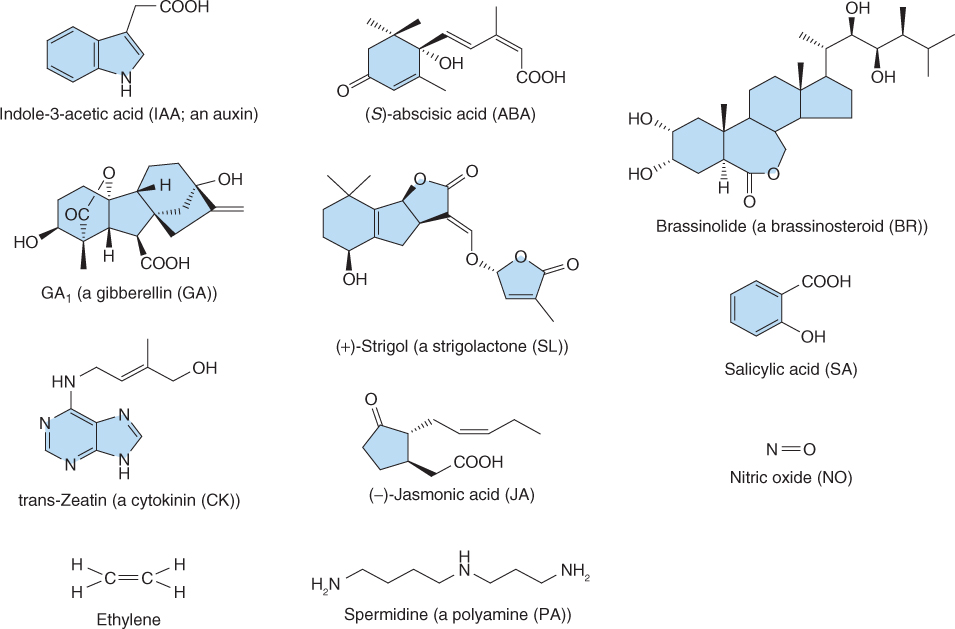
Figure 10.1 Chemical structures, names and abbreviations for the major classes of hormones found in plants.
Chapter 10
Hormones and other signals
Hormones are classically defined as chemical compounds produced in one part of an organ or organism and transported to another where they elicit responses. Charles Darwin and his son Francis were among the first to show that signals produced in one part of the plant affect growth in another. The Darwins exposed seedlings of various grasses and eudicots to unilateral sources of light to investigate growth responses; growth in response to a light source is called phototropism (see Chapter 8). The clearest evidence they obtained for the involvement of a transmissible signal was with canary grass (Phalaris canariensis), where exposing the tip of the seedling coleoptile to unilateral light caused the base to grow toward the source of illumination. In 1880 they wrote: ‘We must, therefore, conclude that when seedlings are freely exposed to a lateral light, some influence is transmitted from the upper to the lower part, causing the latter to bend’.
Work in the early part of the 20th century helped establish that the signal produced by the tips of grass seedlings exposed to unilateral light was chemical in nature, and work by Frits Went in the 1920s led to the development of the first bioassay for plant hormones. Went showed a quantitative relationship between the amount of the chemical substance produced by coleoptile tips of grasses and the bending response in the lower part of the coleoptile. He called the chemical auxin. This bioassay allowed researchers to determine the amount of hormone in a plant extract by measuring the bending response of a de-tipped coleoptile. The coleoptile bioassay led to the identification of the hormone that caused coleoptile bending as indole-3-acetic acid (IAA) (Figure 10.1).
Figure 10.1 Chemical structures, names and abbreviations for the major classes of hormones found in plants.

Bioassays proved to be key tools in early work identifying hormones from plants and animals and their use persisted until the 1960s and 1970s, when advances in chemical analysis allowed for the precise identification and quantification of hormones isolated from tissues. These advances included the development of sensitive gas chromatographs and mass spectrometers in the 1960s and 1970s, instruments that were later combined so that GC-MS (gas chromatography–mass spectrometry) and modern variants of these analytical instruments became the standard methods for analyzing plant natural products.
Representatives of the 11 major classes of plant hormones—abscisic acid (ABA), auxins, brassinosteroids (BRs), cytokinins (CKs), ethylene, gibberellins (GAs), jasmonates (JAs), polyamines (PAs), salicylic acid (SA), strigolactone (SL) and nitric oxide (NO)—are shown in Figure 10.1. Although these hormones differ widely in their chemistry, many aspects of their synthesis and breakdown pathways are shared (Figure 10.2). The isoprenoid pathway serves as the source of the precursors of ABA, BRs, CKs, GAs and SL (see Chapter 15), whereas the auxins, ethylene and PAs arise from the amino acids tryptophan, methionine and arginine/ornithine, respectively. Lipids, specifically the fatty acid α-linolenic acid, are precursors of jasmonates. Two of the hormones, ethylene and NO, are gases and emerging evidence suggests that another gas, hydrogen sulfide, also produced by plants, can act as a potent signaling molecule.
Figure 10.2 Plant hormones can be grouped into three general categories based on their biosynthetic origins: along the isoprenoid pathway (ABA, BR, CK. GA and SL), from amino acids (ethylene, IAA and polyamines)) and from lipids (JA).

The major plant hormones had been well characterized chemically by the end of the 1980s and a combination of chemical and biochemical approaches had elucidated the basic elements of their biosynthesis. The advent of molecular cloning led to major advances in understanding hormone biosynthesis, and the powerful methods of molecular genetics have permitted the identification of the signal transduction pathways for each of the major classes of plant hormone. This has led to the identification of receptors for almost all of the major classes of hormones, progress that was crucially dependent on developments in molecular genetics. As we will see from this chapter, some of the most recent advances have resulted in what can be regarded as major surprises for those working in the field of hormone signaling. For example, we now know that strigolactones, in addition to auxin, regulate axillary bud dormancy. It is likely that more surprises await as we investigate the biology of mobile signals in plants.
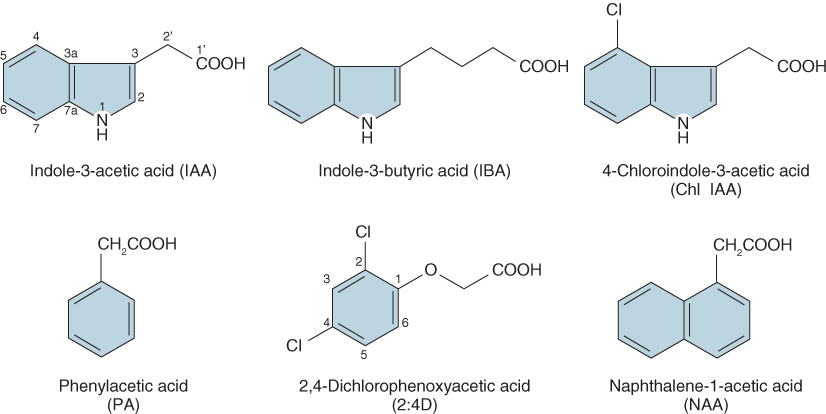
In this chapter we define auxins as compounds that are active in auxin-specific bioassays. The most abundant auxin is IAA, but other indole ring compounds such as indole-3-butyric acid and 4-chlorindole-3-acetic acid, and at least one non-indole auxin, phenylacetic acid, are found in plants (Figure 10.3) and elicit responses in auxin bioassays. Auxins affect almost all aspects of plant development from pollination and fertilization, through vegetative development to flowering. Auxins are key signaling molecules in the tropic responses of plants to gravity, light and touch, and they are known to play a role in the synthesis and action of other plant hormones, which means that their effects on plant growth and development are extensive.
