacteria perform anaerobic (anoxygenic) photosynthesis. They have only one photosystem, which can operate in a cyclic manner to produce a proton potential. In these cases, the electron equivalents for the reduction of CO2 come from H2S, H2 or dissolved organic compounds, depending on the type of bacterium and the resources available to it. The photosynthetic pigments of the anoxygenic photoautotrophs, called bacteriochlorophylls, differ from cyanobacterial and plant chlorophylls in having intense absorbance maxima in the infra-red region of the spectrum. In many of these organisms, photoautotrophic growth represents only one among several metabolic options and production of photosynthetic pigments occurs only in the absence of oxygen.
Another form of autotrophy, chemoautotrophy, is unique to prokaryotes. Analogous to the harvesting of light energy by photoautotrophs, chemoautotrophs derive energy by mediating the oxidation and reduction of inorganic compounds in their environments. Aerobic chemoautotrophs include organisms that oxidize H2, CO, NH4+, NO2−, elemental S, H2S or Fe2+ using O2. Anaerobic chemoautotrophs include organisms that derive energy from the reduction of CO2 to CH4, or of SO4−− to H2S.
Among both photo- and chemoautotrophs, at least three metabolic pathways have been identified by which CO2 is “fixed” (i.e. reduced and incorporated into some common intermediary metabolite): (1) the Calvin cycle; (2) the acetyl CoA pathway; and (3) the reductive tricarboxylic acid cycle (Caldwell, 1995). Most photoautotrophic bacteria (and all green plants) use the Calvin–Benson cycle: carboxylation of ribulose bis-phosphate to yield two molecules of 3-phosphoglycerate, followed by a complex series of reactions to regenerate ribulose bis-phosphate (see Chapter 51). The acetyl-CoA pathway, used by methanogenic archaea and sulfate-reducing bacteria, involves the differential reduction of two CO2 molecules. One is reduced to form a methyl group whereas another is reduced to form a carbonyl unit, which is condensed to the methyl group. The resulting acetyl unit is transferred to co-enzyme A for assimilation into cell material. The reductive tricarboxylic acid pathway begins with carboxylation of succinyl CoA to yield 2-oxo-glutarate, which is in turn carboxylated to yield isocitrate. The transformations of the TCA cycle then continue in reverse, leading to the cleavage of citrate by an ATP-dependent citrate lyase to yield acetyl CoA and oxaloacetate. Prokaryotes using this pathway include the “green” photosynthetic bacteria.
VB Nitrogen Fixation
The only organisms that can fix nitrogen, i.e. convert N2 into ammonia, are prokaryotes. This capability is relatively rare, however, and widely dispersed phylogenetically. It occurs in various aerobic, anaerobic, facultatively anaerobic bacteria (i.e. those able to grow either aerobically or anaerobically, including both heterotrophs and autotrophs) and in methanogenic archaea. N2 reduction is carried out by a large enzyme complex, nitrogenase. It includes an iron-containing component and a molybdenum- and iron-containing component and utilizes ATP plus a reduced low-potential ferredoxin or flavoprotein as the source of electrons. Nitrogenases are intrinsically O2-sensitive and N2-fixing prokaryotes use a variety of strategies to protect these enzymes from oxidative inactivation. For example, facultative anaerobes such as Klebsiella spp. express nitrogenase genes only under anaerobic conditions. Filamentous cyanobacteria, which generate O2 during photosynthesis, sequester nitrogenase in specialized cells called heterocysts, in which the oxygenic photosystem II (PS2) does not operate. Azotobacter spp. are obligate aerobes that can fix nitrogen; they appear to scavenge intracellular O2 by maintaining very high respiration rates.
VI Responding to the Environment
Prokaryotic cells cannot insulate themselves from the chemical and physical properties of their environments. As a result, they must routinely respond to stress and environmental change in order to optimize their growth or simply to survive. The molecular mechanisms underlying these responses are diverse, but all result in changes to the biochemical composition of the cell.
