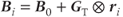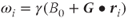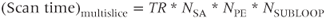Chapter 4
Principles of magnetic resonance imaging – 1
Chapter 2 described the relationship between the frequency of energy that a proton absorbs and the magnetic field strength that it experiences. MRI uses this field dependence to localize these proton frequencies to different regions of space. This idea of using the field dependence to spatially localize the protons earned the 2003 Nobel Prize in Physiology or Medicine for Paul Lauterbur and Sir Peter Mansfield, and helped transform MR from a niche industry serving research labs into a multibillion-dollar industry serving hospitals worldwide.
4.1 Gradient fields
In MRI, the magnetic field is made spatially dependent through the application of magnetic field gradients. These gradients are relatively small perturbations superimposed on the main magnetic field  , with a typical imaging gradient producing a total field variation of less than 1%. They are also linear perturbations to
, with a typical imaging gradient producing a total field variation of less than 1%. They are also linear perturbations to 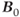 , so that the exact magnetic field is linearly dependent on the location inside the magnet:
, so that the exact magnetic field is linearly dependent on the location inside the magnet:
where 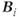 is the magnetic field at location
is the magnetic field at location 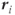 and
and 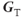 is the total gradient amplitude, mathematically represented as a tensor. Gradients are also applied for short periods of time during a scan and are referred to as gradient pulses.
is the total gradient amplitude, mathematically represented as a tensor. Gradients are also applied for short periods of time during a scan and are referred to as gradient pulses.
The presence of linear magnetic field gradients requires an expanded version of the Larmor equation given in equation (1.1):
where 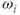 is the frequency of the proton at position
is the frequency of the proton at position 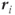 and
and  is a vector representing the total gradient amplitude and direction. The units of measure for
is a vector representing the total gradient amplitude and direction. The units of measure for 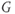 are expressed in millitesla per meter
are expressed in millitesla per meter 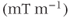 or gauss per centimeter
or gauss per centimeter 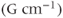 , where
, where 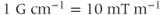 . Equation 4.2 states that, in the presence of a gradient field, each proton will resonate at a unique frequency that depends on its exact position within the gradient field. The MR image is simply a frequency and phase map of the protons generated by unique magnetic fields at each point throughout the image. The displayed image consists of digital picture elements (pixels) that represent volume elements (voxels) of tissue. The pixel intensity is related to the number of protons contained within the voxel weighted by the tissue characteristics, like T1 and T2 relaxation times, for the tissues within the voxel according to the pulse sequence utilized.
. Equation 4.2 states that, in the presence of a gradient field, each proton will resonate at a unique frequency that depends on its exact position within the gradient field. The MR image is simply a frequency and phase map of the protons generated by unique magnetic fields at each point throughout the image. The displayed image consists of digital picture elements (pixels) that represent volume elements (voxels) of tissue. The pixel intensity is related to the number of protons contained within the voxel weighted by the tissue characteristics, like T1 and T2 relaxation times, for the tissues within the voxel according to the pulse sequence utilized.
4.2 Slice selection
The slice orientation is determined by the particular physical gradient or gradients defined as the logical slice selection gradient. The slice orientation is defined so that the gradient orientation is perpendicular or normal to the surface of the slice, so that every proton within the slice experiences the same total magnetic field (to within the bandwidth) regardless of its position within the slice. Orthogonal slices are those in which only the 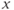 ,
,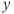 , or
, or  gradient is used as the slice selection gradient. Oblique slices, those not in one of the principal directions, are obtained by applying more than one physical gradient when the RF pulse is broadcast. The total gradient amplitude, whether from one, two, or three physical gradients, determines the slice thickness as shown in equation (4.3). When images are viewed on the monitor or film, the slice selection direction is always perpendicular to the surface; that is, hidden from the viewer (Figure 4.4).
gradient is used as the slice selection gradient. Oblique slices, those not in one of the principal directions, are obtained by applying more than one physical gradient when the RF pulse is broadcast. The total gradient amplitude, whether from one, two, or three physical gradients, determines the slice thickness as shown in equation (4.3). When images are viewed on the monitor or film, the slice selection direction is always perpendicular to the surface; that is, hidden from the viewer (Figure 4.4).