Figure 10.3 Structures of naturally occurring auxins (IAA, IBA, Chl IAA and PA) and two synthetic auxins, one widely used as a herbicide (2:4D) and the other as an inducer of root formation on stem cuttings (NAA).
The effects of auxins in promoting root initiation and inhibiting shoot branching are well-known phenomena that are widely exploited in agriculture and horticulture (Figure 10.4). Shoot cuttings can be induced to form adventitious roots by treating the cut ends of shoots with an auxin. In the case of the Calycanthus ‘Venus’ hybrid (Venus sweetshrub) shown in Figure 10.4A, the cut ends of cuttings were treated with the potassium salt of the auxin indole-3-butyric acid (KIBA). In the absence of auxin, adventitious roots do not develop, but as the concentration of KIBA is raised an increasing number of roots are formed. The growth of axillary buds is also controlled by auxin. In the case of the P. sativum plants shown in Figure 10.4B, axillary buds are normally suppressed, a phenomenon known as apical dominance, but when the shoot apical meristem is removed axillary buds grow. If the decapitated shoot is treated with IAA, axillary bud growth is again suppressed. The hormonal mechanisms underlying apical dominance are described in further detail in Chapter 12.
Figure 10.4 (A) Induction of lateral roots on shoot cuttings of Venus sweetshrub (Calycanthus ‘Venus’ hybrid) by increasing concentrations of the auxin KIBA, the potassium salt of indole-3-butyric acid. (B) Effect of indole-3-acetic acid (IAA) on apical dominance in peas (Pisum sativum). In the experiment shown here, pea seedlings were decapitated and their apices were replaced with lanolin paste (*) alone or with added IAA. When IAA is present (left) axillary buds (arrows) remain dormant and do not elongate; in absence of applied IAA (right), axillary buds are released from dormancy and develop into branches (arrows).

When considering the effectiveness of plant hormones as regulatory molecules it is crucial to know how the pool of active hormone is regulated. For a hormone to be effective it must be synthesized and degraded. Simply synthesizing a hormone would not necessarily be sufficient for it act as a signal because in the absence of a mechanism to reduce its local concentration the signal would always be ‘on’. Turning the signal ‘off’ is therefore as important as turning it on. For auxin, biosynthesis, conjugation, metabolic degradation and polar transport are all mechanisms that control its local concentration (Figures 10.5 and 10.6; see Section 10.2.2).
Figure 10.5 Routes for the degradation and storage of indole-3-acetic acid (IAA). IAA-amido synthase catalyzes the synthesis of amino acid conjugates. Some of these, such as IAA-Ala and IAA-Leu, can be stored and reconverted to active IAA by IAA-imido hydrolase. Others, such as IAA-Asp and IAA-Glu, are irreversibly degraded. Ester conjugates are another storage form of IAA.

Figure 10.6 Two pathways for the synthesis of indole-3-acetic acid (IAA). Both pathways share a common set of reactions from chorismate to indole. At this point IAA may be synthesized from indole by the tryptophan-independent pathway via indole-pyruvic acid. Alternatively indole may be converted to tryptophan and subsequently to IAA via the tryptophan-dependent pathway.

There are two principal pathways of IAA biosynthesis, one from the amino acid tryptophan and the other in a tryptophan-independent pathway (Figure 10.6). A common pathway proceeds from chorismate to indole; at this point it branches. One branch, the tryptophan-independent pathway, proceeds through indole-3-pyruvic acid to IAA. The other branch leads to the formation of tryptophan. At this point, analysis of mutants shows that there are at least four, possibly five, routes for synthesizing IAA from tryptophan, which has made dissection of these pathways difficult. Despite the multiplicity of biosynthetic routes from tryptophan to IAA, experimental evidence indicates that tryptophan is the predominant precursor of IAA in plants.
Catabolism of IAA can also follow several paths, one that results in the formation of conjugates, and others that bring about IAA degradation (Figure 10.5). The formation of some inactive IAA conjugates is reversible and their hydrolysis to active IAA is important in IAA homeostasis. Most of these reactions are under feedback regulation by IAA. Conjugation of IAA is enhanced by high levels of IAA; conversely, hydrolysis of conjugates is promoted by low levels of free IAA. Conjugation of IAA to L-amino acids and to glucose is widespread, and the formation of glucose esters is especially prevalent in seeds. While some IAA–amino acid conjugates can be remobilized and can therefore function as storage products, many of these conjugates are components of irreversible IAA degradation pathways. Degradation of free IAA and its amido conjugates can be carried out by enzymes of the peroxidase class.
The movement of auxins from cell to cell is directional. In the shoot and root, transport occurs basipetally (away from the organ apex). This feature, called polar transport, is unique to auxins and has an important role in many developmental processes including the formation and repair of tracheary elements, apical dominance, initiation of lateral roots (see Chapter 12) and growth responses to light (phototropism,; see Chapter 8) and gravity (gravitropism and the downward bending of the petiole termed epinasty; see Chapter 15).
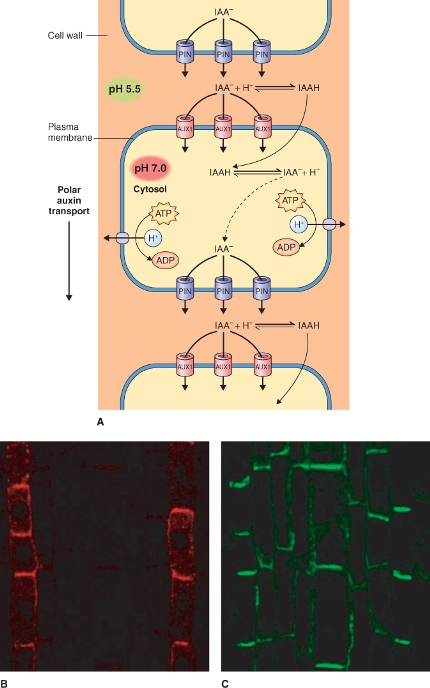
Polar auxin transport is largely driven by gradients in protonated IAA (IAAH) in the apoplast, and anionic forms (IAA−) in the symplasm, which are maintained by specific auxin transporters in the plasma membrane (Figure 10.7). Gradients in IAAH and IAA− are generated by pH differences across the plant plasma membrane (see Chapter 5). IAA is a weak acid with a pKa of 4.7, therefore in the cytosol at a pH of about 7.0 it exists largely as IAA−, whereas in the cell wall at a pH of about 5.5 it is predominantly IAAH. The plasma membrane is relatively permeable to uncharged IAAH but impermeable to IAA−; therefore the ΔpH across the plasma membrane effectively traps IAA− in the cytosol.