VIA Osmotic Stress
Some cellular responses to environmental change are mediated directly by activation of existing proteins, as exemplified by the effects of osmotic stress on bacteria. A sharp increase in the extracellular solute concentration triggers a response in which the osmotic strength of the bacterial cytoplasm also increases (Neidhardt et al., 1990). In E. coli, the initial increase is primarily due to net K+ uptake by transporters in the CM, which seem to sense cell turgor directly. At extremely high external ionic strength, however, K+ accumulation alone does not appear to alleviate the deleterious effects of osmotic stress and the bacterial cells accumulate high concentrations of “compatible solutes” in the cytoplasm. Compatible solutes are charged organic molecules of which the two best studied examples are L-proline and glycine betaine. Typically, these compounds are scavenged from the environment and their accumulation in the cytoplasm permits growth at otherwise inhibitory external salt concentrations.
This osmotic stress response, which is typical of bacteria, indicates the physiological importance of water, solutes and turgor pressure for bacterial cells. In particular, the conservation, sophistication and energetic cost of this response suggest that turgor pressure per se is essential. This may reflect the fact that it provides the physical force that expands the bacterial cell, allowing its growth and subsequent division. Second, the importance of compatible solutes in physiological adaptation to high levels of osmotic stress suggests that these compounds perturb enzymatic and other cellular functions less than inorganic salts do, allowing them to accumulate to high intracellular concentrations with less impact on metabolism.
VIB Acid Stress
Other responses to the environment involve changes in the pattern of gene expression which are often complex and lead to corresponding changes in the protein composition of the prokaryotic cell (Neidhardt et al., 1990). The response of enteric bacteria to pH stress provides an example of this strategy that involves changing the amount of enzymes and other proteins. When the pH of the growth medium decreases sharply, E. coli and Salmonella enterica increase the synthesis rate of about 50 proteins. These acid-induced proteins include catabolic (degradative) enzymes that decarboxylate basic amino acids (arginine, ornithine and lysine decarboxylases). The action of these enzymes on amino acids taken up from the environment generates CO2 and diamines; the latter are exported from the cell and act to neutralize the acidic growth medium. Similarly, acid stress during anaerobic growth on a fermentable sugar increases lactate dehydrogenase and formate-hydrogen lyase; both of these enzymes divert the flow of intermediates through fermentative metabolism in ways that decrease the amount of acid produced (Slonczewski and Foster, 1996).
Many other prokaryotic responses to environmental change similarly involve changing the protein composition of the cell by modulating transcription of certain genes. This general strategy takes advantage of the relatively large biosynthetic capacity of the growing prokaryotic cell, its simple organization and a very short half-life of messenger RNA which, in E. coli (under standard conditions) averages 1.3 min. Studies of specific responses in bacteria have revealed an array of elegant and diverse molecular mechanisms for controlling gene expression, which lie outside the scope of this chapter. One example will be described, however, which involves transmembrane signaling and appears in many environmental responses in bacteria.
VIC Phosphate Metabolism and “Two-Component” Regulatory Systems
When Gram-negative bacteria begin to starve for inorganic phosphate, they activate the transcription of several families of genes, whose products help the cell conserve phosphate and take up more of it from the environment (Wanner, 1993). The regulation process, which typifies a mechanism underlying many signal transduction systems in bacteria, involves the specific interaction and phosphorylation of two proteins, PhoR and PhoB, named after their respective genes in E. coli.
The phosphorylation state of the PhoB protein determines expression of the various genes of the phosphate-starvation response. The phosphorylated form of PhoB (here designated “∗PhoB”) activates transcription of these genes, whereas the unphosphorylated form does not. The relative abundance of ∗PhoB is in turn determined by the conformational state of a CM protein, PhoR (Fig. 50.8). In its “activated” conformation (“∗PhoR”), this protein acts as a PhoB kinase, whereas its “inactive” conformation (“PhoR”) acts as a ∗PhoB phosphatase. Ultimately, the conformational state of PhoR is determined by the availability of inorganic phosphate in the external medium. By a mechanism which is not entirely clear, PhoR can sense the absence of phosphate-periplasmic binding protein complexes in the periplasm and assumes its active conformation under these conditions. The resulting ∗PhoR phosphorylates PhoB, forming ∗PhoB which, in turn, stimulates the transcription of phoA and other phosphate-scavenging genes (see Fig. 50.6). The result is a rapid increase in certain enzymatic activities which serve to alleviate a particular state of the cell, in this case, phosphate deficiency.