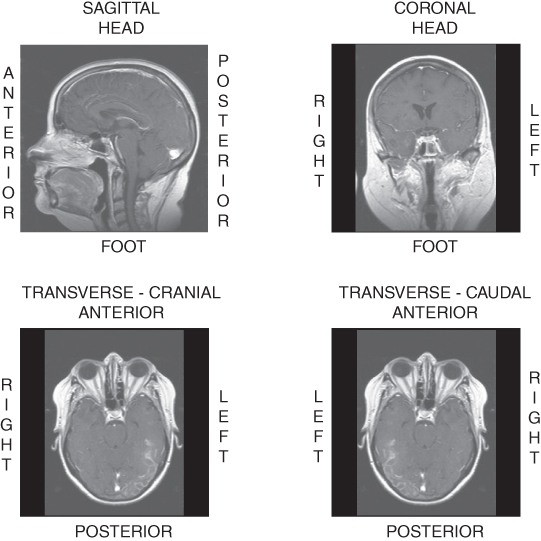
Figure 4.4 Images in standard slice orientations: sagittal, coronal, and transverse or axial. For transverse images, two view directions are possible: cranial and caudal. Image annotations are based on patient axes.
4.3 Readout or frequency encoding
In an MR image, the resolution may be expressed in one of two ways: the spatial resolution and the frequency resolution. The spatial resolution, expressed as the voxel size with units of mm/pixel, is derived from two user parameters, 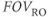 and the number of readout sample points in the acquisition matrix,
and the number of readout sample points in the acquisition matrix, 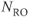 :
:
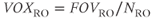
The frequency resolution, with units of Hz/pixel, is based on 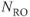 and
and 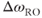 for the image:
for the image:

It is possible to improve the frequency resolution for the measurement independent of the spatial resolution by increasing the total sampling time used to measure the signal. This reduces the Nyquist frequency for the image and the background noise contributing to the measurement. In order to maintain the correct spatial resolution within the image, 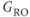 is reduced, in accordance with equation 4.4.
is reduced, in accordance with equation 4.4.
4.4 Phase encoding
The third direction in an MR image is the phase encoding direction. It is visualized along with the readout direction in a two-dimensional (2D) image (see Figure 4.5). The phase encoding gradient, 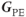 , is perpendicular to both
, is perpendicular to both 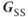 and
and  and is the only gradient that changes amplitude during the data acquisition loop of a standard 2D imaging sequence. Any signal variation detected from one acquisition to the next is assumed to be caused by the influence of
and is the only gradient that changes amplitude during the data acquisition loop of a standard 2D imaging sequence. Any signal variation detected from one acquisition to the next is assumed to be caused by the influence of 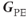 during the measurement.
during the measurement.
Because of the two different physical processes involved,  is not required to be the same as the
is not required to be the same as the 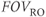 , nor is the voxel size. The resulting pixel dimensions also need not be equal. The readout pixel size divided by the phase encoding pixel size is known as the aspect ratio between the two dimensions. An aspect ratio of 1.0 (100%) means that the pixel size is the same in both directions, a situation referred to as isotropic pixels. An aspect ratio less than 1.0 (<100%) is referred to as anisotropic pixels, with the phase encoding dimension typically larger than the readout dimension.
, nor is the voxel size. The resulting pixel dimensions also need not be equal. The readout pixel size divided by the phase encoding pixel size is known as the aspect ratio between the two dimensions. An aspect ratio of 1.0 (100%) means that the pixel size is the same in both directions, a situation referred to as isotropic pixels. An aspect ratio less than 1.0 (<100%) is referred to as anisotropic pixels, with the phase encoding dimension typically larger than the readout dimension.
4.5 Sequence looping
The previous two sections described the individual steps used for spatial localization of the MR signal to a point within a slice. For most MR applications, information from many slices is measured during the scan in order to acquire images from large volumes of tissue. Several approaches are used for data acquisition that balance the desire for good spatial resolution and contrast-to-noise ratio (signal difference relative to background noise) while maintaining reasonable scan times.