Figure 10.7 Polar auxin transport. (A) Model showing membrane localized transporters that participate in the polar transport of auxin. The cell wall is relatively acidic at around pH 5.5 causing IAA to be protonated, but the cytoplasm has a pH of around 7 causing IAAH to dissociate into IAA− and H+. IAAH can diffuse freely through the plasma membrane while IAA− requires a membrane transporter. The difference in pH between the wall and cytoplasm creates a gradient in IAA concentration that drives its movement into and out of the cell. Localized IAA− transporters establish auxin polar transport. The auxin influx carrier AUX1, located at the apical plasma membrane, allows transport of IAA− into the cell, whereas auxin efflux carriers of the PIN class that transport IAA− out of the cell are preferentially localized at the basal plasma membrane. (B, C) These carriers can be visualized in Arabidopsis root stele cells using immunofluorescence microscopy. This shows localization at the opposite ends of cells of the auxin influx carrier AUX1, stained red (B) and the auxin efflux carrier PIN1, stained green (C).
The discovery that specific transporters play a role in polar auxin transport came from genetic analysis of Arabidopsis mutants. Auxin influx carrier (aux) mutants were discovered in Arabidopsis plants that are resistant to high concentrations of applied auxin. Sequence analysis suggests that AUX1 belongs to a family of proteins similar to prokaryotic amino acid permeases. Given that IAA and tryptophan are chemically related it seems reasonable to suggest that AUX1 is indeed involved in auxin transport, acting as a H+/IAA− symporter, but direct evidence is lacking. Auxin influx transporters are asymmetrically localized in the plasma membrane at the apical end of the cell (Figure 10.7B).
Another class of asymmetrically distributed transporters called PIN proteins was first found in the Arabidopsis pin1 mutant shown in Figure 10.8. This mutant was called pin because of the needle-like shape of the inflorescence stem that lacks leaves, buds or flowers. The pin phenotype could be replicated by treatment of wild-type Arabidopsis with inhibitors of auxin transport; this suggested that PIN proteins participate in auxin efflux from cells. At least eight PIN genes have been isolated from Arabidopsis. A number of these have been implicated in tropic responses to light and gravity, responses that are known to involve auxin (see Chapters 8 and 15). Several of the PIN proteins are asymmetrically localized in the basal plasma membrane of cells where they are responsible for the polar efflux of auxin (Figure 10.7C).
Figure 10.8 Phenotypes of the wild-type and pin formed/pin1 double mutant of Arabidopsis. Note that the inflorescence stem of the mutant lacks discernible leaves or lateral buds, giving this mutant its pin-like phenotype. When this gene was cloned it was found to encode a transmembrane protein, PIN, that is now known to be an auxin efflux carrier.

A third set of auxin transporters is a group of the ABC transporter class of membrane proteins (see Chapter 5) that were discovered in loss-of-function mutants in Arabidopsis. Several members of the B subclass of the ABC transporter family have been shown to transport auxin, and they can function in IAA− efflux across the plasma membrane and tonoplast. Unlike PIN proteins they do not show asymmetrical distribution in cells so they are probably not involved in polar transport. Transporters of this class require energy in the form of ATP to transport IAA−.
The low pH of the apoplast, the permeability of the plasma membrane to IAAH and the dissociation of IAAH at the neutral pH of the cytosol lead to the trapping of IAA− in the cytosol. Polar transport of auxin results from the localization of AUX1 at the apical end of the cell, which brings about a directional influx of auxin, and the polar efflux of IAA− through PINs localized at the basal end of the cell. A significant driving force for the efflux of IAA− from the cell is the negative membrane potential of most cells, often between −200 and −300 mV.
It takes less than 10 minutes for auxin to induce changes in growth of target tissue and some aspects of the mechanism by which auxin brings about such rapid changes are now well understood. At least two types of auxin receptor have been isolated, the auxin-binding protein (ABP) and the F-box protein TIR1 (Transport Inhibitor Response 1), a component of the AUX/IAA-SCFTIR1 E3 ubiquitin ligase complex that is described below. ABP has a KDEL/HDEL endoplasmic reticulum (ER) retention signal and is therefore a largely ER resident protein (see Chapter 5), although some ABP traffics to the plasma membrane. How ABP functions in either the ER or plasma membrane is not well understood, but it is known that the affinity of ABP for IAA is much stronger at an acidic pH. Since the lumen of the ER has a pH near neutrality and the extracellular face of the plasma membrane is about pH 5.5, it is likely that ABP functions as an IAA receptor only when it is transported to the plasma membrane.
In contrast to ABP, a great deal is known about the role of the AUX/IAA-SCFTIR1 complex in auxin signaling. SCF complexes are members of the CULLIN/RING E3 ubiquitin ligase family of E3 ubiquitin ligases (see Figure 5.36C). These complexes are components of the ubiquitin–proteasome system (UbPS; see Chapter 5). SCFs are named for the three most important constituent subunits, SKP1, CULLIN and F-box protein (Figure 10.9) of the complex in mammals; plants have homologous proteins. The F-box protein recognizes the substrate to be ubiquitinated. CULLIN forms a dimer with the RING domain protein RBX1 and CULLIN-RBX1 transfers ubiquitin to the target protein. The SCF complex involved in auxin signaling is localized in the nucleus and includes the SKP1 protein ASK (Arabidopsis SKP1-like), CULLIN and the F-box protein TIR1 (Figure 10.10).
Figure 10.9 Model of a generic SKP1/CULLIN/F-box (SCF) E3 ligase. An SCF E3 ligase complex includes four proteins: CULLIN, SKP1, F-box and the RING finger protein RBX1. RBX1 binds an E2 ubiquitin conjugating enzyme. The F-box protein recognizes a target protein and positions it for ubiquitination by E2. Up to seven ubiquitin molecules (Ub) may be added to the target molecule, which is then degraded by the 26S proteasome. Target specificity is conferred by the F-box protein.
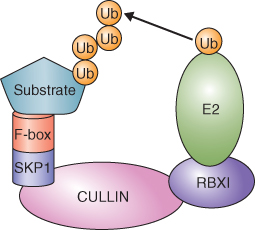
Figure 10.10 Model showing components of the AUX/IAA-SCFTIR1 E3 ligase complex involved in IAA perception and signaling. The F-box protein TIR1 is the IAA receptor and it forms a complex with the repressor proteins of the AUX/IAA class. ASK, the Arabidopsis SKP1 homolog, CULLIN and the RING domain protein RBX1 bring the AUX/IAA target proteins and E2 ligase together allowing AUX/IAA to be ubiquitinated (Ub) and subsequently broken down by the proteasome.
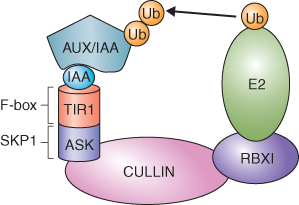
Figure 10.11 Model showing signal transduction during the response to auxin. In the absence of IAA, AUX/IAAs repress auxin-regulated genes by complexing with auxin response factors (ARFs). In the presence of IAA, AUX/IAA proteins become ubiquitinated by SCFTIR1 E3 ligase complex and are broken down by the proteasome. ARF can then function, often forming ARF–ARF dimers that allow the transcription of auxin-responsive genes.