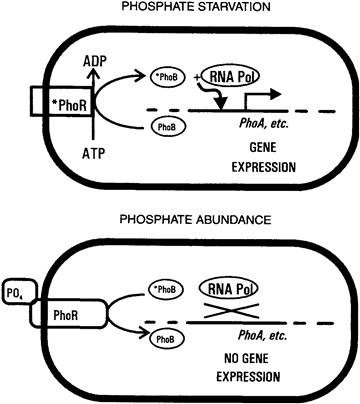
FIGURE 50.8 Essential features of two-component regulatory systems. A simplified version of phosphate-regulated gene expression is shown. The PhoR protein senses the extracellular availability of phosphate, either directly through an externally exposed domain, or indirectly via interaction with the high-affinity phosphate transporter (Neidhardt et al., 1990; Wanner, 1993). When the external phosphate concentration is low, PhoR becomes activated; this activated PhoR phosphorylates itself at the expense of intracellular ATP and then phosphorylates the PhoB protein. When the external phosphate concentration is high, PhoR becomes “de-activated”; in this form it removes phosphate groups from phosphorylated PhoB protein. PhoB is a soluble transcription factor for a specific set of genes, but stimulates transcription only when phosphorylated.
VID Other Two-Component Regulatory Systems of Bacteria
Protein pairs resembling PhoR/PhoB mediate a wide variety of regulatory responses of bacteria to various types of environmental change (Table 50.3). The PhoR protein, a transmembrane signal transducer, typifies the component I, or sensor-kinase, of these systems. With a few exceptions, this is a protein of the cytoplasmic membrane that senses the status of some environmental parameter and accordingly adopts either of two conformations. In its “active” conformation, the component I has two characteristic phosphotransferase activities: (1) it phosphorylates one of its own histidine residues, using ATP; (2) it transfers the phosphate to an aspartyl residue on a cognate component II or response regulator. These two activities result in the net transfer of a phosphate group from ATP to the specific component II, which is usually a transcriptional regulator that activates transcription of a certain set of genes, but only in its phosphorylated form.
TABLE 50.3. Examples of Two-Component Regulatory Systems
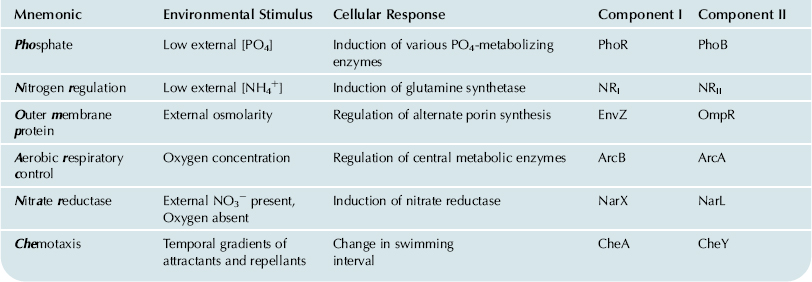
The similarities in interactions and biochemical properties of various components I and II (Table 50.4) are underscored by regions of sequence homology. Although they respond to diverse signals, all components I have two highly conserved sequence motifs at their carboxy termini. Similarly, all components II have sequence homology near their amino termini and various subfamilies may share regions of additional homology elsewhere in the amino acid sequence. The conserved regions of components I recognize and interact with the conserved regions of components II as an essential part of the signal-transmission process. The non-conserved regions among the components I and II presumably define their specificity for the external stimulus detected and for the particular cellular response, respectively.
TABLE 50.4. General Features of Two-Component Regulators
| Feature | Component I | Component II |
| Cellular location | Membrane | Cytoplasm |
| Cellular function | Sense and signal an external condition | Control gene expression |
| Biochemical activities | Autophosphorylation at histidine residue | Binding to regulatory regions of genes |
| Phosphorylation of cognate component II | Stimulating transcription of specific genes | |
| may dephosphorylate cognate component II | May inhibit transcription of other specific genes | |
| Sequence conservation | Two highly conserved regions near carboxyl end | One highly conserved region near amino end (subfamilies have additional regions of homology near carboxyl end) |
VIE Other Environmental Responses and their Mechanisms
VIE1 Iron Uptake
In oxidizing environments, iron occurs as the ferric ion and this form is extremely insoluble in neutral or basic solution. Bacteria have a characteristic strategy for scavenging ferric iron, which involves specialized iron chelators called siderophores, and transport systems specific for the iron–siderophore complexes (Earhart, 1996).