In order to accomplish efficient data collection with minimal computer processing, most MRI techniques use some form of repetitive execution, which is achieved using computer instructions known as loops. This allows common instructions such as fixed amplitude gradient pulses (e.g., readout or slice selection gradient pulses) to be programmed one time yet provide a convenient method for modifying variable quantities such as phase encoding gradient amplitudes or RF pulse center frequencies.
There are several ways to differentiate measurement techniques. One method is based on the volume of tissue excited that is used to generate the signal. The most common technique is 2D multislice imaging, in which a narrow volume of tissue (typically 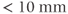 ) is excited by a slice-selective RF pulse and generates the echo signal. The TR specified by the user is the time between successive excitation pulses for a given slice. The total number of lines of data collected for each slice depends on the number of phase encoding steps,
) is excited by a slice-selective RF pulse and generates the echo signal. The TR specified by the user is the time between successive excitation pulses for a given slice. The total number of lines of data collected for each slice depends on the number of phase encoding steps, 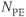 , and the number of signals added together for signal averaging,
, and the number of signals added together for signal averaging, 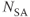 . The sequence kernel time, or minimum TR per slice, is the actual time required for the measurement hardware to perform all the steps necessary to acquire raw data from a single excitation, typically one line from one slice. Often, the sequence kernel time is much shorter than TR, allowing excitation and detection of many slices to be performed within one TR time period.
. The sequence kernel time, or minimum TR per slice, is the actual time required for the measurement hardware to perform all the steps necessary to acquire raw data from a single excitation, typically one line from one slice. Often, the sequence kernel time is much shorter than TR, allowing excitation and detection of many slices to be performed within one TR time period.
Traditionally, multislice scanning acquired one line of data from each slice during each TR time period (Figure 4.10a). This approach set a lower limit for TR. By subdividing the slice loop into subloops so that a subset of slices is acquired, a shorter TR and greater contrast control is possible. The total scan time is TR times the total number of lines times the number of subloops:
Two multislice loop structures are commonly used. Traditional multislice looping uses 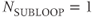 (Figure 4.10a), so that one line of data is acquired for each slice prior to measurement of a second line of data from any slice. The maximum number of slices is limited by TR. This provides the most efficient data collection process for a given TR and is useful when TR is relatively long. At the midpoint of a scan using this looping scheme, each image has
(Figure 4.10a), so that one line of data is acquired for each slice prior to measurement of a second line of data from any slice. The maximum number of slices is limited by TR. This provides the most efficient data collection process for a given TR and is useful when TR is relatively long. At the midpoint of a scan using this looping scheme, each image has 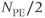 lines of raw data, each with the requested number of acquisitions.
lines of raw data, each with the requested number of acquisitions.
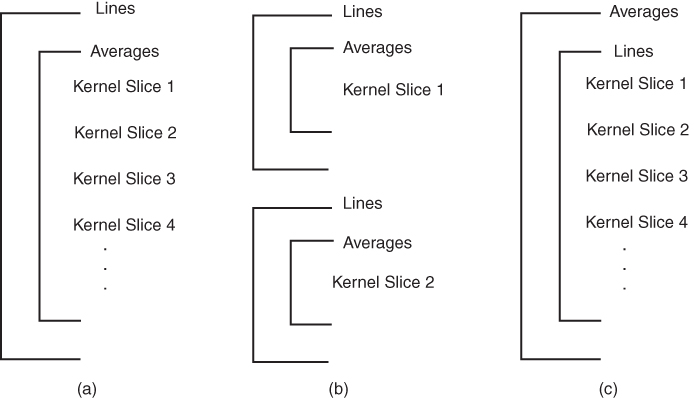
Figure 4.10 Two-dimensional slice loop structures. Three slice loop structures are commonly used. (a) Traditional multislice looping. The slice loop is the innermost loop ( 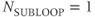 not indicated). Each slice is excited and signal-detected prior to any slice being excited a second time for purposes of signal averaging or phase encoding (lines). This loop structure is the most common. (b) Sequential slice looping. All information for a given slice is acquired prior to any excitation for a different slice. In this figure,
not indicated). Each slice is excited and signal-detected prior to any slice being excited a second time for purposes of signal averaging or phase encoding (lines). This loop structure is the most common. (b) Sequential slice looping. All information for a given slice is acquired prior to any excitation for a different slice. In this figure, 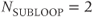 . (c) Long-term averaging. All lines for all slices are acquired before performing signal averaging.