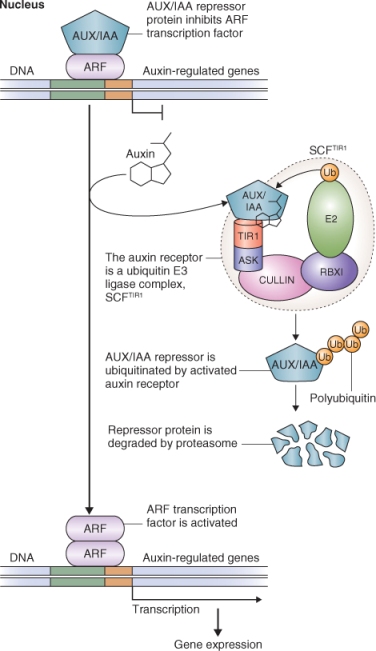
Transcription of auxin-responsive genes is regulated by many factors that interact with their promoters, including transcription factors such as the auxin response factor (ARF) family of proteins. ARFs bind to specific auxin response elements in the promoters of auxin-responsive genes and activate transcription of these genes. In the absence of auxin, the transcription of auxin-responsive genes is downregulated by transcriptional repressors called AUX/IAAs, which repress the activity of ARFs, and often by co-repressor proteins (Figure 10.11).
Figure 10.12 A rice (Oryza sativa) plant (white arrow) infected with the fungus Gibberella fujikuroi showing a highly elongated leaf and shoot.

TIR1 functions as an IAA receptor. IAA initiates the expression of auxin-inducible genes by binding to TIR1. This allows TIR1 to bind AUX/IAAs and target them for ubiquitination by the SCFTIR1 complex and subsequent degradation by the 26S proteasome (Figure 10.11). Degradation of AUX/IAAs allows the transcription of auxin-responsive genes.
The gibberellins (GAs) belong to a large family of natural products produced by plants and fungi. They are tetracyclic diterpenes having 19 or 20 carbon atoms, and to date 136 GAs have been isolated and characterized. The GAs are identified by numbers in the order of their discovery, i.e., GA1, GA2, etc. Not all GAs are biologically active in plants; the two most common active GAs are GA1 and GA4. Most other GAs isolated from plants and fungi are metabolic intermediates in the synthesis of active GAs or their degradation products.
Gibberellins were discovered in the 1920s by Japanese plant pathologists studying the ‘foolish seedling’ disease of rice (Figure 10.12). Foolish rice seedlings resulted from infection by the fungus Gibberella fujikuroi and they grew much taller than uninfected plants. It was quickly concluded that the infecting fungus produced a chemical that stimulated rice growth, and this was isolated and chemically characterized in the 1950s and called gibberellic acid (GA3; see Figure 10.1).
The isolation of GA3 led to work on its physiology, and results showing dramatic effects on plant growth generated immense interest in the GAs. The effects of GAs on shoot growth are clearly illustrated by the experiment shown in Figure 10.13A. Arabidopsis plants with mutation in the GA1-3 gene have lower GA concentration and a very dwarf phenotype that can be rescued by spraying ga1-3 plants with GA3. In addition to having a profound effect on shoot growth, GAs control many developmental processes, including leaf growth, orientation and senescence, flowering, fruit and seed formation, seed germination and early seedling growth. Among the commercial uses of the GAs are the stimulation of fruit set and fruit growth in table grapes. Although the effects of GA on shoot growth suggested opportunities to increase plant biomass, technologies that reduce the GA content of plants have become far more valuable in increasing crop yield. Plants have been bred and engineered to reduce stem height and these new varieties have been instrumental in increasing grain yield, as seen in the Rht (reduced height) mutants of wheat (Figure 10.13B). Short straw wheat plants are often referred to as being part of the Green Revolution. These plants divert more photosynthate from stem growth to grain yield. They have the added trait of reduced lodging which occurs when wind causes wheat fields to be flattened and makes harvesting difficult or impossible (Figure 10.13C). The control of stature in cereal and other species is further discussed in Chapter 12.
Figure 10.13 Mutations in gibberellin (GA) biosynthesis and signaling affect many aspects of plant growth and development. (A) The ga1-3 mutant of Arabidopsis (center) lacks a key enzyme in GA biosynthesis and consequently has extremely low GA levels; mutant plants have a dwarf phenotype and delayed flowering. This mutant can be rescued by the application of gibberellic acid (GA3) (right) and will grow to the same height as wild-type plants (WT) (left). (B) These wheat (Triticum aestivum) plants bear different alleles of the reduced height gene (Rht) that affects GA signaling. (C) This field of wheat shows the effect of wind and rain on plants of normal and reduced height: normal plants, on the left, have experienced lodging (flattening), while reduced height plants, on the right, are lodging-resistant.

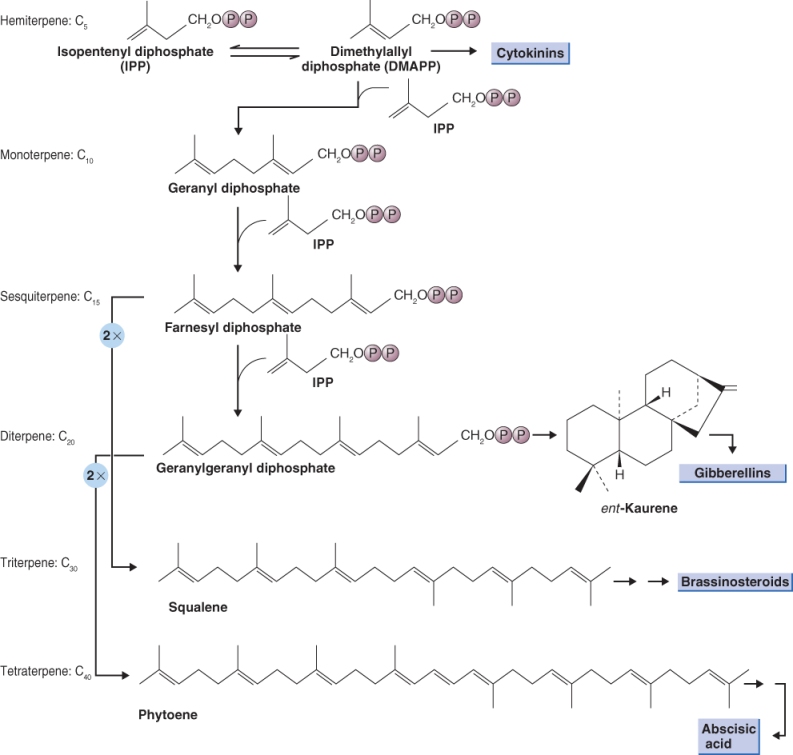
Gibberellins are present in vegetative tissue at very low concentration, rarely more than 1 ng GA per gram of fresh tissue weight, which made unraveling their biosynthesis difficult. GAs are synthesized from isopentenyl diphosphate, which is synthesized in the chloroplast (see Chapter 15). Early steps in the synthesis of GAs are biochemically similar to those for abscisic acid, brassinosteroids and cytokinins (see Figure 10.2). The starting point for the synthesis of all these hormones is the same intermediate, namely the hemiterpene isopentenyl diphosphate (IPP) which isomerizes to dimethylallyl diphosphate (DMAPP; Figure 10.14). IPP and DMAPP are five-carbon compounds that can be synthesized by two distinct pathways using two different precursors. IPP destined for ABA and GA synthesis and DMAPP for the side-chain of CKs are synthesized in the plastid along the DOXP pathway (see Figure 15.23), whereas the IPP required for BRs is synthesized along the mevalonic acid (MVA) pathway in the cytosol (Figure 10.14).