Nearly 100 distinct siderophores produced by microorganisms have been identified; all are relatively small molecules and they fall into two classes, i.e. those that use catecholate groups to complex the ferric ion vs those that use hydroxamate groups. Bacteria that have been examined intensively, such as E. coli, have been found to produce several siderophores of both classes, which they release into the environment under limiting-iron conditions. The siderophores complex ferric iron specifically and with extremely high affinity (for ferric enterobactin, Kd = 10−52 M) and bacteria use specialized, high-affinity transport systems to recover the resulting complexes. In the case of Gram-negative bacteria, transport is complicated by the fact that the complexes, though relatively small, are nevertheless too large to pass through the OM pores formed by porins. Gram-negative bacteria therefore have specialized OM receptors, which appear to function as gated pores that allow the complexes to enter the periplasm. This initial transport step involves a form of energetic coupling whose mechanism is still somewhat unclear, but requires a specific protein (TonB) that energizes transport of a few other solutes across the OM. Transport across the CM involves another siderophore-specific transporter (Earhart, 1996).
Once the iron–siderophore complex is inside the bacterial cytoplasm, the cell is faced with two additional challenges. First, the iron must be released from the siderophore; this is made difficult by the extreme affinity of the two components and appears to be solved by chemically modifying one or both of them. In some cases, the siderophore component of the complex appears to be destroyed enzymatically in the bacterial cytoplasm by hydrolases. The alternative strategy, reduction of the complexed ferric ion to ferrous ion, has also been detected, although some question remains as to whether reduction precedes release. Finally, it should be noted that excess (i.e. free) ferrous iron in the cytoplasm presents a serious threat, particularly to aerobic organisms, because it reacts with H2O2 to produce the extremely reactive hydroxyl radical. This problem is addressed by close regulation of the iron uptake systems and by synthesis of iron-storage proteins to sequester unused iron intracellularly (Earhart, 1996).
VIE2 Magnetotaxis
Certain motile bacteria have been found that swim along the lines of an externally applied magnet field. Several species of these magnetotactic bacteria have been isolated and maintained as pure cultures in the laboratory, but most have resisted cultivation, although they can be observed in samples from natural environments. The difficulty of culturing magnetotactic bacteria stems, in part, from the fact that all known examples are microaerophiles, meaning that they require oxygen at low concentrations but are killed by it at atmospheric levels. Accordingly, these organisms typically occur at interfaces between oxic and anoxic layers of natural waters, as occur in marshes or swamps. In these settings, magnetotaxis along the vertically inclined geomagnetic lines of force, perhaps combined with other environmental cues, may guide the cells to an appropriate position in a vertical oxygen gradient.
The dominant mechanism of magnetotaxis among these bacteria seems to be passive, in the sense that the cell aligns itself with the magnetic field while rotation of the helical flagellum drives it forward (Lefevre et al., 2009). This mechanism requires a strong magnetic dipole that is stably aligned (i.e. fixed) within the cell and parallel to the main axis of the flagellum. In magnetotactic bacteria, the dipole is provided by a short chain of single-domain magnetic crystals, called magnetosomes, each of which is enclosed in a phospholipid membrane. Other mechanistic details of magnetosome synthesis, including what determines the magnetic polarity of the chain and how the parallel orientation of internal magnet and flagellum is maintained, remain to be resolved (Jogler and Schüler, 2009).
VII The Physiology of Pathogenesis
Bacteria capable of causing disease represent a tiny minority of known species. These pathogens are distinguished from the vast majority of bacteria by adaptations that allow them to invade their animal or plant host, elude multiple defense mechanisms (including the immune system), reproduce within host fluids and tissues, and damage host function in some way, thereby causing disease. The host defenses against the pathogens, as well as the mechanisms by which the pathogens damage the host, often involve specialized physiological functions of the host and bacterial cells, respectively.
One of the first host defenses encountered by a bacterium after invading the body is often a phagocyte, a specialized motile cell that circulates in the blood and engulfs bacteria. After a bacterial cell has been internalized (see Fig. 50.6), the resulting cytoplasmic vesicle fuses with a lysosome to form a phagolysosome. In addition to the low pH and digestive enzymes typical of lysosomes, phagolysosomes generate high concentrations of nitrous oxide, hypochlorous acid and reactive oxygen species, including hydrogen peroxide, singlet oxygen, superoxide and the hydroxyl radical. Synthesis of these strong oxidants is triggered when a phagocyte is activated and is accompanied by a sudden demand for oxygen, called the respiratory burst. The reactive compounds produced in this process, combined with the low pH inside the phagolysosome, normally kill the ingested bacterium.
