Chapter 4. First Steps in the Cloud
In the previous two chapters, we took you through the essentials of genomics and computing technology. Our goal was to make sure you have enough of a grounding in both domains regardless of whether you’re coming to this more from one side or the other—or perhaps even from another domain altogether; if so, welcome! And hang in there.
We realize that those first two chapters might have felt very passive since there were no hands-on exercises involved. So here’s the good news: you’re finally going to get to do some hands-on work. This chapter is all about getting you oriented and comfortable with the GCP services that we use throughout this book. First, we walk you through creating a GCP account and running simple commands in Google Cloud Shell. After that, we show you how to set up your own VM in the cloud, get Docker running on it, and set up the environment that you’ll use in Chapter 5 to run GATK analyses. Finally, we show you how to configure the IGV to access data in Google Cloud Storage. After you have all that set up, you’ll be ready to do some actual genomics.
Setting Up Your Google Cloud Account and First Project
You can sign up for an account on GCP by navigating to https://cloud.google.com and following the prompts. We are purposely light on the details here because the interface for account setup has been known to change. At a high level, though, your goals are to establish a new Google Cloud account, set up a billing account, accept the free trial credits (if you’re eligible), and create a new project that links to your billing account.
If you don’t already have a Google identity of some kind, you can create one with your regular email account; you don’t need to use a Gmail account. Keep in mind also that if your institution uses G Suite, your work email might already be associated with a Google identity even if the domain name is not gmail.com.
After you’ve signed up, make your way to the GCP console, which provides a web-based graphical interface for managing cloud resources. You can access most of the functionality offered in the console through a pure command-line interface. In the course of the book, we show you how to do some things through the web interface and some through the command line, depending on what we believe is most convenient and/or typical.
Creating a Project
Let’s begin by creating your first project, which is necessary to organize your work, set up billing, and gain access to GCP services. In the console, go to the “Manage resources” page and then, at the top of the page, select Create Project. As shown in Figure 4-1, you need to give your project a name, which must be unique within the entire GCP. You can also select an organization if your Google identity is associated with one (which is usually the case if you have an institutional/work G Suite account), but if you just created your account, this might not be applicable to you at the moment. Having an organization selected means new projects will be associated with that organization by default, which allows for central management of projects. For the purposes of these instructions, we assume that you’re setting up your account for the first time and there isn’t a preexisting organization linked to it.
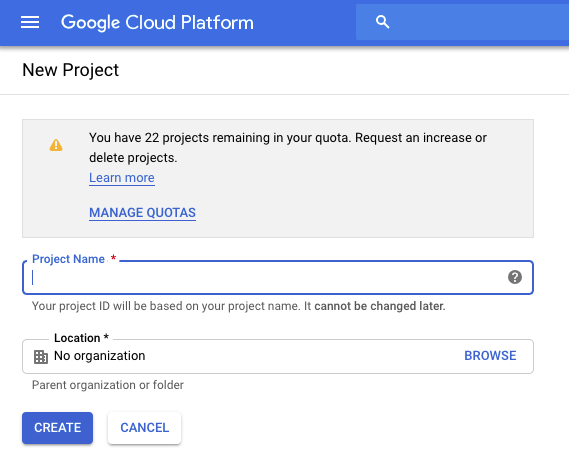
Figure 4-1. Creating a new project.
Checking Your Billing Account and Activating Free Credits
If you followed the sign-up process outlined in the previous section and activated your free trial, the system will have set up billing information for you as part of the overall account creation process. You can check your billing information in the Billing section of the console, which you can also access at any time from the sidebar menu.
If you’re eligible for the free credits program, one of the panels on the billing overview page will summarize the number of credits and days you have left to spend them. Note that if yours is displaying a blue Upgrade button, as shown in Figure 4-2, your trial has not yet started and you need to activate it in order to take advantage of the program. You might also see a “Free trial status” banner at the top of your browser window with a blue Activate button. Someone at GCP is working really hard to not let you walk away from free money, so click either of those buttons to start the process and receive your free credits.
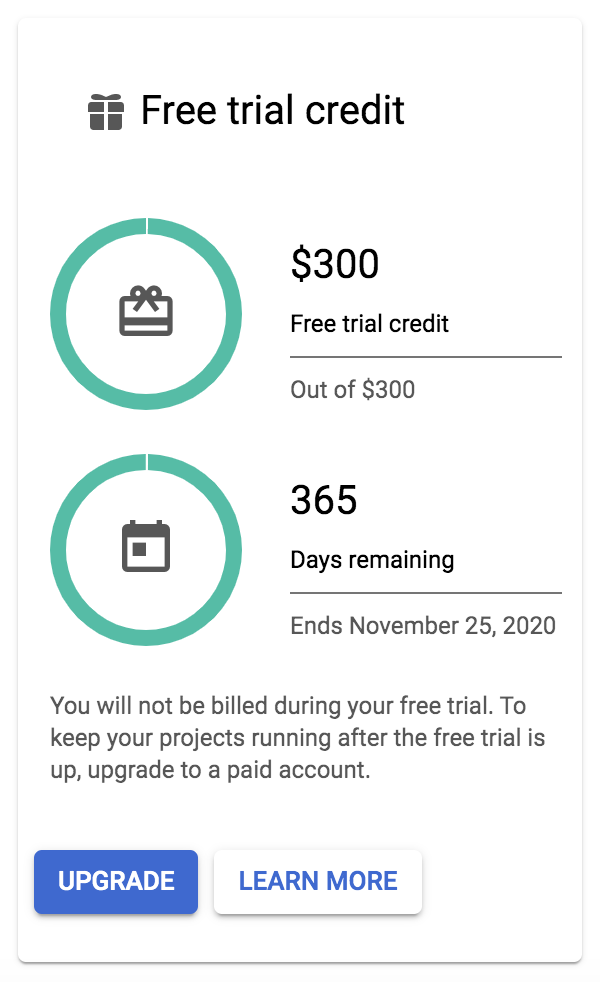
Figure 4-2. The panel in the Billing console summarizing free trial credits availability.
More generally, the billing overview page provides summaries of how much money (or credits) you have spent so far as well as some basic forecasting. That being said, it’s important to understand that the system does not show you costs in real time: there is some lag time between the moments when you use chargeable resources and when the costs are updated on your billing page.
Many people who make the move to the cloud report that keeping track of their spending is one of the most difficult parts of the process. It’s also the one that causes them the most anxiety because it can be very easy to spend large sums of money pretty quickly in the cloud if you’re not careful. One feature offered by GCP that we find particularly useful in this respect is the “Budgets & alerts” settings, as depicted in Figure 4-3. This allows you to set email alerts that will notify you (or whoever is the billing administrator on your account) when you exceed certain spending thresholds. To be clear, this won’t stop anything from running or prevent you from starting any new work that would push you over the threshold, but at least it will let you know where you stand.
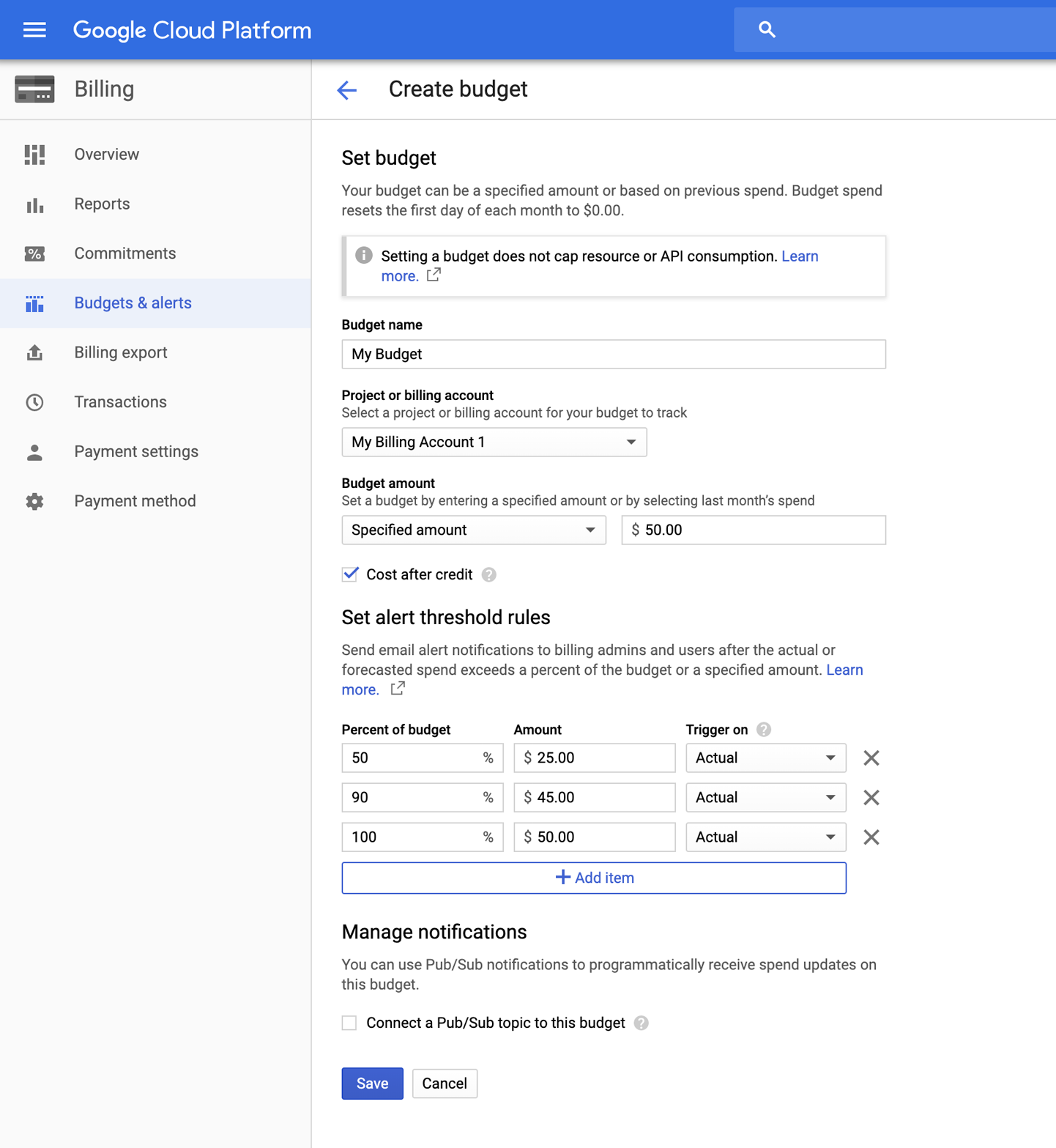
Figure 4-3. Budget and alert threshold administration.
To access the billing notifications feature, on the main menu on the GCP console, choose Billing, select the billing account you just created, and then look for the Budgets and alerts option. After you select it, you will be able to set up a new budget using the Create budget form shown in Figure 4-3. You can create multiple budgets and set multiple triggers for different percentages of the budget if you want warnings as you approach your budget amount. But as we just mentioned, keep in mind that it is still only a notification service and will not prevent you from incurring additional charges.