Figure 10.14 Overview of the biosynthetic steps occurring in plastids that lead to the synthesis of all or parts of the cytokinins, gibberrelins, brassinosteroids and abscisic acid.
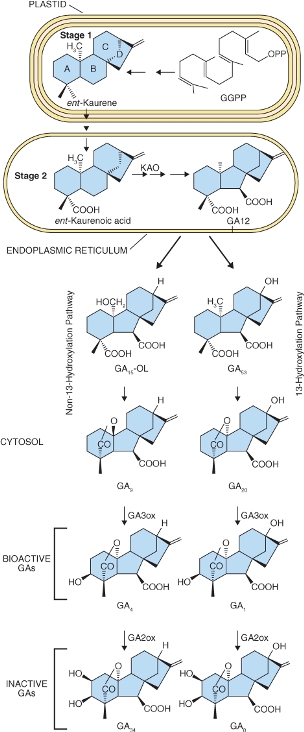
The immediate diterpene precursor of GA is geranylgeranyl diphosphate which is converted to ent-kaurene in a two-step reaction also occurring in the plastid (Figure 10.15). The conversion of ent-kaurene to kaurenoic acid and GA12 is catalyzed by two multifunctional enzymes of the cytochrome P450 class. It begins on the plastid envelope and is completed in the lumen of the ER. GA12 can follow one of two cytosolic pathways to bioactive GAs. In the 13-hydroxylation pathway GA12 is converted to GA53, then by a series of steps to bioactive GA1. The non-13-hydroxylation pathway from GA12 leads to GA4 (Figure 10.15). The immediate precursors of GA1 and GA4 are inactive GA20 and GA9 respectively; both become active when hydroxylated at carbon 3 by the soluble dioxygenase enzyme GA-3 oxidase (GA3ox). Hydroxylation of GA1 and GA4 at carbon 2 is catalyzed by GA-2 oxidase (GA2ox), also a soluble dioxygenase, leading to inactivation of these hormones by forming GA8 from GA1 and GA34 from GA4 (Figure 10.15).
Figure 10.15 Gibberellin biosynthesis showing the major steps in the pathways and their subcellular localization. The 13-hydroxylation pathway produces bioactive GA1 and the non-13-hydroxylation pathway produces bioactive GA4. GGPP, geranylgeranyl pyrophosphate); KAO, kaurenoic acid oxidase; PPO, pyrophosphate.
Although GAs are present in plant tissues at extremely low levels, changes in their concentration must be strictly controlled if they are to be effective as regulatory hormones. The levels of GAs are subject to feed-forward and feedback mechanisms that bring about hydroxylation at the three- and two-carbon atoms of the GA molecule. In two examples of feedback regulation, the GA20ox and GA3ox that convert GA12 to GA9, and GA9 and GA4, respectively, are inhibited by high concentrations of bioactive GAs (Figure 10.16). Sequential hydroxylation at carbons 20 and 3 of GA12 leads to the formation of active GA4 and GA1. When the levels of bioactive GAs are high, plants show a strong response to the presence of GA, and the synthesis of GA-20 and GA-3 oxidases is inhibited, thereby blocking the synthesis of additional active GAs. The dwarfing gene le studied by Gregor Mendel in Pisum sativum (see Figure 12.50) is now known to encode a GA3ox. Mutations in Le bring about a reduction in the amount of bioactive GA1 and result in a dwarf phenotype, establishing a causal relationship between GA1 concentrations and stem growth in this species.
Figure 10.16 Biosynthesis of the bioactive gibberellins, GA4 and GA1, is regulated by feedback ( ) and feed-forward (→) mechanisms. The activities of GA20ox and GA3ox are downregulated by bioactive GA4 and GA1, an example of a feedback-regulated step. On the other hand, the activity of GA2ox, which produces inactive GA34 and GA8, is upregulated by these bioactive GAs, an example of feed-forward regulation.
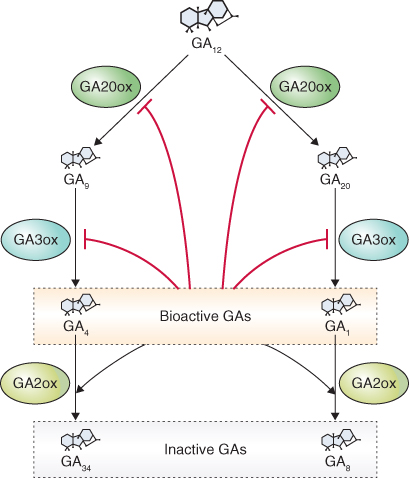
It was thought that only GA4 and GA1, the principal bioactive GAs, were hydroxylated at carbon 2, but we now know that earlier GA precursors can also be hydroxylated at carbon 2. This converts them to inactive GAs that cannot become active GAs when further metabolized. An enzyme that oxidizes the double bond at carbon 16,17 also brings about inactivation of GAs. This reaction is an epoxidation and is restricted to those GAs such as GA9 and GA4 that lack OH at carbon 13; 13-hydroxy GAs such as GA53, GA20 and GA1 are not inactivated by this pathway.
Molecular genetic approaches have identified the GA receptor in plants and the mechanism by which GAs regulate responses at the transcriptional level. The GA receptor was first identified in a rice mutant that was insensitive to GA. The mutant was called Gibberellin Insensitive Dwarf1 (GID1) and the gene responsible for this phenotype was called GID1. Only one copy of GID1 is present in rice. Purification of the GID1 protein showed that it has a high affinity for biologically active GAs but not for inactive GA8 and GA34, providing strong evidence that it is a GA receptor. GID1 is also found in Arabidopsis but in this species there are three homologs of the gene.
As is the case with auxin perception and signaling, the ubiquitin–proteasome system (UbPS) pathway is a key component of GA signal transduction. The basic elements of GA signaling via the receptor GID1 that lead to proteolysis of repressor proteins are illustrated in Figure 10.17. Unlike the auxin receptor TIR1, GID1 is a soluble, nuclear protein and is not part of the SCF E3 ligase complex. The F-box protein involved in GA signaling is called GID2 in rice and SLY1 in Arabidopsis. Thus the E3 ligase for GA signaling is SCFSLY1/GID2. In the presence of bioactive GA and the receptor GID1, SLY1/GID2 recognizes a class of repressor proteins called DELLA domain proteins and this results in their ubiquitination and proteolysis by the proteasome (Figure 10.17).