However, certain pathogenic bacteria can survive in the phagolysosome, escape it and multiply in the cytoplasm of the phagocyte. In some cases, the strategy involves killing the phagocyte by secreting a toxin termed a leukocidin. Other bacterial countermeasures against phagocytosis include synthesis of cell-associated carotenoid pigments, which quench singlet oxygen, and synthesis of an external capsule (see Section II), which inhibits the initial engulfment of the bacterial cell.
Various pathogenic bacteria also secrete toxins at other stages of pathogenesis which target specific functions of host cells and tissues in causing disease. Several pathogens, for example, colonize the small intestine and produce an enterotoxin that induces diarrhea. Vibrio cholerae, which causes cholera, remains one of the most threatening of these bacteria globally, but pathogenic strains of Bacillus cereus, E. coli and Salmonella enterica also cause serious disease. Once established in the host, these bacteria produce enterotoxins that bind to specific gangliosides in the plasma membranes of intestinal epithelial cells. The cholera toxin covalently modifies and inactivates a GTP-binding protein that normally regulates an adenylate cyclase complex in the plasma membrane. Loss of this regulatory function causes overproduction of cAMP in the epithelial cell which, in turn, activates a number of cAMP-stimulated cellular functions, one of which is export of chloride ion. As large amounts of chloride are pumped into the lumen of the intestine, water follows by passive diffusion across the plasma membrane (O’Brien and Holmes, 1996). The massive secretion of salts and water into the lumen of the intestine results in a severe diarrhea, which may be lethal if water and salts are not replaced at an adequate rate.
Another class of damage-inducing proteins from pathogenic bacteria includes the hemolysins, named according to the fact that these proteins promote the lysis of erythrocytes (red blood cells). The most common technique for detecting hemolysin production involves incorporating whole blood into the agar plating medium used to grow bacterial colonies. If the bacterium produces a hemolysin, a clear zone, or halo, will form around the colony, caused by lysis of all the red blood cells within a short distance of the colony. Although they differ in some properties, hemolysins are generally small, secreted proteins which bind to the plasma membrane of the blood cells as individual subunits. The membrane-bound subunits then oligomerize to form a large pore in the plasma membrane, through which the cytoplasm escapes.
Bacterial toxins affecting yet a different host function, muscle contraction, are produced by two anaerobic, spore-forming pathogens: Clostridium tetani and Clostridium botulinum. Both species induce paralysis through disruption of the nervous system, but the two corresponding neurotoxins differ in the form of paralysis and the underlying mechanism. Cl. tetani infects host tissue, typically through deep puncture wounds, and produces tetanus toxin. This protein accumulates at the ends of inhibitory neurons where it blocks the inhibition of muscle contraction. The resulting uncontrolled contraction leads to a general spastic paralysis which can result in death by asphyxia. In contrast, the neurotoxin of Cl. botulinum is normally produced during growth outside the host, but can be lethal if ingested, even in very small quantities. Botulism toxin binds to stimulatory motor neurons and prevents release of acetylcholine at the neuromuscular junction. This blocks muscle contraction, leading to a flaccid paralysis.
VIII Prokaryotes Living in Extreme Environments
The physiological diversity and specialization of prokaryotes is illustrated dramatically by organisms which not merely tolerate chemical and physical extremes, but actually require them for normal cellular function. This heterogeneous group includes many species of archaea that have been discovered only in recent years as their natural habitats have been subjected to the appropriate sampling and cultivation methods.