Running Basic Commands in Google Cloud Shell
Now that you’ve established your account, set up billing, and created your project, the next step is to log in to your first VM. For our exercises here, we use Google Cloud Shell, which does not require any configuration to get started and is completely free, although it comes with a few important limitations that we discuss in a moment.
Logging in to the Cloud Shell VM
To create a secure connection to a Cloud Shell VM using the SSH protocol, in the upper-right corner of the console, click the terminal icon:

This launches a new panel in the bottom on the console; if you want, you can also pop the terminal out to its own window. This gives you shell access to your own Debian-based Linux VM provisioned with modest resources, including 5 GB of free storage (mounted at $HOME) on a persistent disk. Some basic packages are preinstalled and ready to go, including the Google Cloud SDK (aka gcloud), which provides a rich set of command-line-based tools for interacting with GCP services. We’ll use it in a few minutes to try out some basic data management commands. In the meantime, feel free to explore this Debian VM, look around, and see what tools are installed.
Note
Be aware that weekly usage quotas limit how much time you can spend running the Cloud Shell; as of this writing, it’s 50 hours per week. In addition, if you don’t use it regularly (within 120 days, as of this writing), the contents of the disk that provides you with free storage might end up being deleted.
When you log in to Cloud Shell for the first time, it prompts you to specify a Project ID using the aforementioned gcloud utility:
Welcome to Cloud Shell! Type "help" to get started. To set your Cloud Platform project in this session use “gcloud config set project [PROJECT_ID]”
You can find your Project ID on the Home page of the console, as shown in Figure 4-4.
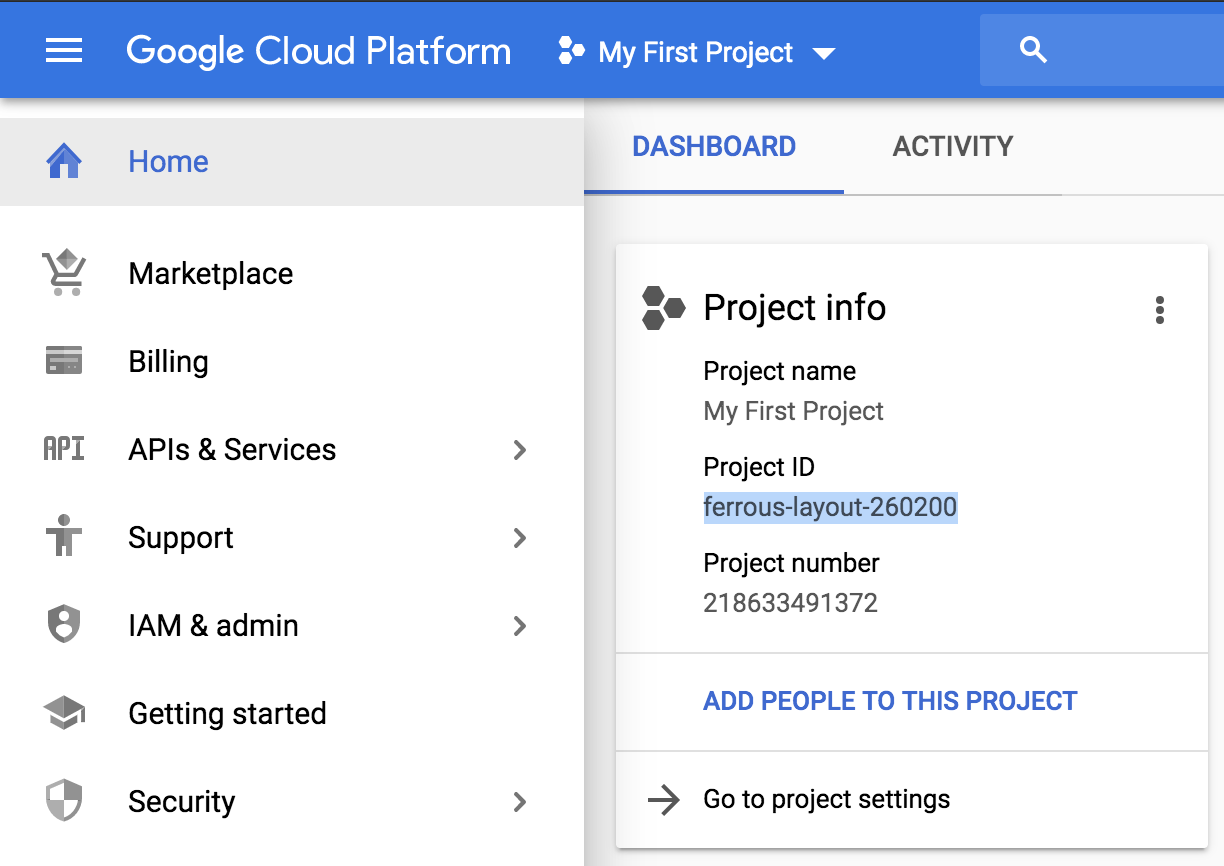
Figure 4-4. Location of the Project ID in the GCP console.
When you have your Project ID, run the following command in the Cloud Shell, substituting your own Project ID for the one shown here:
genomics_book@cloudshell:~$ gcloud config set project ferrous-layout-260200 Updated property [core/project]. genomics_book@cloudshell:~ (ferrous-layout-260200)$
Notice that your command prompt now includes your Project ID. It is quite long, so going forward, we’ll show only the last character in the prompt—in this case, the dollar sign ($)—when we demonstrate running commands. For example, if we list the contents of the working directory using the ls command, it will look like this:
$ ls README-cloudshell.txt
And, hey, there’s already something here: a README file, which, as the name indicates, really wants you to read it. You can do so by running the cat command:
$ cat README-cloudshell.txt
This displays a welcome message that summarizes some usage instructions and recommendations for getting help. And with that, you’re ready to use Cloud Shell to begin interacting with basic GCP services. Let’s get cracking!
Using gsutil to Access and Manage Files
Now that we have access to this extremely simple-to-launch and free (if fairly limited) VM, let’s use it to see whether we can access the bundle of example data provided with this book. The data bundle resides in Google Cloud Storage (GCS), which is a form of object store (i.e., it’s used for storing files) with units of storage called buckets. You can view the contents of GCS buckets and perform basic management tasks on them via the web through the storage browser section of the GCP console, but the interface is fairly limited. The more powerful approach is to use the gcloud tool, gsutil (Google Storage Utilities), from the command line. You can access buckets through their GCS path, which is just their name prefixed with gs://.
As an example, the path for the public storage bucket for this book is gs://genomics-in-the-cloud. You can list the contents of the bucket by typing the following command in your cloud shell:
$ gsutil ls gs://genomics-in-the-cloud gs://genomics-in-the-cloud/hello.txt gs://genomics-in-the-cloud/v1/
There should be a file called hello.txt. Let’s use the gsutil version of the Unix command cat, which allows us to read the content of text files to see what this hello.txt file contains:
$ gsutil cat gs://genomics-in-the-cloud/hello.txt HELLO, DEAR READER!
You can also try copying the file to your storage disk:
$ gsutil cp gs://genomics-in-the-cloud/hello.txt . Copying gs://genomics-in-the-cloud/hello.txt... / [1 files][ 20.0 B/ 20.0 B] Operation completed over 1 objects/20.0 B.
If you list the contents of your working directory by using ls again, you should now have a local copy of the hello.txt file:
$ ls hello.txt README-cloudshell.txt
While we’re playing with gsutil, how about we do something that will be useful later: create a storage bucket of your own, so that you can store outputs in GCS. You’ll need to substitute my-bucket in the command shown here because bucket names must be unique across all of GCS:
$ gsutil mb gs://my-bucket
If you didn’t change the bucket name or you tried a name that was already taken by someone else, you might get the following error message:
Creating gs://my-bucket/... ServiceException: 409 Bucket my-bucket already exists.
If this is the case, just try something else that’s more likely to be unique. You’ll know it worked when you see the Creating name... in the output and then get back to the prompt without any further complaint from gsutil. When that’s done, you’re going to create an environment variable that will serve as an alias for your bucket name. That way, you’ll save yourself some typing and you’ll be able to copy and paste subsequent commands without having to substitute the bucket name every time:
$ export BUCKET="gs://my-bucket"
You can run the echo command on your new variable to verify that your bucket name has been stored properly:
$ echo $BUCKET gs://my-bucket
Now, let’s get you comfortable with using gsutil. First, copy the hello.txt file to your new bucket. You can do either directly from the original bucket:
$ gsutil cp gs://genomics-in-the-cloud/hello.txt $BUCKET/
Or, you can do it from your local copy; for example, if you made modifications that you want to save:
$ gsutil cp hello.txt $BUCKET/
Finally, as one more example of basic file management, you can decide that the file should reside in its own directory in your bucket:
$ gsutil cp $BUCKET/hello.txt $BUCKET/my-directory/
As you can see, the gsutil commands are set up to be as similar as possible to their original Unix counterparts. So, for example, you’ll also be able to use -r to make the cp and mv commands recursive to apply to directories. For large file transfers, you can use a few cloud-specification optimizations to speed up the process, like the gsutil -m option, which parallelizes file transfers. Conveniently, the system will usually inform you in the terminal output when you could take advantage of such optimizations, so you don’t need to go and memorize the documentation before getting underway.
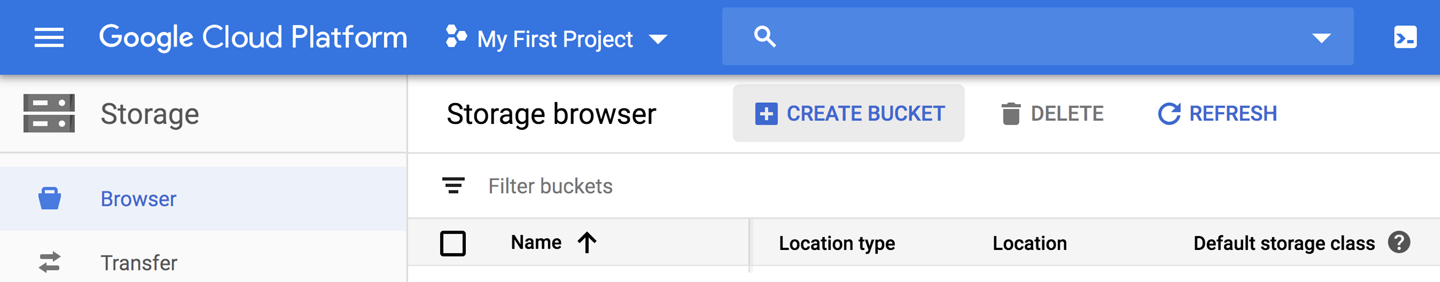
Figure 4-5. GCP console storage browser.
This opens a reasonably simple configuration form. The most important thing to do here is to choose a good name because the name you choose must be unique across all of Google Cloud—so be creative! If you choose a name that is already in use, the system will let you know when you click Continue in the configuration form, as demonstrated in Figure 4-6.