Figure 10.17 Model of the gibberellin (GA) signal transduction pathway in rice showing the central role of DELLA domain proteins as repressors of transcription of GA-regulated genes. DELLA proteins are degraded by the proteasome after they are ubiquitinated by the E3 ligase SCFSLY1. GA binds to the GA receptor GID1. The hormone–receptor complex then interacts with SLY1, the F-box protein of SCFSLY1 and together they bind DELLA domain proteins allowing them to be ubiquitinated for subsequent proteolysis by the proteasome. Removal of DELLA proteins allows the transcription of GA-regulated genes to proceed. Among the genes that are upregulated by DELLA are those involved in GA perception (GID1) and biosynthesis (GA20ox and GA3ox). An example of a gene downregulated by DELLA domain proteins is GA2ox.
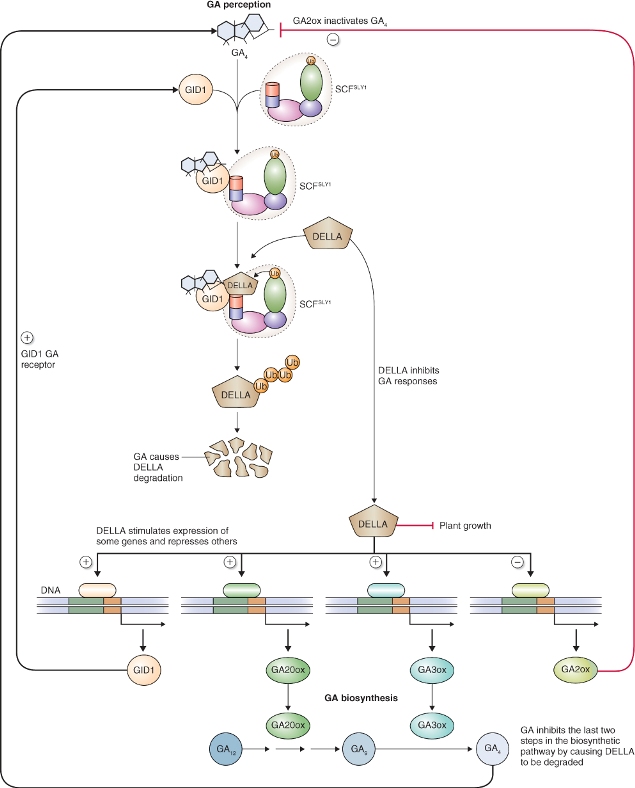
Although DELLA domain proteins are referred to as repressor proteins, the transcription of some genes is promoted by DELLAs. Furthermore, DELLA domain proteins may not act directly as repressors, but function by interacting with other proteins to bring about repression or de-repression of genes indirectly. Examples of such interactions are shown in Figure 10.17. DELLA domain proteins have been shown to affect a suite of genes related to the synthesis of GA as well as those controlling growth. In addition to their involvement in GA responses, DELLA proteins are known to mediate a wide variety of plant responses to light and other abiotic factors including temperature. DELLA domain proteins interact with PIF (phytochrome-interacting factor) proteins leading to the regulation of transcription of both GA- and light-regulated genes (see Chapters 8 and 12). Treatment of plants with GA brings about degradation of DELLAs by the UbPS, which in turn leads to the expression or repression of genes which results in the promotion of a wide variety of GA responses including altered growth and development (see Figure 10.13), synthesis and secretion of hydrolytic enzymes by the cereal aleurone (see Figure 6.32), breaking of dormancy (see Chapters 6 and 17) and induction of flowering (see Chapter 16).
Cytokinins (CKs) were discovered in the 1930s as compounds that, in concert with auxin, would support cell division and shoot development in plant tissue grown in sterile culture (Figure 10.18). When plant cells are grown on a medium that contain both CK and IAA, they form masses of undifferentiated cells called callus. If callus is transferred to a medium with high CK and low auxin concentration, it produces shoots. If transferred to a medium containing only auxin, callus produces roots. The first CK, called kinetin, was isolated from autoclaved herring sperm DNA. Subsequently several CKs have been isolated from plant tissue. Plant tissue culture became the basis of the standard bioassay in the search for compounds that would stimulate shoot development, and compounds that were active in this bioassay were called cytokinins. CKs have a wide range of effects on plant growth and development. They play roles in shoot and root organogenesis, control lateral bud growth, and regulate senescence and seed germination.
Figure 10.18 The production of roots and shoots by Arabidopsis callus tissue. Callus subcultured on a medium containing the auxin indole butyric acid (IBA) produces only roots (left); but on a medium containing a high ratio of the cytokinin trans-zeatin (tZ) to IBA it produces shoots (right).

Kinetin and benzyladenine, another synthetic cytokinin (Figure 10.19), do not occur naturally in plants but a range of natural CKs have now been isolated including isopentenyl adenine (iPA), trans-zeatin (tZ), cis-zeatin (cZ) and dihydrozeatin (DZ) (Figure 10.19). With the exception of diphenylurea, all native and synthetic CKs are derivatives of the purine base adenine.
Figure 10.19 Structures of some naturally occurring and synthetic cytokinins. The naturally occuring cytokinins may be divided into two categories, isoprenoid cytokinins and aromatic cytokinins, based on the side-chain attached to the 6 position on the purine base adenine. Synthetic cytokinins such as benzyladenine are also 6-substituted purines, but diphenylurea lacks a purine ring structure.
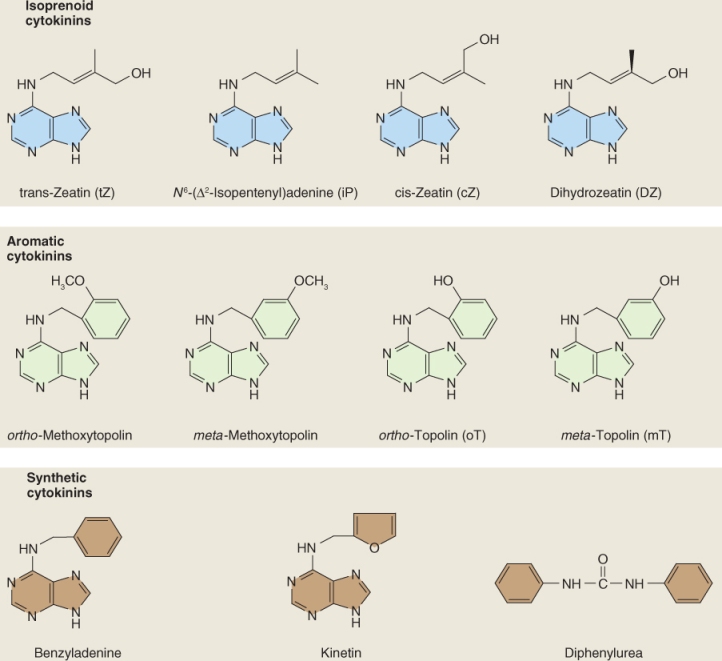
The naturally occurring CKs fall into two groups based on the side-chain at the 6 position of the purine moiety. The most abundant naturally occurring CKs, including iPA, tZ, cZ and DZ, have an isopentenyl side-chain at this position; other less abundant CKs have an aromatic group (Figure 10.19). Of the isoprenoid CKs, iPA and tZ are more active and generally more abundant than cZ or DZ; however, the former are more susceptible to degradation by the enzyme cytokinin oxidase (see Section 10.4.2) than either cZ or DZ. The aromatic CKs are much less abundant than the isoprenoid CKs and less is known about their physiological role.