. (c) Long-term averaging. All lines for all slices are acquired before performing signal averaging.
The other approach uses  equal to the number of slices (Figure 4.10b), a so-called sequential slice technique. In this technique, all information for a slice is acquired before acquiring any information for another slice. Only one line of data is measured during each TR time period. This allows the use of very short TR times when acquiring large numbers of slices. At the midpoint of a scan using this looping scheme, all the data for one-half the requested number of slices has been acquired.
equal to the number of slices (Figure 4.10b), a so-called sequential slice technique. In this technique, all information for a slice is acquired before acquiring any information for another slice. Only one line of data is measured during each TR time period. This allows the use of very short TR times when acquiring large numbers of slices. At the midpoint of a scan using this looping scheme, all the data for one-half the requested number of slices has been acquired.
There is also variability in the order of the main loops for 2D multislice imaging. Traditional looping as in Figure 4.10a acquires all signals for a specified phase encoding step (all slices, all averages) before acquiring a signal for a different phase encoding step. This allows all signal averaging for a given raw data line to be done within a short period of time and allows initial steps of image reconstruction to be performed. The other variation acquires a complete raw data set for a slice before beginning any signal averaging (Figure 4.10c). This increases the elapsed time between successive signals being averaged together, reducing any possible contamination between the signals, but renders the image more susceptible to gross patient motion.
The other category of measurement technique is a 3D volume acquisition, which is, in essence, a double phase encoding technique. For 3D volume imaging, tissue volumes of 30–150 mm are excited as compared to 3–10 mm in 2D imaging. In addition, a second phase encoding table is applied in the slice selection direction to “partition” or subdivide the volume into individual slices. Each echo is acquired following application of encoding gradients in both the phase encoding and slice selection directions, one amplitude from each.
The advantages of 3D volume acquisition techniques are that the slices within a volume are contiguous and that the detected signal is based on the total volume of excitation rather than the effective slice thickness. The 3D volume acquisitions have two primary disadvantages. Because of the potentially long scan times, they are usually gradient echo or echo train spin echo sequences and are limited to one or two volumes. Image processing is also longer since an additional Fourier transformation and other processing steps must be performed in order to produce an image.
For 3D volume MRI techniques, in each excitation volume the number of slices is determined by the number of partitions,  . The total scan time is:
. The total scan time is:

There are two possibilities for the order of the two encoding loops. Traditional looping has the partitions loop inside the phase encoding loop (Figure 4.10a), typical of 3D gradient echo techniques. Since the partitions loop normally has fewer entries, this enables initial image processing to be performed while data collection continues. The other order has the phase encoding loop inside the partitions loop (Figure 4.11b). This is more typical for 3D echo train spin echo, where the phase encoding loop is segmented in nature (see Chapter 5).
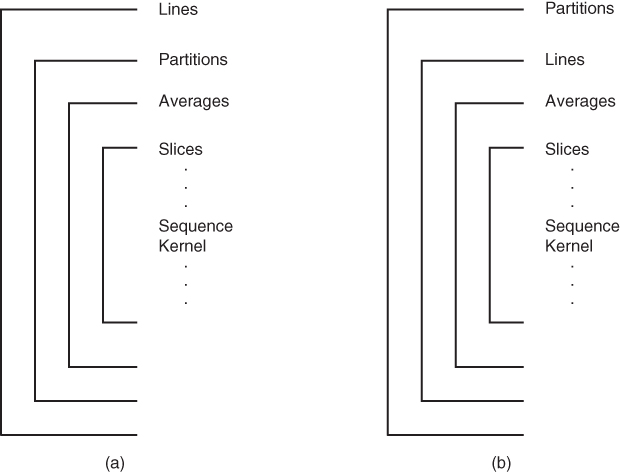
Figure 4.11 Three-dimensional slice loop structures. A fourth loop, the partitions loop, is added. Two slice loop structures are commonly used: (a) partitions-in-lines: the partitions gradient amplitudes are varied most frequently; (b) lines-in-partitions: the lines gradient amplitudes are varied most frequently.