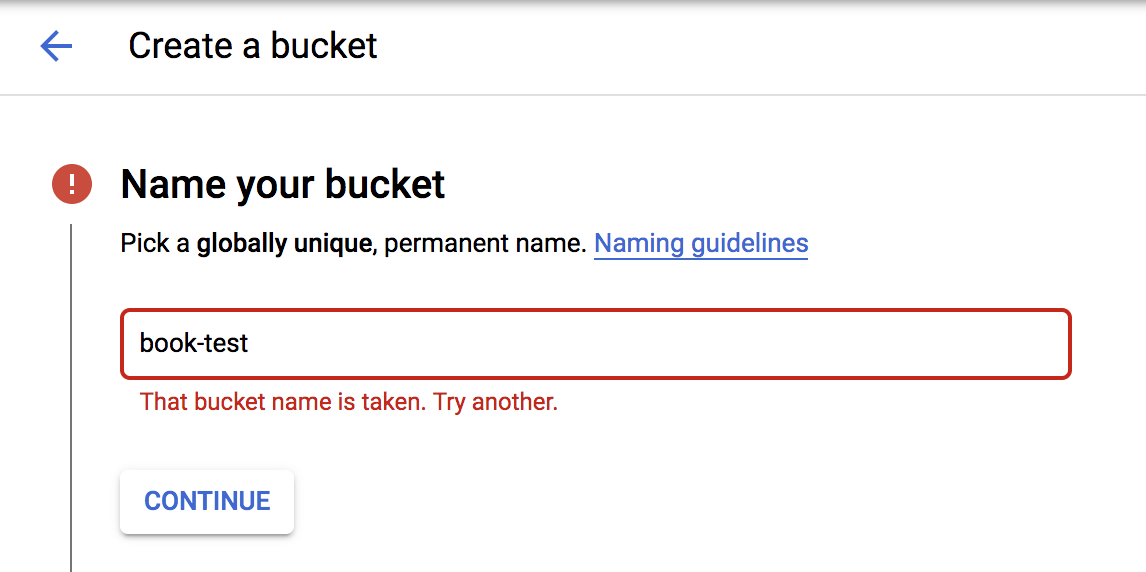
Figure 4-6. Naming your bucket.
When you have a unique name, the system will let you proceed to the next step by expanding the menu options. These allow you to customize the storage location and access controls for your bucket, but for the time being, feel free to just accept the defaults and click Create. Doing so will take you back to the list of buckets, which should at this point include your newly created one. You can click its name to view its contents—but of course it’s still empty, so the view won’t be particularly exciting, as illustrated in Figure 4-7.
The interface offers a few basic management options like deleting buckets and files as well as uploading files and folders. Note that you can even drag and drop files and folders from your local machine into the bucket contents window, which is stunningly easy (go ahead, try it), but it’s not something you can expect to do very often in the course of your genomics work. In the real world, you’re more likely to use the gsutil command-line utility. One of the advantages of using the command-line path is that you can save those commands as a script, for provenance and so that your steps can be reproduced if needed.
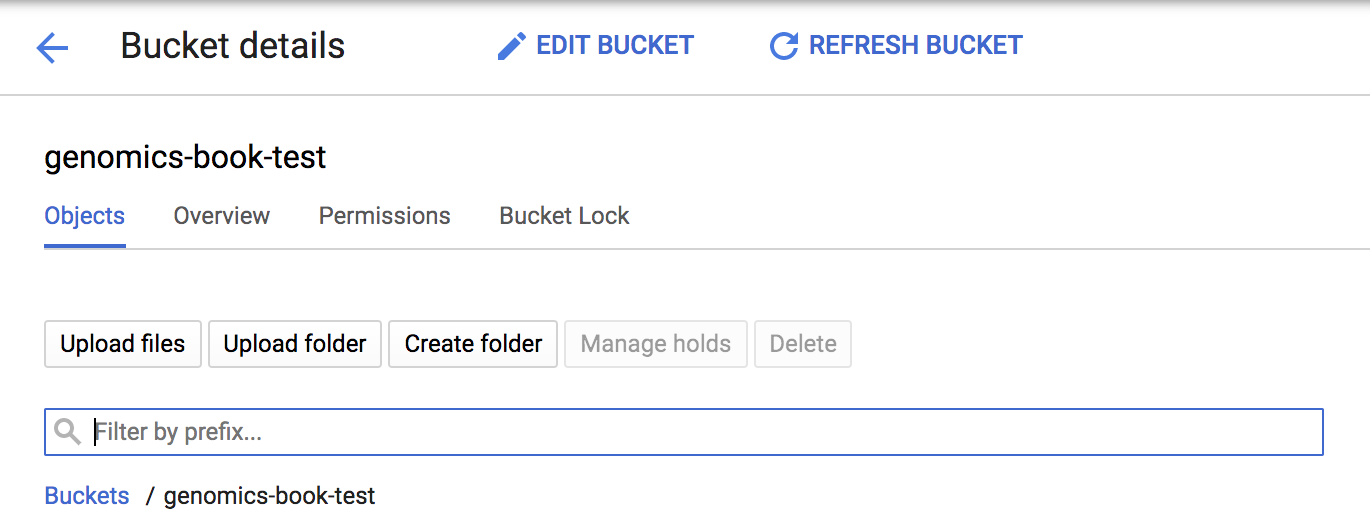
Figure 4-7. Viewing the contents of your bucket.
Pulling a Docker Image and Spinning Up the Container
Cloud Shell is the gift that keeps on giving: the Docker application (which we introduced in Chapter 3) comes preinstalled, so you can go ahead and get started with that, too! We’re going to use a simple Ubuntu container to illustrate basic Docker functionality. Although a Docker image is available for GATK—and that’s what we’re going to use for a good chunk of the next few chapters—we’re not going to use it here because it’s rather large, so it takes a little while to get going. We wouldn’t actually be able to run any realistic analyses with it in the free Cloud Shell because of the small amount of CPU and memory resources allocated for this free VM.
Note
The first thing to do to learn how to use Docker containers in this context is to...well, avoid the online Docker documentation! Seriously. Not because it’s bad, but because the majority of those documents are written mainly for people who want to run web applications in the cloud. If that’s what you want to do, more power to you, but you’re reading the wrong book. What we’re providing here are tailored instructions that will teach you how to use Docker to run research software in containers.
As just noted, we’re going to use a very generic example: an image containing the Ubuntu Linux OS. It’s an official image that is provided as part of the core library in a public container image repository, Docker Hub, so we just need to state its name. You’ll see later that images contributed by the community are prefixed by the contributor’s username or organization name. While still in your Cloud Shell terminal (it doesn’t matter where your working directory is), run the following command to retrieve the Ubuntu image from the Docker Hub library of official (certified) images:
$ docker pull ubuntu Using default tag: latest latest: Pulling from library/ubuntu 7413c47ba209: Pull complete 0fe7e7cbb2e8: Pull complete 1d425c982345: Pull complete 344da5c95cec: Pull complete Digest: sha256:d91842ef309155b85a9e5c59566719308fab816b40d376809c39cf1cf4de3c6a Status: Downloaded newer image for ubuntu:latest docker.io/library/ubuntu:latest
The pull command fetches the image and saves it to your VM. The version of the container image is indicated by its tag (which can be anything the image creator wants to assign) and by its sha256 hash (which is based on the image contents). By default, the system gives us the latest version that is available because we did not specify a particular tag; in a later exercise, you’ll see how to request a specific version by its tag. Note that container images are typically composed of several modular slices, which are pulled separately. They’re organized so that the next time you pull a version of the image, the system will skip downloading any slices that are unchanged compared to the version you already have.
Now let’s start up the container. There are three main options for running it, but the tricky thing is that there is usually only one correct way to do it as its author intended, and it’s difficult to know what that is if the documentation doesn’t specify it (which is soooo often the case). Confused? Let’s walk through the cases to make this a bit more concrete, and you’ll see why we’re putting you through this momentary frustration and mystery—it’s to save you potential misery down the road.
- First option
-
$ docker run ubuntu
- Result
-
A short pause, then your command prompt comes back. No output. What happened? Docker did in fact spin up the container, but the container wasn’t configured to do anything under those conditions, so it basically shrugged and shut down again.
- Second option
-
Run it with a command appended:
$ docker run ubuntu echo "Hello World!" Hello World!
- Result
-
It echoed
Hello World!, as requested, and then shut down again. OK, so now we know that we can pass commands to the container, and if it’s a command that is recognized by something in there, it will be executed. Then, when any and all commands have been completed, the container will shut down. A bit lazy, but reasonable. - Third option
-
Run it interactively by using the
-itoption:$ docker run -it ubuntu /bin/bash root@d84c079d0623:/#
- Result
-
Aha! A new command prompt (Bash in this case)! But with a different shell symbol:
#instead of$. This means that the container is running and you are in it. You can now run any command that you would normally use on an Ubuntu system, including installing new packages if you like. Try running a few Unix commands such aslsorls -lato poke around and see what the container can do. Later in the book, particularly in Chapter 12, we go into some of the implications of this, including practical instructions for how to package and redistribute an image you’ve customized in order to share your own analysis in a reproducible way.
When you’re done poking around, type exit at the command prompt (or press Ctrl+D) to terminate the shell. Because this is the main process the container was running, terminating it will cause the container to shut down and return to the Cloud Shell itself. To be clear, this will shut down the container and any commands that are currently running.
If you’re curious: yes, it is possible to step outside of the container without shutting it down; this is called detaching. To do so, press Ctrl+P+Q instead of using the exit command. You’ll then be able to jump back into the container at any time—provided that you can identify it. By default, Docker assigns your container a universally unique identifier (UUID) as well as a random human-readable name (which tend to sound a bit silly). You can run docker ps to list currently running containers or docker ps -a to list containers that have been created. This displays a list of containers indexed by their container IDs that should look something like this:
$ docker ps -a CONTAINER ID IMAGE COMMAND CREATED STATUS PORTS NAMES c2b4f8a0c7a6 ubuntu "/bin/bash" 5 minutes ago Up 5 minutes vigorous_rosalind 9336068da866 ubuntu "echo ’Hello World!’" 8 minutes ago Exited (0) 8 minutes ago objective_curie
We’re showing that two entries correspond to the last two invocations of Docker, each with a unique identifier, the CONTAINER ID. We see the container with ID c2b4f8a0c7a6 that is currently running was named vigorous_rosalind and has a status of Up 5 minutes. You can tell that the other container, objective_curie, is not running because its status is Exited (0) 8 minutes ago. The names we see here were randomly assigned (We swear! What are the odds?), so they’re admittedly not terribly meaningful. If you have multiple containers running at the same time, this can become a bit confusing, so you’ll want a better way to identify them. The good news is that you can give them a meaningful name by adding --name=meaningful_name immediately after docker run in your initial command, substituting meaningful_name with the name that you want to give the container.