The biosynthesis of tZ is shown in Figure 10.20. The transfer of an isopentenyl side-chain from dimethylallyl diphosphate (DMAPP) to ATP or ADP is catalyzed by the enzyme isopentenyl transferase (IPT). This step of tZ biosynthesis occurs in the plastid because DMAPP is synthesized in plastids (see Figure 10.14) and IPT enzymes are localized there. IPT genes constitute a small family (seven in Arabidopsis that are involved in CK synthesis). Mutations in IPT genes result in phenotypes with altered branching, enhanced root growth and reduced levels of iPA and tZ. The expression of some IPT genes is downregulated by CKs, suggesting a feedback loop in CK synthesis similar to that found in the GA biosynthesis pathway. The pattern of IPT gene expression in plant tissues indicates that the synthesis of CKs occurs in the apical region of shoots and roots as well as in developing seeds.
Figure 10.20 Major steps in the cytokinin biosynthetic pathway. CK biosynthesis begins with the addition of an isopentenyl side-chain from dimethylallyl diphosphate (DMAPP) to ATP or ADP. The product of this reaction is converted to zeatin triphosphate by a cytochrome P450 enzyme, followed by sequential removal of two additional phosphate groups and ribose to produce trans-zeatin. A single step formation of trans-zeatin can occur catalyzed by LOG, the product of the LONELY GUY gene in Arabidopsis.
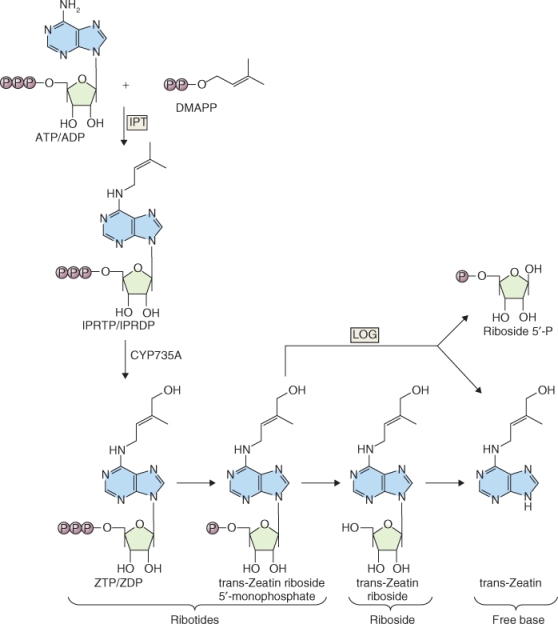
The next step in tZ biosynthesis is the hydroxylation of the isopentenyl side-chain by a member of the cytochrome P450 monooxygenase (CYP) family of enzymes. These reactions occur largely on the membranes of the ER and are regulated by several hormones, indicating that CYPs are points of cross-talk between the various hormone-related pathways. For example, CKs bring about the upregulation of CYP genes whereas IAA and ABA reduce the level of their expression. CKs are biologically active only as the free base. The formation of the free base from the ribotide, trans-zeatin riboside-5′-monophosphate, can occur in a single step or in a two-step process. The single-step formation of a free base removes the phosphorylated ribose from the ribotide, releasing free tZ and is catalyzed by the product of the LONELY GUY gene in Arabidopsis. The two-step process requires hydrolytic cleavage of the phosphate from the ribotide by a phosphohydrolase to form the riboside followed by removal of ribose by a nucleosidase (Figure 10.20).
The possibility that CKs can be produced by the hydrolysis of tRNA has been widely explored. Specific IPT activities that can transfer isopentenyl side-chains to particular adenine residues in tRNA have been found in animal and plant cells. It has also been shown that if purified tRNA is hydrolyzed, the CK released shows biological activity. Whether tRNA contributes a physiologically important amount of CK to the endogenous pool of hormone has not been resolved.
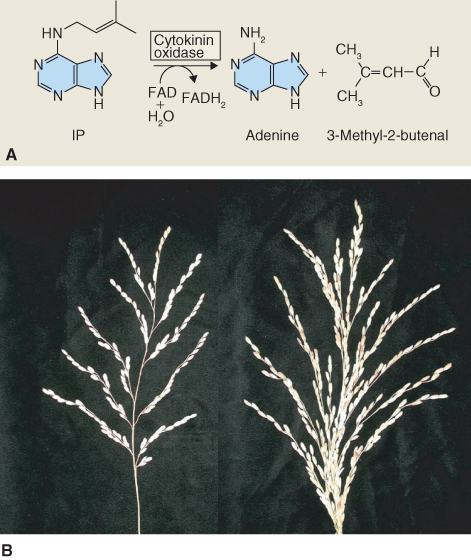
As with the other hormones discussed so far, regulation of the breakdown of CKs is key to their function as signaling molecules. There are two basic pathways by which CKs are catabolized. The first occurs by oxidation by CKX (cytokinin oxidase), a FAD oxidoreductase enzyme, which cleaves the isopentenyl side-chain of CK and releases adenine and 3-methylbutenal (Figure 10.21A). Evidence that a reduced activity of CKX causes developmental effects has been found in experiments on rice, where there is natural variation in CKX expression. Rice plants with reduced CKX expression show much larger panicle size and consequently higher grain yield (Figure 10.21B).
Figure 10.21 (A) Cytokinin oxidase is a flavin-containing oxidase that irreversibly degrades many cytokinins, including isopentenyladenine (IP) as shown here. (B) Wild-type rice (Oryza sativa) (left) and a mutant with lowered expression of cytokinin oxidase (right). Note that the mutant has a larger panicle size and consequently larger grain yield.
CK can also be inactivated by the addition of sugars or amino acids. Glucosylation can occur either at N-7 or N-9 positions of the adenine moiety, or on the OH group of the isopentenyl side-chain. N-glucosylation is irreversible and results in inactivation of CK but O-glucosylation can be reversed. Alanine can be added to the N-9 position; this reaction is also reversible. Reversible modification of CK by the addition of sugars or amino acids may be an important mechanism for storage, since these modified CKs are located in the vacuole. Free CK can be released from the storage form, for example during seed germination.
Molecular genetic approaches have provided a comprehensive model of how CKs influence development at the cellular level, but several aspects of this pathway remain to be elucidated. The CK receptor is related to bacterial two-component histidine kinase family receptor proteins (Figure 10.22A). These receptors consist of a histidine kinase sensor with input and transmitter domains, and a response regulator with receiver and output domains. Ligand binding to the input domain of the sensor results in activation of its transmitter domain and autophosphorylation of the His residue. The sensor then transphosphorylates an Asp residue on the response regulator, and in bacteria this leads to activation of transcription.
Figure 10.22 Models of simple and complex two-component histidine kinase systems. (A) In the simple two-component system the histidine kinase responds to the presence of a signal by autophosphorylation on a His (H) residue of its transmitter domain. The phosphoryl group is transferred to an Asp (D) residue on the receiver domain of the response regulator. (B) In the more complex phosphorelay model the histidine kinase receptor has both sensor and receiver domains that become sequentially phosphorylated. The phosphoryl group is then transferred from the histidine kinase to a histidine phosphotransferase (Hpt, called AHP in Arabidopsis), and finally to the response regulator ARR (Arabidopsis response regulator).