Most extreme halophiles, organisms that thrive in concentrated brines, such as those that occur in natural salt lakes or artificial evaporation ponds, are archaea. Well-studied species include the rod-shaped Halobacterium salinarum (formerly halobium) and the irregularly shaped Haloferax volcanii. H. salinarum grows best in media containing 3–4 M NaCl and will not grow at salt concentrations less than 1.5 M. The cell maintains an internal K+ concentration of upto 5 M and contains Cl− as the major counterion. Accordingly, most H. salinarum enzymes function best in extremely high salt concentrations and denature at low salt concentrations (Kushner, 1985). The native structure of these halophilic enzymes tends to incorporate an unusually high number of acidic amino acid residues on the protein surface. Other notable features of the H. salinarum cell include gas vesicles in the cytoplasm and patches of a special retinal protein, bacteriorhodopsin, in the cytoplasmic membrane. The gas vesicles make the cells buoyant and thus help them maintain contact with two sources of energy: O2 and light. Oxidation of organic compounds in the environment normally supports growth, but if oxygen becomes limiting, extra energy can be supplied by bacteriorhodopsin. This is a membrane-bound retinal protein that functions as a light-driven ion pump. When the cell is illuminated, it pumps protons from the cytoplasm; this contributes to the cell’s PMF and thus, to the production of ATP, during O2 deprivation (Kushner, 1985). Related proteins, collectively termed proteorhodopsins have more recently been identified in a number of archaea and also bacteria and some of these rhodopsins may have cellular functions distinct from ATP production (Fuhrman et al., 2008).
Extreme acidophiles, organisms which require low pH for optimal cell growth, include both bacteria and archaea. Thiobacillus spp. are Gram-negative bacteria that derive energy from the oxidation of reduced sulfur compounds. Since the oxidized end-product is usually H2SO4, these bacteria acidify their environment in the normal course of their metabolism. Some Thiobacillus cells grow optimally at about pH 2 but maintain a cytoplasmic pH near 6.5. At least one species, Tb. ferrooxidans, can also oxidize Fe2+; this metabolic strategy is aided by the fact that auto-oxidation of Fe2+, which would compete with the biologically mediated process, occurs very slowly at low pH values.
The most extreme acidophile known to date is a moderately thermophilic archaeon, Picrophilus oshimae. This organism, isolated from the soil of a Japanese geothermal field, grows optimally in dilute sulfuric acid at about pH 0.9. P. oshimae cells can grow at pH 0 but not at pH values above 3.5 and at pH values greater than 5, the cells lyse (Schleper et al., 1995).
Bacteria that require low temperatures are termed psychrophiles and typically occur in polar marine environments. Enzymes of these organisms may denature, and the cells may die, at room temperature. In several cases, the minimum temperature for growth has not been determined, but lies below about −10°C. The membrane phospholipids of these bacteria contain unusually high proportions of unsaturated and short-chain fatty acids (Morita, 1975).
At the other extreme, prokaryotes that grow optimally at 80°C or higher temperatures are termed extreme thermophiles or hyperthermophiles (Stetter et al., 1990). The cultivated species consist primarily of archaea isolated from geothermal habitats such as terrestrial hot springs or submarine thermal vents. Aerobic hyperthermophiles include Sulfolobus spp., which require acidic conditions as well as high temperatures. The combination of high temperature and low pH required for optimal growth (about 80°C and pH 3) is extremely effective at denaturing proteins. The Sulfolobus cell helps protect its cytoplasmic proteins, which appear to be generally heat-stable but not acid-stable (Grogan, 1996), by maintaining its cytoplasmic pH at about 6 (Schäfer et al., 1990). Physical properties of the unique lipids of Sulfolobus spp. probably help maintain the resulting ΔpH. The ether-linked phytanyl chains of these lipids span the entire membrane, forming a stable monolayer. Such membranes exhibit very low rates of proton leakage at high temperature, compared to ester-linked phospholipid bilayers (Van de Vossenberg et al., 1995) and this seems to represent an important prerequisite for efficient function of these cells.
The most thermophilic organisms known are anaerobic archaea isolated from submarine vents, where hydrostatic pressure permits liquid water to be superheated. Pyrolobus fumarii, the most thermophilic organism available in culture, grows optimally at about 106°C (Blöchl et al., 1997). The very existence of cells with such growth requirements raises fundamental questions regarding their need to stabilize nucleic acids, proteins and other molecules necessary for all life. The thermostability of enzymes of most hyperthermophiles appears to be intrinsic, i.e. not due to counter-ions or other solutes. Biophysical analyses indicate that several classes of interactions of the amino acid residues contribute to this intrinsic structural stability (Jaenicke and Böhm, 1998).