To enter the container, simply run docker attach c2b4f8a0c7a6 (substituting your container ID), press Enter, and you will find yourself back at the helm (your keyboard might be labeled Return instead of Enter). You can open a second command tab in Cloud Shell if you’d like to be able to run commands outside the container alongside the work you’re doing inside the container. Note that you can have multiple containers running at the same time on a single VM—that’s one of the great advantages of the container system—but they will be competing for the CPU and memory resources of the VM, which in Cloud Shell are rather minimal. Later in this chapter, we show you how to spin up VMs with beefier capabilities.
Mounting a Volume to Access the Filesystem from Within the Container
Having completed the previous exercise, you are now able to retrieve and run an instance of any container image shared in a public repository. Many commonly used bioinformatics tools, including GATK, are available preinstalled in Docker containers. The idea is that knowing how to use them out of a Docker container means you won’t need to worry about having the correct OS or software environment. However, there’s still one trick that we need to show you in order to make that really work for you: how to access your machine’s filesystem from within the container by mounting a volume.
What does that last bit mean? By default, when you’re inside the container, you can’t access any data that resides on the filesystem outside of the container. The container is a closed box. There are ways to copy things back and forth between the container and your filesystem, but that becomes tedious really fast. So we’re going to follow the easier path, which is to establish a link between a directory outside the container in a way that makes it appear as if it were within the container. In other words, we’re going to poke a hole in the container wall, as shown in Figure 4-8.

Figure 4-8. Mounting a directory from your Google Cloud Shell VM into a Docker container: Ubuntu container used in this chapter (left); GATK container introduced in Chapter 5 (right).
As an example, let’s create a new directory called book in our Cloud Shell VM’s home directory, and put the hello.txt file from earlier inside it:
$ mkdir book $ mv hello.txt book/ $ ls book hello.txt
So this time, let’s run the command to spin up our Ubuntu container by using the -v argument (where v is for volume), which allows us to specify a filesystem location and a mount point within the container:
$ docker run -v ~/book:/home/book -it ubuntu /bin/bash
The -v ~/book_data:/home/book part of the command links the location you specified to the path /home/book directory within the Docker container. The /home part of the path is a directory that already exists in the container, whereas the book part can be any name you choose to give it. Now, everything in the book directory on your filesystem can be accessed from within the Docker container’s /home/book directory:
# ls home/book hello.txt
Here, we’re using the same name for the mount point as for the actual location we’re mounting because it’s more intuitive that way, but you could use a different name if you wanted. Note that if you give your mount point the name of a directory or file that already exists with that path in the container, it will “squash” the existing path, meaning that path will not be accessible for as long as the volume is mounted.
A few other Docker tricks are good to know, but for now, this is enough of a demonstration of the core Docker functionality that you’re going to use in Chapter 5. We go into the details of more sophisticated options as we encounter them.
Setting Up Your Own Custom VM
Now that you’ve successfully run some basic file-management commands and got the hang of interacting with Docker containers, it’s time to move on to bigger and better things. The Google Cloud Shell environment is excellent for quickly getting started with some light coding and execution tasks, but the VM allocated for Cloud Shell is really underpowered and will definitely not cut the mustard when it comes to running real GATK analyses in Chapter 5.
In this section, we show you how to set up your own VM in the cloud (sometimes called an instance) using Google’s Compute Engine service, which allows you to select, configure, and run VMs of whatever size you need.
Creating and Configuring Your VM Instance
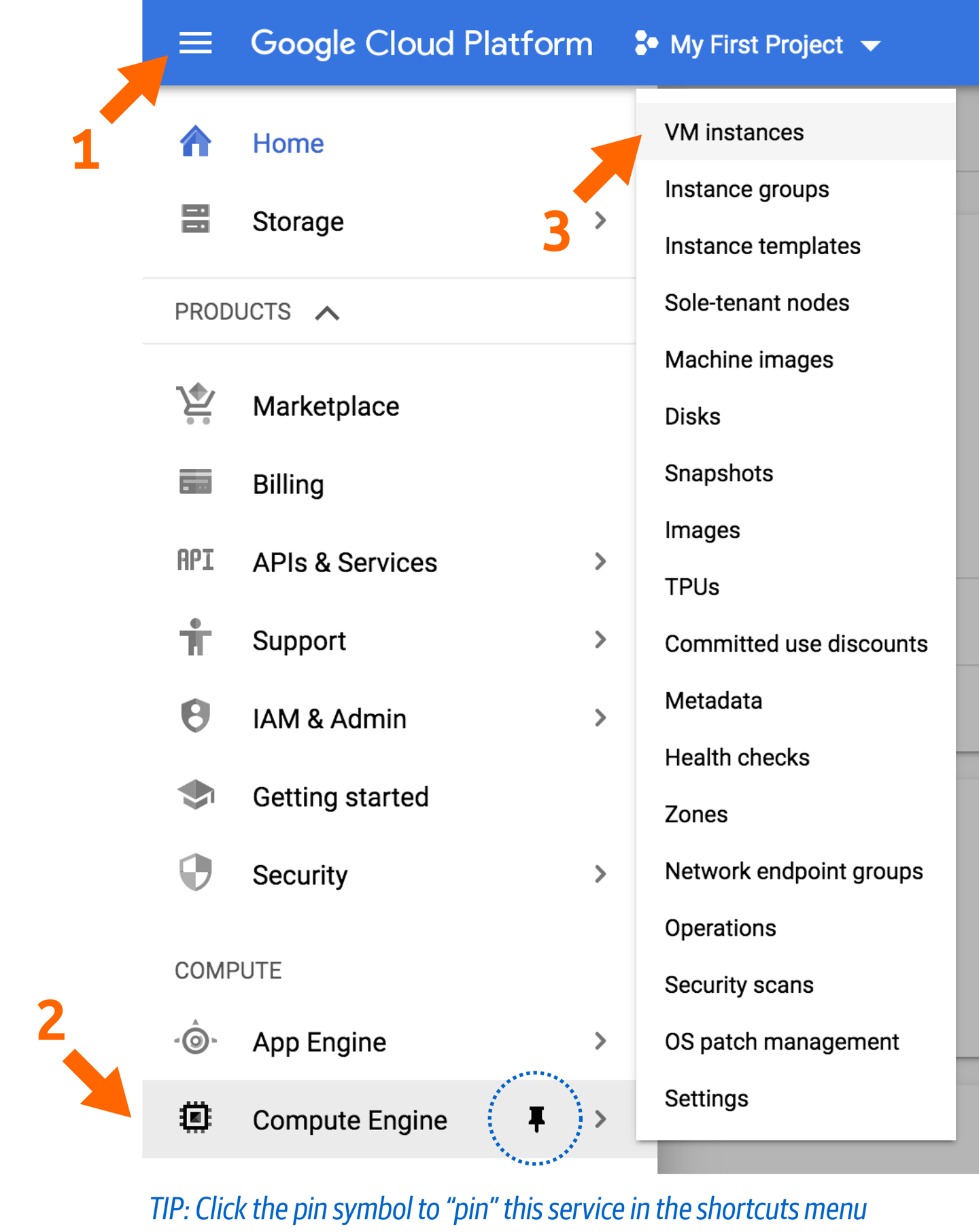
First, go to the Compute Engine or access the page through the sidebar menu on the left, as shown in Figure 4-9.
Click the VM Instances link in this menu to go to an overview of running images. If this is a new account, you won’t have any running. Notice at the top that there’s an option for Create Instance. Click that, and let’s walk through the process of creating a new VM with just the resources you need.
Next, in the top menu bar, click Create Instance, as shown in Figure 4-10. This brings up a configuration form, as shown in Figure 4-11.
Figure 4-10. Create a VM instance.
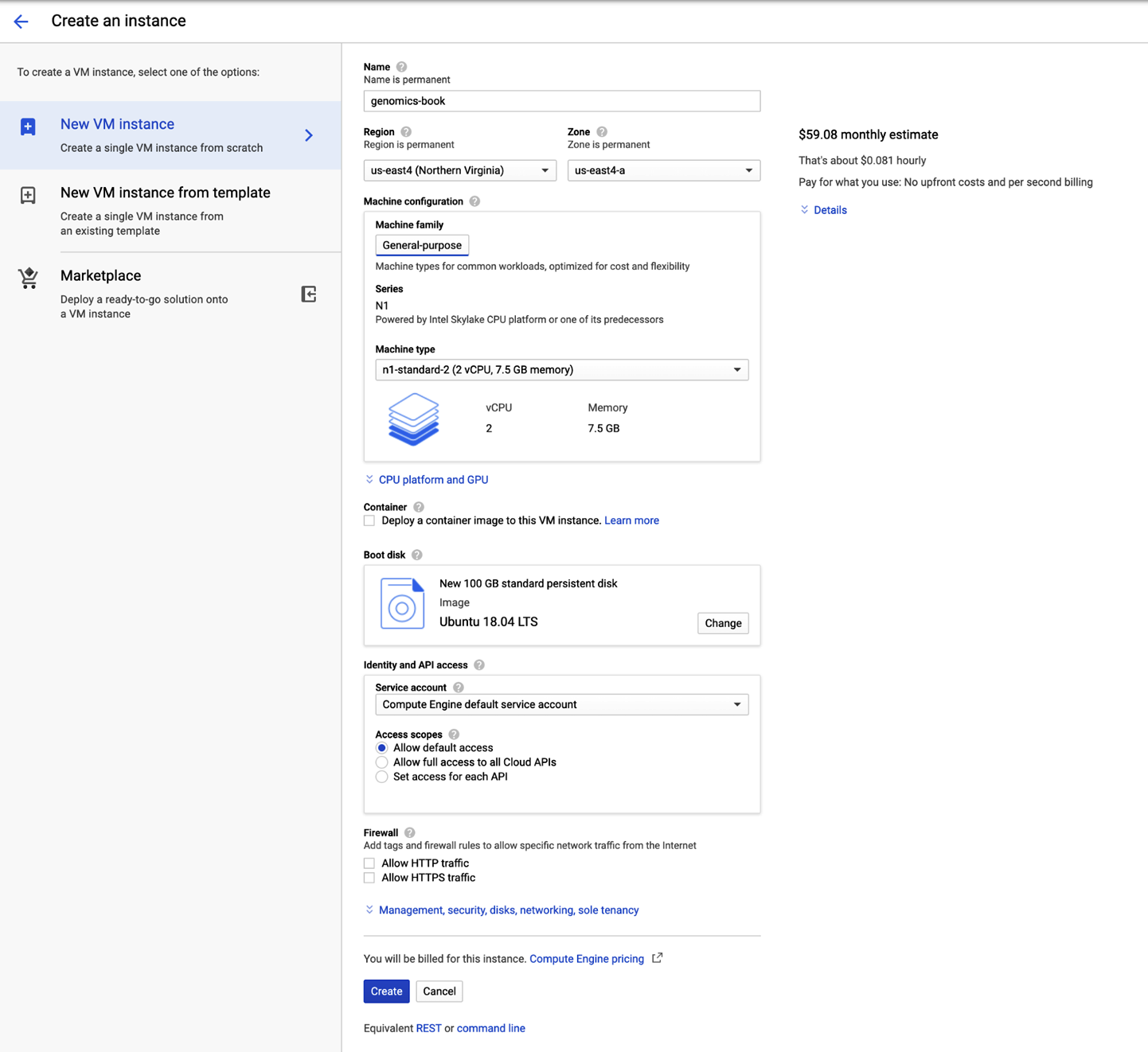
Figure 4-11. The VM instance configuration panel.
Follow the step-by-step instructions in the subsections that follow to configure the VM. There are tons of options and the process can be quite confusing if you don’t have experience with the terminology, so we mapped out the simplest path through the configuration form that will allow you to run all of the command exercises in the first few chapters of this book. Please make sure that you use exactly the same settings as shown here unless you really know what you’re doing.
Name your VM
Give your VM a name; for example, genomics-book, as shown in Figure 4-12. This must be unique within your project, but unlike bucket names, it does not need to be unique across GCP. Some people like to use their username so that others with access to the project can instantly identify who created the resource.

Figure 4-12. Name your VM instance.
Choose a region (important!) and zone (not so important)
There are different physical locations for the cloud. Like most commercial cloud providers, GCP maintains datacenters in many parts of the world and provides you with the option to choose which one you want to use. Regions are the top-level geographical distinction, with names that are reasonably descriptive (like us-west2, which refers to a facility in Los Angeles). Each region is further divided into two or more zones designated by single letters (a, b, c, etc.), which correspond to separate datacenters with their own physical infrastructure (power, network, etc.), though in some cases they might share the same building.
This system of regions and zones plays an important role in limiting the impact of localized problems like power outages, and all major cloud providers use some version of this strategy. For more on this topic, see this entertaining blog post by Kyle Galbraith about how cloud regions and zones (in his case, on AWS) could play an important role in the event of a zombie apocalypse.
Note
The ability to choose specific regions and zones for your projects is increasingly helpful for dealing with regulatory restrictions on where human-subjects data can be stored because it allows you to specify a compliant location for all storage and compute resources. However, some parts of the world are not yet well covered by cloud services or are covered differently by the various cloud providers, so you might need to factor in available datacenter locations when choosing a provider.
To choose a region for your project, you can consult the full list of available Google Cloud regions and zones and make a decision based on geographic proximity. Alternatively, you can use an online utility that measures how close you effectively are to each datacenter in terms of network response time, like http://www.gcping.com. For example, if we run this test from the small town of Sunderland in western Massachusetts (results in Table 4-1), we find that it takes 38 milliseconds to get a response from the us-east4 region located in Northern Virginia (698 km away), versus 41 milliseconds from the northamerica-northeast1 region located in Montreal (441 km away). This shows us that geographical proximity does not correlate directly with network region proximity. As an even more striking example, we find that we are quite a bit “closer” to the europe-west2 region in London (5,353 km away), with a response time of 102 milliseconds, than to the us-west2 region in Los Angeles (4,697 km away) which gives us a response time of 180 milliseconds.
| Region | Location | Distance (km) | Response (ms) |
|---|---|---|---|
| us-east4 | Northern Virginia, US | 698 | 38 |
| northamerica-northeast1 | Montreal | 441 | 41 |
| europe-west2 | London | 5,353 | 102 |
| us-west2 | Los Angeles | 4,697 | 180 |
This brings us back to our VM configuration. For the Region, we’re going to be using us-east4 (Northern Virginia) because it’s closest to the one of us who travels least (Geraldine), and for the Zone we just randomly choose us-east4-a. You need to make sure that you choose your region based on the preceding discussion, both for your own benefit (it will be faster) and to avoid clobbering that one datacenter in Virginia in the unlikely event that all 60,000 registered users of the GATK software begin working through these exercises at the same time—though that’s one way to test the vaunted “elasticity” of the cloud.
Select a machine type
This is where you can configure the resources of the VM you’re about to launch. You can control RAM as well as CPUs. For some instance types (available under Customize) you can even select VMs with GPUs, which are used to accelerate certain programs. The hitch is that what you select here will determine how much you’ll be billed per second of the VM’s uptime; the bigger and beefier the machine, the more it will cost you. The right side of the page should show how the hourly and monthly cost changes when you change the machine type. Note also that you’re billed for how long the VM is online, not for how much time you spend actually using it. We cover strategies for limiting costs later, but keep that in mind!
Here, select n1-standard-2; this is a fairly basic machine that’s not going to cost much at all, as shown in Figure 4-13.
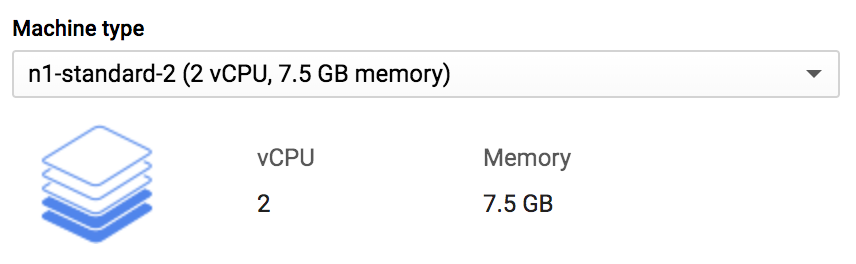
Figure 4-13. Selecting a machine type.
Specify a container? (nope)
We’re not going to fill this out. This is useful if you want to use a very specific setup using a custom container image that you’ve preselected or generated yourself. In fact, we could have preconfigured a container for you and skipped a bunch of setup that’s coming next. But then you wouldn’t have the opportunity to learn how to do those things for yourself, would you? So, for now, let’s just skip this option.
Customize the boot disk
Like Machine Type, this is another really useful setting. You can define two things here: the OS that you want to use and the amount of disk space you want. The former is especially important if you need to use a particular type and version of OS. And, of course, the latter is important if you don’t want to run out of disk space halfway through your analysis.
By default, the system proposes a particular flavor of Linux OS, accompanied by a paltry 10 GB of disk space, as shown in Figure 4-14. We’re going to need a bigger boat.
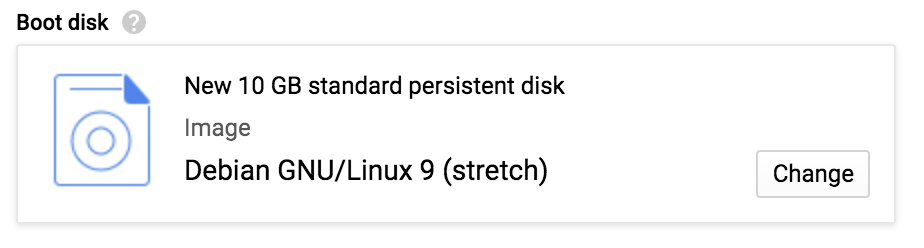
Figure 4-14. Choosing a boot disk size and image.
To access the settings menu for this, click Change. This opens a new screen with a menu of predefined options. You can also make your own custom images, or even find more images in Google Cloud Marketplace.
For our immediate purposes, we prefer Ubuntu 18.04 LTS, which is the most recent version of Ubuntu’s long-term release, as of this writing. It might not be as bleeding edge as Ubuntu 19.04, but the LTS, which stands for long-term support, guarantees that it’s being maintained for security vulnerabilities and package updates for five years from release. This Ubuntu image has a ton of what we already need, ready to go and installed, including various standard Linux tools and the GCP SDK command-line tools, which we will rely on quite heavily.
Select Ubuntu in the Operating System menu, then select Ubuntu 18.04 LTS in the version menu, as shown in Figure 4-15.
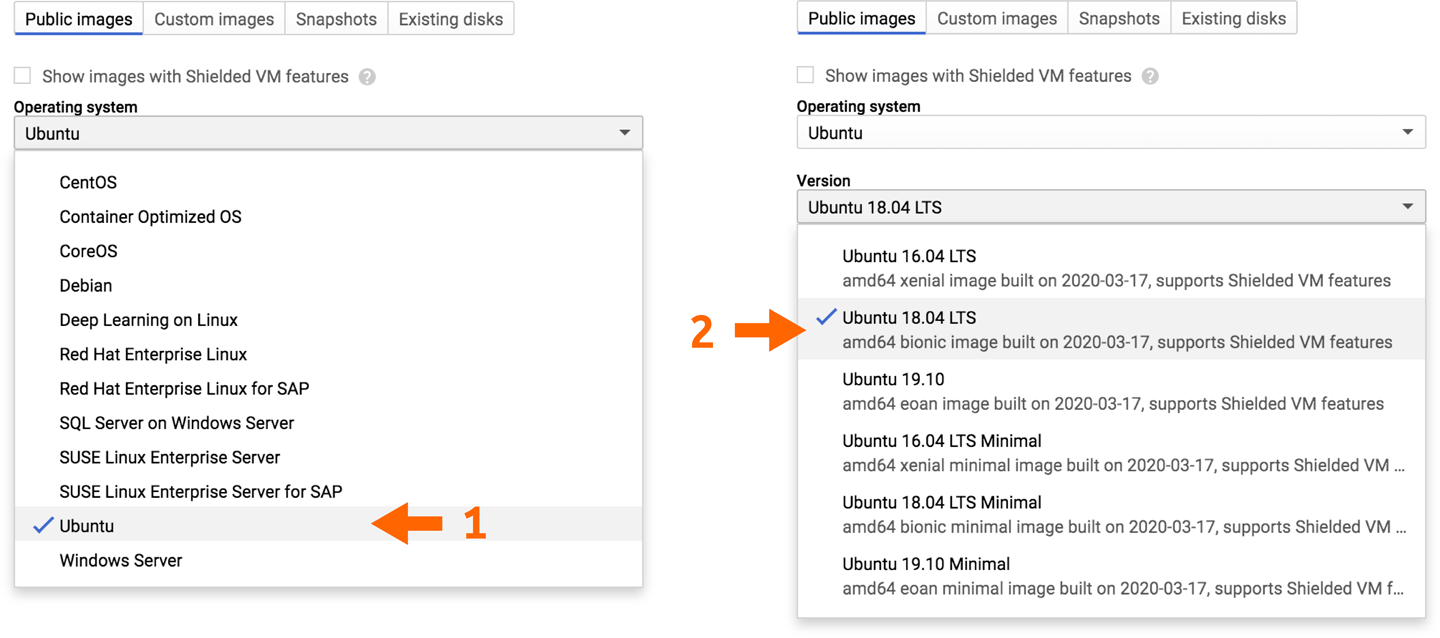
Figure 4-15. Selecting a base image.
At the bottom of the form, you can change the Boot disk Size to give yourself more space. As shown in Figure 4-16, go ahead and select 100 GB instead of the default 10 GB (the data we’re going to be working with can easily take up a lot of space). You can bump this up quite a bit more, depending on your dataset size and needs. Although you can’t easily adjust it after the VM launches, you do have the option of adding block storage volumes to the running instance after launch—think of it as the cloud equivalent of plugging in a USB drive. So if you run out of disk space, you won’t be totally stuck.