IX Conclusions
The smallest and simplest living cells are prokaryotic, consist of a nucleoid and cytoplasm enclosed in a cytoplasmic membrane. Most free-living prokaryotes also have a cell wall of some type and many bacteria have a second lipid membrane outside the cell wall, whereas a few have additional, internal structures. Though structurally simple, prokaryotic cells are evolutionarily diverse and include two distinct lineages, the Bacteria and the Archaea, each with a taxonomic status equal to all eukaryotic organisms. The cells of bacteria and archaea are functionally complex, mediating nutrient uptake, energy conversion and conservation, growth, secretion, genome replication, cell division and regulation of gene expression in response to environmental change. Prokaryotes encompass a wide range of metabolic strategies, including many not represented among eukaryotes. Several alternatives of CO2 fixation and photosynthesis are represented among bacteria and archaea, of which only one, that of the cyanobacteria, has been adopted by green plants. Some prokaryotes grow chemoautotrophically, some can fix N2 and some can “respire” anaerobically, using electron acceptors other than O2 in their environments.
Bacteria depend on solute-specific transport proteins located in the CM to accumulate nutrients and to move inorganic ions into and out of the cell. Those proteins involved in active transport display a high affinity for the external solute and expend energy to accumulate it to high intracellular levels. A common type of active transport in Gram-negative bacteria utilizes soluble proteins which bind the solute in the periplasm and deliver it to the corresponding membrane-bound transporter. Some membrane transporters mediate homeostatic responses to environmental stress. Others communicate with “two-component” systems to regulate bacterial gene expression. In most cases, bacteria change their macromolecular compositions in response to changes in external conditions. This form of physiological adaptation can occur quickly, due to the high biosynthetic capacity of prokaryotic cells relative to cell mass.
A few bacteria are specialized to evade the defense systems of multicellular organisms and cause damage to their host. The host defenses of mammals include cells specialized for the destruction of bacteria via controlled generation of reactive chemicals, whereas the damage to the host caused by pathogenic bacteria is, in many cases, mediated by protein toxins which target specific proteins or cell types.
The specialization and functional diversity of prokaryotic cells becomes apparent in extreme environments. Certain bacteria require low external pH or low temperature to survive and grow. Certain archaea require extremely high salt concentration or extremely high temperature for normal cellular function. Because archaea differ radically from E. coli and other well-studied microorganisms, pinpointing the physiological and biochemical basis of their adaptation to harsh environments remains a challenging area of research.
BIBLIOGRAPHY
1. Blöchl E, Rachel R, Burggraf S, Hafenbrandl D, Jannasch H, Stetter KO. Pyrolobus fumarii, gen and sp.nov., represents a novel group of archaea, extending the upper temperature limit for life to 113°C. Extremophiles. 1997;1:14–21.
2. Boucher Y. Lipids: biosynthesis, function, and evolution. In: Cavicchioli R, ed. Archaea: Molecular and Cellular Biology. Washington, DC: American Society for Microbiology; 2007;:341–353.
3. Caldwell DR. Microbial Physiology and Metabolism. Dubuque, IA: William C. Brown Publishers; 1995.
4. Costerton JW, Cheng K-J, Geesey GG, et al. Bacterial biofilms in nature and disease. Ann Rev Microbiol. 1987;41:435–464.
5. Dierstein R, Schumacher A, Drews G. On insertion of pigment-associated polypeptides during membrane biogenesis in Rhodopseudomonas capsulata. Arch Microbiol. 1981;128:376–383.
6. Earhart CF. Uptake and metabolism of iron and molybdenum. In: Neidhardt FC, ed. Escherichia coli and Salmonella: Cellular and Molecular Biology. Washington, DC: American Society for Microbiology; 1996;:1079–1090.
7. Fuerst JA. Intracellular compartmentation in planctomycetes. Ann Rev Microbiol. 2005;59:299–328.
8. Fuhrman JA, Schwalbach MS, Stingl U. Proteorhodopsins: an array of physiological roles?. Nat Rev Microbiol. 2008;6:488–494.
9. Furlong C. Osmotic-shock-sensitive transport systems. In: Neidhardt FC, ed. Escherichia coli and Salmonella: Cellular and Molecular Biology. Washington, DC: American Society for Microbiology; 1987;:768–796.
10. Grogan DW. Organization and interactions of cell envelope proteins of the extreme thermoacidophile Sulfolobus acidocaldarius. Can J Microbiol. 1996;42:1163–1171.
11. Gunsalus RP. Control of electron flow in Escherichia coli: co-ordinated transcription of respiratory pathway genes. J Bacteriol. 1992;174:7069–7074.
12. Harold FM, Maloney PC. Energy transduction by ion currents. In: Neidhardt FC, ed. Escherichia coli and Salmonella: Cellular and Molecular Biology. Washington, DC: American Society for Microbiology; 1996;:283–306.
13. Jaenicke R, Böhm G. The stability of proteins in extreme environments. Curr Opin Struct Biol. 1998;8:738–748.
14. Jogler C, Schüler D. Genomics, genetics, and cell biology of magnetosome formation. Annu Rev Microbiol. 2009;63:501–521.
15. Kletzin A. General characteristics and important model organisms. In: Cavicchioli R, ed. Archaea: Molecular and Cellular Biology. Washington, DC: American Society for Microbiology; 2007;:14–92.
16. Kushner DJ. The Halobactericeae. In: Socatch JR, Ornston LN, eds. The Bacteria: vol VIII Archaebacteria. Orlando: Academic Press, Inc; 1985;:171–214.
17. Lefevre CT, Song T, Yonnet JP, Wu LF. Characterization of bacterial magnetotactic behaviors by using a magnetospectrophotometry assay. Appl Environ Microbiol. 2009;75:3835–3841.
18. Morita RY. Psychrophilic bacteria. Bacteriol Rev. 1975;89:144–167.
19. Neidhardt FC, Ingraham JL, Schaechter M. Physiology of the Bacterial Cell. Sunderland, MA: Sinauer Associates, Inc; 1990.
20. Nikaido H, Vaara M. Molecular basis of bacterial outer membrane permeability. Microbiol Rev. 1985;49:1–32.
21. O’Brien AD, Holmes RK. Protein toxins of Escherichia coli and Salmonella. In: Neidhardt FC, ed. Escherichia coli and Salmonella: Cellular and Molecular Biology. Washington, DC: American Society for Microbiology; 1996;:2788–2802.
22. Pettijohn DE. The nucleoid. In: Neidhardt FC, ed. Escherichia coli and Salmonella: Cellular and Molecular Biology. Washington, DC: American Society for Microbiology; 1996;:158–166.
23. Schäfer G, Anemüller S, Moll R, Meyer W, Lübben M. Electron transport and energy conservation in the archaebacterium Sulfolobus acidocaldarius. FEMS Microbiol Rev. 1990;75:335–348.
24. Schleper C, Pühler G, Holz I, et al. Picrophilus gen nov., fam nov: a novel heterotrophic, thermoacidophilic genus and family comprising archaea capable of growth around pH 0. J Bacteriol. 1995;177:7050–7059.
25. Schwartz RM, Dayhoff MO. Origin of prokaryotes, eukaryotes, mitochondria, and chloroplasts. Science. 1978;199:395–403.
26. Schwarz U, Asmus A, Frank H. Autolytic enzymes and cell division of Escherichia coli. J Mol Biol. 1969;41:419–429.
27. Skulachev VP. The latest news from the sodium world. Biochim Biophys Acta. 1994;1187:216–221.
28. Slonczewski JL, Foster JW. pH-regulated genes and survival at extreme pH. In: Neidhardt FC, ed. Escherichia coli and Salmonella: Cellular and Molecular Biology. Washington, DC: American Society for Microbiology; 1996;:283–306.
29. Stetter KO, Fiala G, Huber G, Huber R, Segerer A. Hyperthermophilic microorganisms. FEMS Microbiol Rev. 1990;75:117–124.
30. Strom M, Lory S. Structure–function and biogenesis of the type IV pili. Ann Rev Microbiol. 1993;47:565–596.
31. Van De Vossenberg J, Ubbink-Kok T, Elferink M, Driessen A, Konings WN. Ion permeability of the cytoplasmic membrane limits the maximum growth temperature of bacteria and archaea. Molec Microbiol. 1995;18:925–932.
32. Walsby AE. Gas vesicles. Microbiol Rev. 1994;58:94–144.
33. Wanner BL. Gene regulation by phosphate in enteric bacteria. JCell Biochem. 1993;51:47–54.
34. White D. The Physiology and Biochemistry of Prokaryotes. 2nd ed. New York: Oxford University Press; 2000.
35. Woese CR, Kandler O, Wheelis M. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci USA. 1990;87:4576–4579.
36. Yanofsky C, Crawford IP. The tryptophan operon. In: Neidhardt FC, ed. Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. Washington, DC: American Society for Microbiology; 1987;:1453–1472.