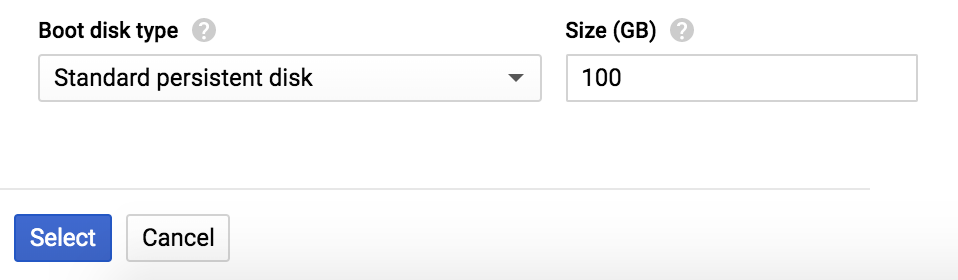
Figure 4-16. Setting the boot disk size.
After you’ve done all this, click Select; this closes the screen and returns you to the instance creation form where the “Boot disk” section should match the screenshot in Figure 4-17.
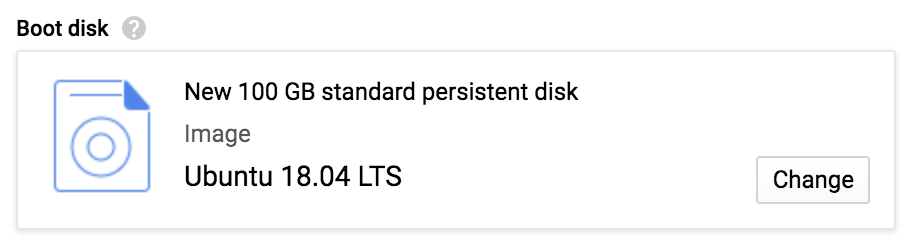
Figure 4-17. The updated boot disk selection.
At the bottom of the form, click Create. This returns you to the page that lists Compute Engine VM instances, including your newly created VM instance. You might see a spinning icon in front of its name while the instance is being created and booted up, and then a green circle with a checkmark will appear when it is running and ready for use, as shown in Figure 4-18.
Figure 4-18. Viewing the VM status.
Logging into Your VM by Using SSH
There are several ways that you can access the VM after it’s running, which you can learn about in the GCP documentation. We’re going to show you the simplest way to do it, using the Google Cloud console and the built-in SSH terminal. It’s hard to beat: as soon as you see a green checkmark in the Google Cloud console, you can simply click the SSH option to open a drop-down menu, as illustrated in Figure 4-19. Select the option “Open in a browser window,” and a few seconds later you should see an SSH terminal open to this VM.
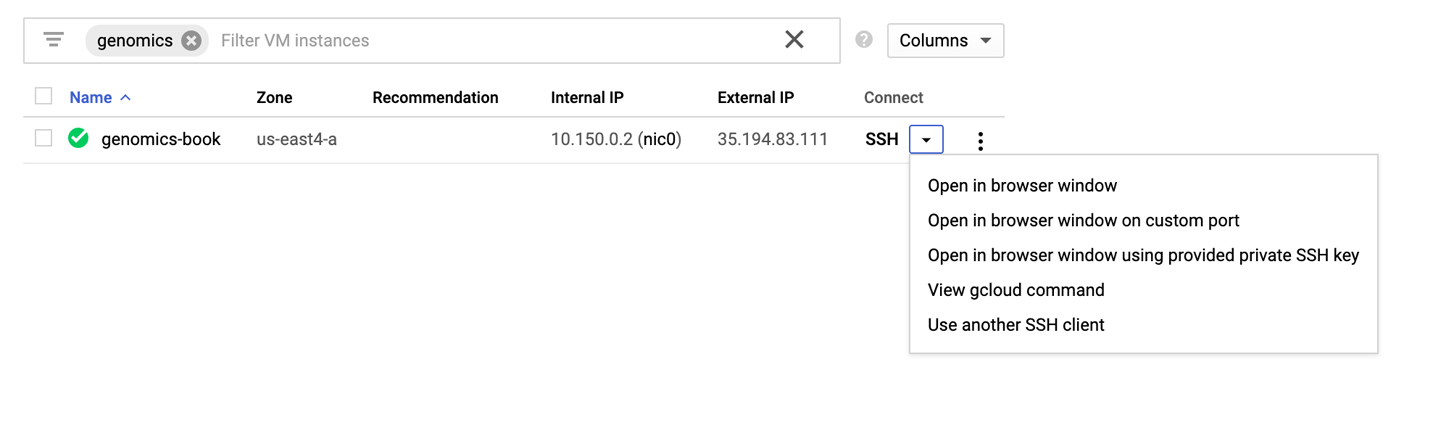
Figure 4-19. Options for SSHing into your VM.
This opens a new window with a terminal that allows you to run commands from within the VM instance, as shown in Figure 4-20. It might take a minute to establish the connection.
Figure 4-20. VM instance terminal.
Feel free to look around and get to know your brand-new VM; you’re going to spend a lot of time with it in the course of the next few chapters (but, like, in a good way).
Checking Your Authentication
You’re probably itching to run something interesting, but let’s begin by making sure your account credentials are set up properly so you can use the GCP command-line tools, which come preinstalled on the image we chose. In the SSH terminal, run the following command:
$ gcloud init Welcome! This command will take you through the configuration of gcloud. Your current configuration has been set to: [default] You can skip diagnostics next time by using the following flag: gcloud init --skip-diagnostics Network diagnostic detects and fixes local network connection issues. Checking network connection...done. Reachability Check passed. Network diagnostic passed (1/1 checks passed). Choose the account you would like to use to perform operations for this configuration: [1] XXXXXXXXXXX-compute@developer.gserviceaccount.com [2] Log in with a new account Please enter your numeric choice:
The line that starts with [1] shows you that by default, GCP has you logged in under a service account: the domain is @developer.gserviceaccount.com. This is fine for running tools within your VM, but if you want to be able to manage resources, including copying files out to GCS buckets, you need to do so under an account with the relevant permissions. It is possible to grant this service account all the various permissions that you’ll need for these exercises, but that would lead us a bit further into the guts of GCP account administration than we’d like to go at this juncture—we want to get you doing genomics work ASAP! So instead, let’s just use the original account that you used to create the project at the beginning of this chapter, given that it already has those permissions as a project owner.
To log in with that account, press 2 at the prompt. This triggers some interaction with the program; GCP will warn you that using your personal credentials on a VM is a security risk because if you give someone else access to the VM, they will be able to use your credentials:
You are running on a Google Compute Engine virtual machine. It is recommended that you use service accounts for authentication. You can run: $ gcloud config set account `ACCOUNT` to switch accounts if necessary. Your credentials may be visible to others with access to this virtual machine. Are you sure you want to authenticate with your personal account? Do you want to continue (Y/n)?
The solution: don’t share access to your personal VM.1
If you type Y for yes, the program will give you a link:
Go to the following link in your browser: https://accounts.google.com/o/oauth2/auth?redirect_uri=<...> Enter verification code:
When you click the link or copy and paste it into your browser, you are presented with a Google login page. Log in with the same account you used for the GCP to get your authentication code and then copy and paste that back into your terminal window. The gcloud utility will confirm your login identity and ask you to select the project ID you want to use from the list of projects you have access to. It will also offer the option to set your preferred compute and storage zone, which should match what you set earlier when you created the VM. If you’re not seeing what you expect in the project ID list, you can always double-check the resource management page in the GCP console.
Copying the Book Materials to Your VM
Throughout the next few chapters, you’re going to run real GATK commands and workflows on your VM, so you need to retrieve the example data, source code, and a couple of software packages. We’ve bundled most of that in a single place: a Cloud Storage bucket called genomics-in-the-cloud. The only piece that is separate is the source code, which we provide in GitHub.
First, you’re going to copy the data bundle from the bucket to your VM using gsutil, the GCP storage utility that we already used earlier in the Cloud Shell portion of this chapter. In your VM’s terminal window, make a new directory called book, and then run the gsutil command to copy the book data bundle to the storage space associated with your VM:
$ mkdir ~/book $ gsutil -m cp -r gs://genomics-in-the-cloud/v1/* ~/book/
This will copy about 10 GB of data to your VM’s storage, so it might take a few minutes even with the -m flag enabling parallel downloads. As you’ll see later, it is possible to run some analysis commands directly on files in Cloud Storage without copying them first, but we want to keep things as simple as possible in the beginning.
Now, go ahead and retrieve the source code from the public repository on GitHub. We’re making the code available there because it’s a highly popular platform for sharing code under version control, and we’re committed to providing long-term maintenance for the code we use in the book. To get a copy on your VM, first use cd to move into the newly created book directory and then use the git clone command to copy the contents of the repository:
$ cd ~/book $ git clone https://github.com/broadinstitute/genomics-in-the-cloud.git code
This creates a directory (~book/code) that includes all the sample code we use throughout the book. Not only that, but it will be set up as an active Git repository, so you can get the latest changes by running the git pull command in the code directory, as follows:
$ cd ~/book/code $ git pull
With that, you should now have the latest and greatest version of the book code. To find out what has changed since the original publication, check out the README text file in the code directory.
Installing Docker on Your VM
You’re going to be working with Docker on your VM, so let’s make sure that you can run it. If you simply run the command docker in the terminal, you’ll get an error message because Docker does not come preinstalled on the VM:
$ docker Command 'docker' not found, but can be installed with: snap install docker # version 18.09.9, or apt install docker.io See 'snap info docker' for additional versions.
The error message helpfully points out how to remedy the situation using a preinstalled package called snap, but we’re actually going to use a slightly different way of installing Docker: we’re going to download and run a script from the Docker website that will largely automate the installation process. This way, you’ll know what to do if you find yourself needing to install Docker somewhere that doesn’t have a built-in package manager option.
Run the following command to install Docker on the VM:
$ curl -sSL https://get.docker.com/ | sh
# Executing docker install script, commit: f45d7c11389849ff46a6b4d94e0dd1ffebca
32c1 + sudo -E sh -c apt-get update -qq >/dev/null
...
Client: Docker Engine - Community
Version: 19.03.5
...
If you would like to use Docker as a non-root user, you should now consider
adding your user to the "docker" group with something like:
sudo usermod -aG docker genomics_book
Remember that you will have to log out and back in for this to take effect!
WARNING: Adding a user to the "docker" group will grant the ability to run
containers which can be used to obtain root privileges on the
docker host.
Refer to https://docs.docker.com/engine/security/security/#docker-daemon-
attack-surface for more information.
This might take a little while to complete, so let’s take that time to examine the command in a bit more detail. First, we’re using a convenient little utility called curl (short for Client URL) to download the installation script from the Docker website URL we provided, with a few command parameters (-sSL) that instruct the program to follow any redirection links and save the output as a file. Then, we use the pipe character (|) to hand that output file over to a second command, sh, which means “run that script that we just gave you.” The first line of output lets you know what it’s doing: Executing docker install script (we omitted parts of the preceding output for brevity).
When it finishes, the script prompts you to run the usermod command in the example that follows in order to grant yourself the ability to run Docker commands without using sudo each time. Invoking sudo docker can result in output files being owned by root, making it difficult to manage or access them later, so it’s really important to do this step:
$ sudo usermod -aG docker $USER
This does not produce any output; we’ll test in a minute whether it worked properly. First, however, you need to log out of your VM and then back in again. Doing so will make the system reevaluate your Unix group membership, which is necessary for the change you just made to take effect. Simply type exit (or press Ctrl+D) at the command prompt:
$ exit
This closes the terminal window to your VM. Go back to the GCP console, find your VM in the list of Compute Engine instances, and then click SSH to log back in again. This probably feels like a lot of hoops to jump through, but hang in there; we’re getting to the good part.
Setting Up the GATK Container Image
When you’re back in your VM, test your Docker installation by pulling the GATK container, which we use in the very next chapter:
$ docker pull us.gcr.io/broad-gatk/gatk:4.1.3.0 4.1.3.0: Pulling from us.gcr.io/broad-gatk/gatk ae79f2514705: Pull complete 5ad56d5fc149: Pull complete 170e558760e8: Pull complete 395460e233f5: Pull complete 6f01dc62e444: Pull complete b48fdadebab0: Pull complete 16fb14f5f7c9: Pull complete Digest: sha256:e37193b61536cf21a2e1bcbdb71eac3d50dcb4917f4d7362b09f8d07e7c2ae50 Status: Downloaded newer image for us.gcr.io/broad-gatk/gatk:4.1.3.0 us.gcr.io/broad-gatk/gatk:4.1.3.0
As a reminder, the last bit after the container name is the version tag, which you can change to get a different version than what we’ve specified here. Note that if you change the version, some commands might no longer work. We can’t guarantee that all code examples are going to be future-compatible, especially for the newer tools, some of which are still under active development. As noted earlier, for updated materials, see this book’s GitHub repository.
The GATK container image is quite large, so the download might take a little while. The good news is that next time you need to pull a GATK image (e.g., to get another release), Docker will pull only the components that have been updated, so it will go faster.
Note
Here we’re pulling the GATK image from the Google Container Repository (GCR) because GCR is on the same network as the VM we’re running on, so it will be faster than pulling it from Docker Hub. However, if you’re working on a different platform, you might find it faster to pull the image from the GATK repository on Docker Hub. To do so, change the us.gcr.io/broad-gatk part of the image path to just broadinstitute.
Now, remember the instructions you followed earlier in this chapter to spin up a container with a mounted folder? You’re going to use that again to make the book directory accessible to the GATK container:
$ docker run -v ~/book:/home/book -it us.gcr.io/broad-gatk/gatk:4.1.3.0 /bin/bash
You should now be able to browse the book directory that you set up in your VM from within the container. It will be located under /home/book. Finally, to double-check that GATK itself is working as expected, try running the command gatk at the command line from within your running container. If everything is working properly, you should see some text output that outlines basic GATK command-line syntax and a few configuration options:
# gatk
Usage template for all tools (uses --spark-runner LOCAL when used with a Spark tool)
gatk AnyTool toolArgs
Usage template for Spark tools (will NOT work on non-Spark tools)
gatk SparkTool toolArgs [ -- --spark-runner <LOCAL | SPARK | GCS> sparkArgs ]
Getting help
gatk --list Print the list of available tools
gatk Tool --help Print help on a particular tool
Configuration File Specification
--gatk-config-file PATH/TO/GATK/PROPERTIES/FILE
gatk forwards commands to GATK and adds some sugar for submitting spark jobs
--spark-runner <target> controls how spark tools are run
valid targets are:
LOCAL: run using the in-memory spark runner
SPARK: run using spark-submit on an existing cluster
--spark-master must be specified
--spark-submit-command may be specified to control the Spark submit command
arguments to spark-submit may optionally be specified after --
GCS: run using Google cloud dataproc
commands after the -- will be passed to dataproc
--cluster <your-cluster> must be specified after the --
spark properties and some common spark-submit parameters will be translated
to dataproc equivalents
--dry-run may be specified to output the generated command line without running it
--java-options 'OPTION1[ OPTION2=Y ... ]'' optional - pass the given string of options to
the java JVM at runtime.
Java options MUST be passed inside a single string with space-separated values
We discuss what that all means in loving detail in Chapter 5; for now, you’re done setting up the environment that you’ll be using to run GATK tools over the course of the next three chapters.
Stopping Your VM…to Stop It from Costing You Money
The VM you just finished setting up is going to come in handy throughout the book; you’ll come back to this VM for many of the exercises in the next few chapters. However, as long as it’s up and running, it’s costing you either credits or actual money. The simplest way to deal with that is to stop it: put it on pause whenever you’re not actively using it.
You can restart it on demand; it just takes a minute or two to get it back up and running, and it will retain all environment settings, the history of what you ran previously, and whatever data you have in local storage. Note that you will be charged a small fee for that storage even while the VM is not running and you’re not getting charged for the VM itself. In our opinion, this is well worth it for the convenience of being able to come back to your VM after an arbitrary amount of time and just pick up your work where you left off.
To stop your VM, in the GCP console, go to the VM instances management page, as shown previously. Find your instance and click the vertical three-dot symbol on the right to open the menu of controls, and then select Stop, as shown in Figure 4-21. The process might take a couple of minutes to complete, but you can safely navigate away from that page. To restart your instance later on, just follow the same steps but click Start in the control menu.
Figure 4-21. Stopping, starting, or deleting your VM instance.
Alternatively, you can delete your VM entirely, but keep in mind that deleting the VM will delete all locally stored data too, so make sure you save anything you care about to a storage bucket first.
Configuring IGV to Read Data from GCS Buckets
Just one more small step remains before you move on to the next chapter: we’re going to install and configure a genome browser called Integrated Genome Viewer (IGV) that can work directly with files in GCP. That will allow you to examine sequence data and variant calls without needing to copy the files to your local machine.
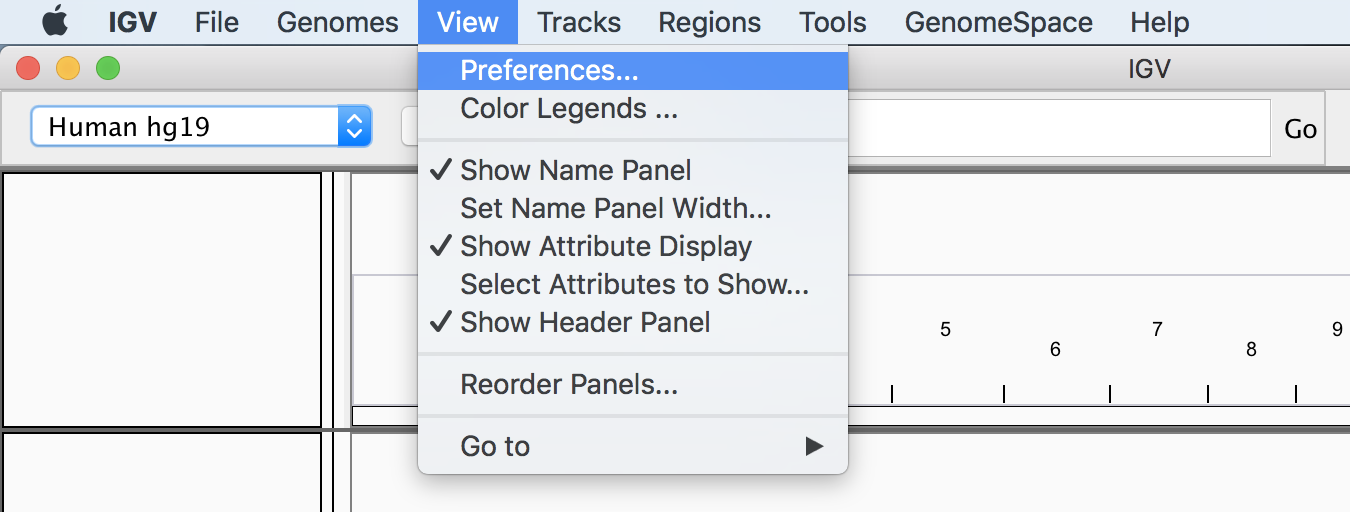
First, if you don’t have it installed yet on your local machine, get the IGV program from the website and follow the installation instructions. If you already have a copy, consider updating it to the latest version; we are using 2.7.2 (macOS version). Once you have the application open, choose View > Preferences from the top menu bar, as shown in Figure 4-22.
This opens the Preferences pane, shown in Figure 4-23.
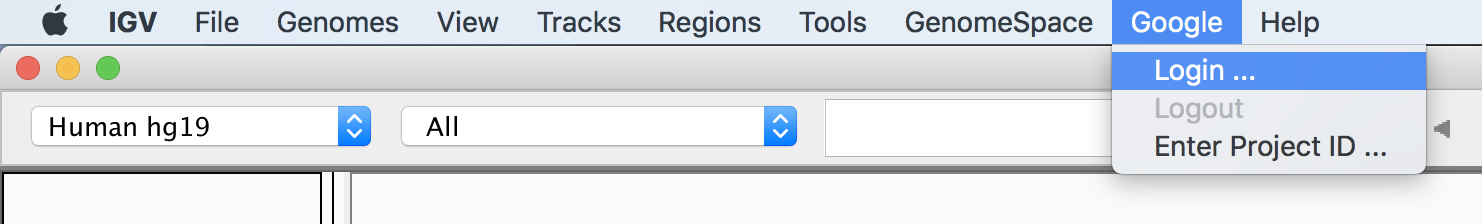
In the Preferences pane, Select the “Enable Google access” checkbox, click Save, and then quit IGV and reopen it to force a refresh of the top menu bar. You should now see a Google menu item that was not there previously; click it and select Login, as shown in Figure 4-24, to set up IGV with your Google account credentials.
Figure 4-23. The IGV Preferences pane.
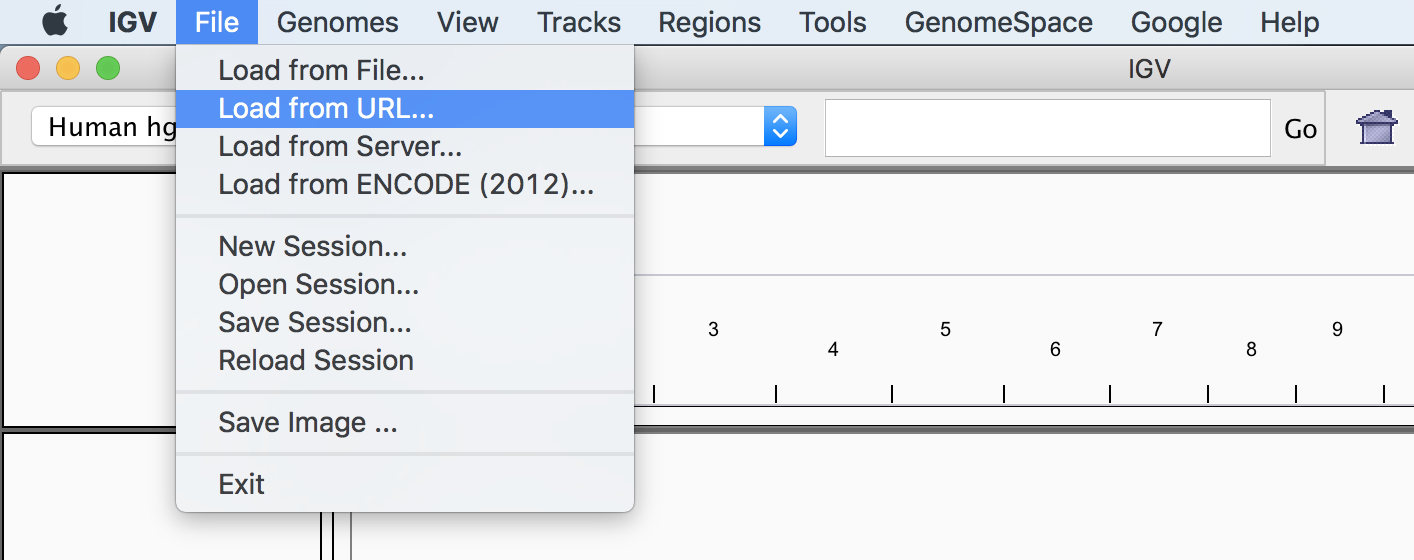
This will take you to a Google login page in your web browser; follow the prompts to allow IGV to access relevant permissions on your Google account. When this is complete, you should see a web page that simply says OK. Let’s switch back to IGV and test that it works. From the top-level menu, click Files > Load from URL, as shown in Figure 4-25, making sure not to select one of the other options by mistake (they look similar, so it’s easy to get tripped up). Make sure also that the reference drop-down menu in the upper-left corner of the IGV window is set to “Human hg19.”
Note
If you’re confused about what is different between the human references, see the notes in “The Reference Genome as Common Framework” about hg19 and GRCh38.
Finally, enter the GCS file path for one of the sample BAM files we provide in the book data bundle in the dialog window that pops up (e.g., mother.bam, as shown in Figure 4-26), and then click OK. Remember, you can get a list of files in the bucket by using gsutil from your VM or from Cloud Shell, or you can browse the contents of the bucket by using the Google Cloud console storage browser. If you use the browser interface to get the path to the file, you’ll need to compose the GCS file path by stripping off the first part of the URL before the bucket name; for instance, remove https://console.cloud.google.com/storage/browser and replace that with gs://. Do the same for the BAM’s accompanying index file, which should have the same filename and path but ends in .bai.2
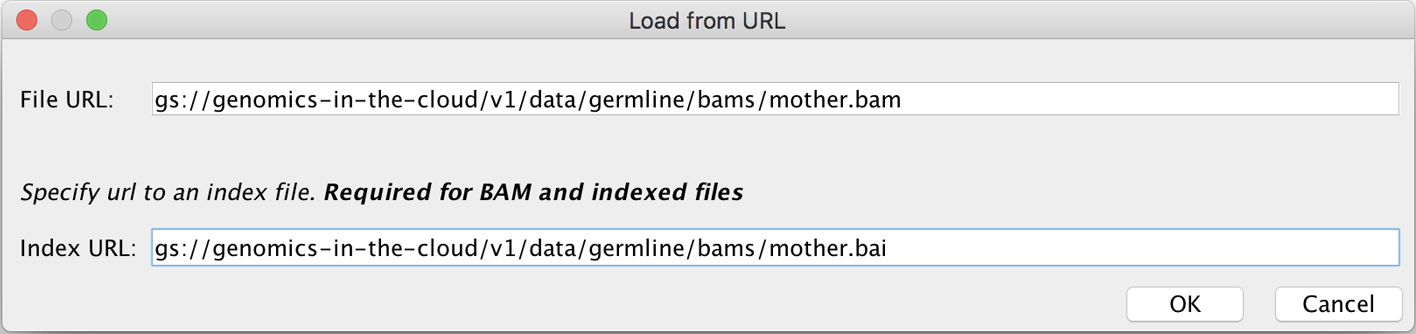
Figure 4-26. The Load from URL dialog box.
This will make the data available to you in IGV as a new data track, but by default nothing will be loaded in the main viewer. To check that you can view data, in the search window, enter the genomic coordinates 20:9,999,830-10,000,170 and then click Go. These coordinates will take you to the 10 millionth DNA base ±170 on the 20th human chromosome, as shown in Figure 4-27, where you’ll see the left-side edge of the slice of sequence data that we provide in this sample file. We explain in detail how to interpret the visual output of IGV in Chapter 5, when we use it to investigate the result of a real (small) analysis.
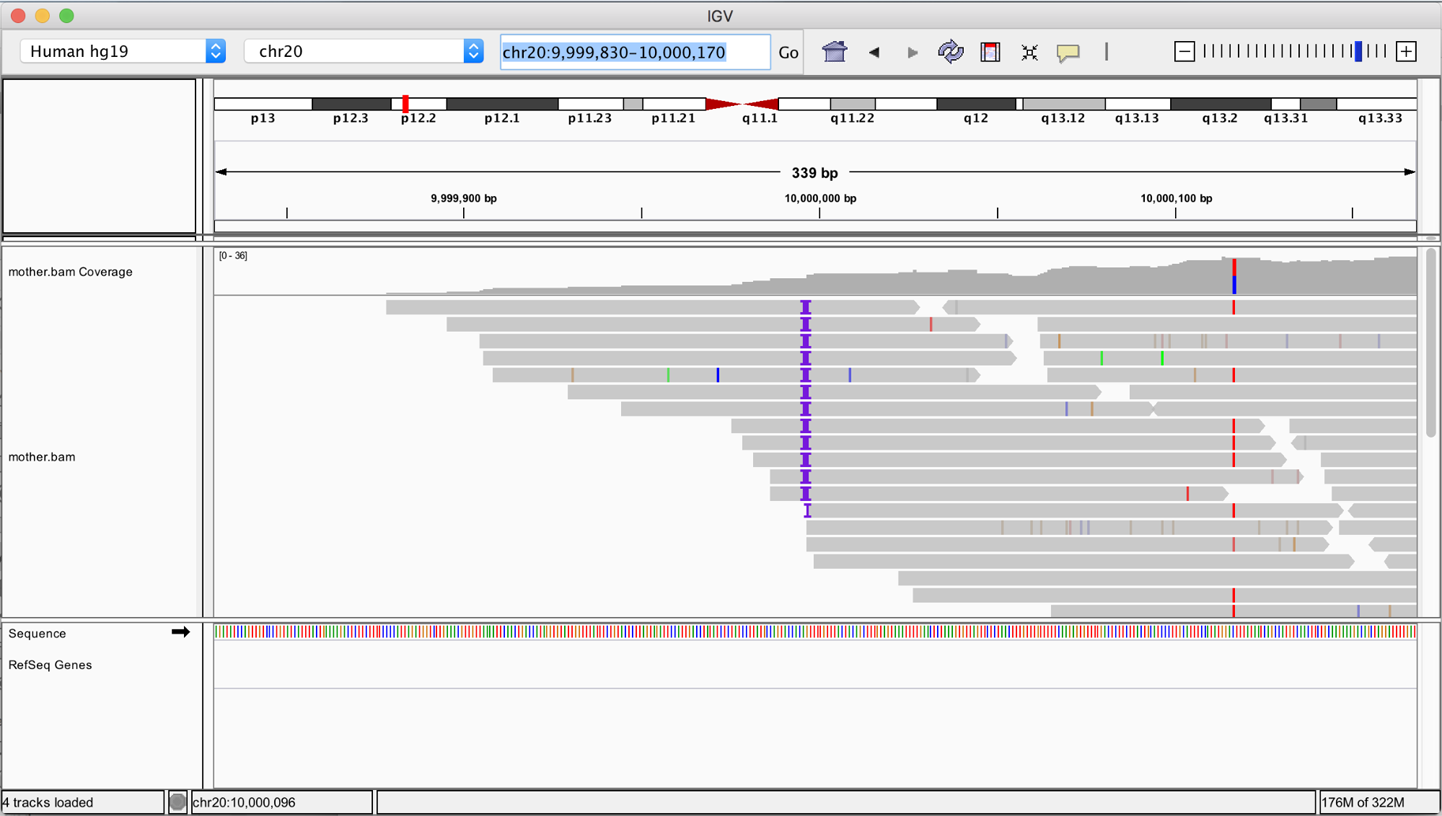
Figure 4-27. IGV view of a BAM file located in a GCS bucket.
IGV retrieves only small slices of data at a time, so the transfer should be very fast unless you have a particularly slow internet connection. Do keep in mind, however, that GCP, like all commercial cloud providers, will charge an egress fee for transferring data out of the cloud. On the bright side, it’s a small fee, proportional to the amount of data you transfer. So the cost of viewing slices of data in IGV is trivial—on the order of fractions of pennies—and it is definitely preferable to what it would cost to transfer the entire file for offline browsing!
You can view the contents of other data files, like VCF files, using the same set of operations, as long as the files are stored in a GCP bucket. Unfortunately, it means that this won’t work for files that are on the local storage of your VM, so anytime you want to examine one of those, you’ll need to copy it to a bucket first. You’re going to get really friendly with gsutil in no time.
Oh, one last thing while you have IGV open: click the little yellow callout bubble in the IGV window toolbar, which controls the behavior of the detail viewer, as shown in Figure 4-28. Do yourself a favor and switch the setting from Show Details on Hover to Show Details on Click. Whichever action you choose will trigger the appearance of little dialog that gives you detailed information about any part of the data that you either click or hover over; for example, for a sequence read, it will give you all the mapping information as well as the full sequence and base qualities. You can try it out now with the data you just loaded. As you’ll see, the detail display functionality in and of itself is very convenient, but the “on Hover” version of this behavior can be a bit overwhelming when you’re new to the interface; hence, our recommendation to switch to “on Click.”
Figure 4-28. Changing the behavior of the detail viewer from “on Hover” to “on Click.”
Wrap-Up and Next Steps
In this chapter, we showed you how to get started with GCP resources, from creating an account, using the super-basic Cloud Shell, and then graduating to your own custom VM. You learned how to manage files in GCS, run Docker containers, and administer your VM. Finally, you retrieved the book data and source code, finished setting up your custom VM to work with the GATK container, and set up IGV to view data stored in buckets. In Chapter 5, we get you started with GATK itself, and before you know it, you’ll be running real genomics tools on example data in the cloud.
1 Keep in mind that if you create accounts for other users in your GCP project, they will be able to SSH to your VMs as well. It is possible to further restrict access to your VMs in a shared project, but that is beyond the simple introduction we’re presenting here.
2 For example, https://console.cloud.google.com/storage/browser/genomics-in-the-cloud/v1/data/germline/bams/mother.bam becomes gs://genomics-in-the-cloud/v1/data/germline/bams/mother.bam.