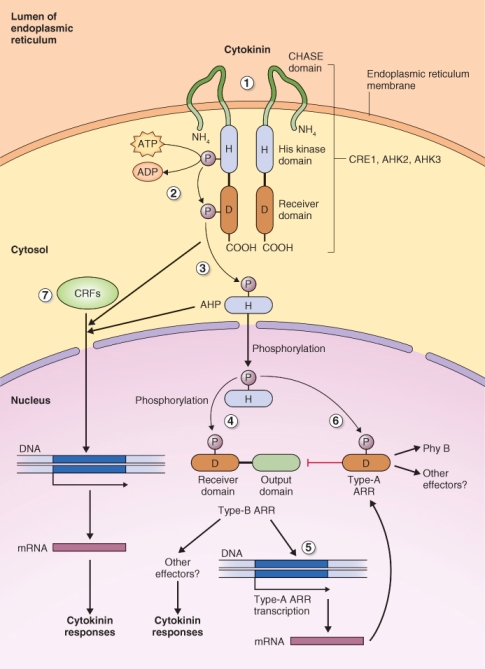
In plants the CK receptor is more complex and is called a phosphorelay two-component system (Figure 10.22B). In this case a hybrid sensor has both sensor and receiver domains that contain His and Asp domains, respectively. Following autophosphorylation, the sensor transfers a phosphoryl group to a histidine phosphotransfer protein (Hpt), called AHP in Arabidopsis, which in turn transfers the phosphoryl group to an Asp residue on a response regulator, called ARR (Arabidopsis response regulator). Figure 10.23 shows the basic outlines of the CK signal transduction pathway. Three CK receptors have been isolated from Arabidopsis: CRE1 (Cytokinin Receptor1), AHK2 (Arabidopsis Hybrid sensor Kinase2) and AHK3. The CK receptor has a membrane-spanning domain and functions as a dimer. Although it has been proposed that this receptor is localized to the plasma membrane, more recent evidence suggests that it may be found in the ER. After CK binds to the receptor, a phosphoryl group is transferred from the receptor to one of many cytoplasmically localized AHP proteins. At least five authentic AHP genes have been identified in Arabidopsis and they appear to play overlapping roles in CK signaling because deleting one or more of these genes does not entirely abolish responses to CK. Triple and quadruple mutants show a much reduced response to CK, suggesting that these His phosphotransfer proteins may function additively.
Figure 10.23 The cytokinin signal transduction pathway. (1) CK binds to the CHASE domain of its receptor (CRE1, AHK2, AHK3) at the ER, activating the histidine kinase domain of this protein (2). The phosphoryl group is transferred from the histidine kinase domain of CRE1 to the receiver domain and (3) it in turn transfers phosphate to histidine phosphotransfer protein AHP. (4, 6) AHP moves to the nucleus where it transfers its phosphoryl group to Type A and B Arabidopsis response regulators (ARRs). Phosphorylation of the type- B ARR leads to (5) transcription of type A ARR genes. ARRs bring about induction of the various CK responses. (7) Cytokinin response factors (CRFs) can also be activated by CRE1 or AHP. Activated CRFs move to the nucleus where they trigger transcription of cytokinin-responsive genes.
AHPs transfer their phosphoryl group to two potential targets, Type A and B ARRs, each of which can regulate subsets of the cell's response to CK; the ARRs are activated and stabilized by this phosphorylation. Eleven Type B ARRs have been identified in Arabidopsis and these are all transcriptional activators that affect CK homeostasis, shoot and root development, cell expansion and senescence. Type B ARRs also regulate the transcription of Type A ARRs.
Phosphorylated Type A ARRs negatively regulate their own phosphorylation by AHPs causing a feedback loop. Type A ARRs are represented by ten genes in Arabidopsis, and loss-of-function mutants have shown that eight of these ARRs negatively regulate CK signaling, although the precise mechanism by which they function is not known. Arabidopsis plants with mutations in Type A ARRs 3, 4, 5, 6, 8 and 9 show enhanced expression of CK primary response genes, indicating that these six ARRs play a role in suppressing CK signaling. At least two Type A ARRs act to positively regulate CK signaling, especially the effects of this hormone on circadian rhythms and phytochrome function.
Cytokinin response factors (CRFs) are another family of proteins that helps mediate the CK signal in cells (Figure 10.23). There are six CRF genes in Arabidopsis and the expression of three of these is enhanced by Type B ARRs. CRFs accumulate in the nucleus following CK treatment. Measurement of gene expression indicates that nuclear localization is required for CRF function, and deletion of these genes results in the reduced expression of a large number of CK-regulated genes.
The role of ethylene in the regulation of plant growth and development was discovered in the early part of the 20th century when it was observed that street trees lost their leaves prematurely when they were growing near gas lamps, which release ethylene, a component of natural gas. Even though it was known as early as the mid-1930s that plants had the ability to produce ethylene, it was not until the advent of gas chromatography and the ability to measure low ethylene concentrations that plant biologists recognized that ethylene is an endogenous regulator of plant development.
Ethylene affects many aspects of plant development. In addition to its influence on abscission of leaves and other organs, it stimulates fruit ripening, promotes germination and affects cell expansion (see Chapters 12 and 18). Changes in cell expansion brought about by exposure to ethylene were described in the mid-19th century when the symptoms now known as the triple response were described (Figure 10.24). The response in seedlings includes bending and thickening of the shoot and pronounced curling of the apical hook. The triple response is a diagnostic of the response of plants to ethylene, a phenotype that has proven invaluable for selecting mutants in the ethylene signaling pathway.
Figure 10.24 The triple response to ethylene of dark-grown pea (Pisum sativum) and mung bean (Vigna radiata) seedlings exposed to ethylene. (A) Control pea seedlings (0) and pea seedlings treated with increasing ethylene concentrations. Ethylene-treated plants have shorter, thicker epicotyls and, at intermediate ethylene concentration, the epicotyl grows horizontally rather than vertically. (B, C) Mung bean seedlings treated with ethylene show the classic triple response: inhibition of hypocotyl and root elongation, increase in hypocotyl width and extreme curvature of the apical hook.

Ethylene is a simple, gaseous, unsaturated hydrocarbon with the empirical formula C2H4 (see Figure 10.1). It is synthesized by almost all plant tissues, but its rate of synthesis is enhanced by wounding and other stresses and during fruit ripening, senescence and abscission (see Chapter 18). Not all ripening fruits produce large amounts of ethylene; indeed, this feature is limited to those fruits referred to as climacteric fruits. These fruits undergo a burst in respiration as they ripen, called the respiratory climacteric, which is stimulated by ethylene. Examples of fruits showing a respiratory climacteric are shown in Table 18.2. Tropical fruits, such as banana, mango and papaya, which are imported into countries in North America and Europe, are mostly climacteric fruits and can over-ripen during transportation from the tropics. As we discuss below, there has been considerable research on strategies to postpone ripening, particularly focused on ethylene.
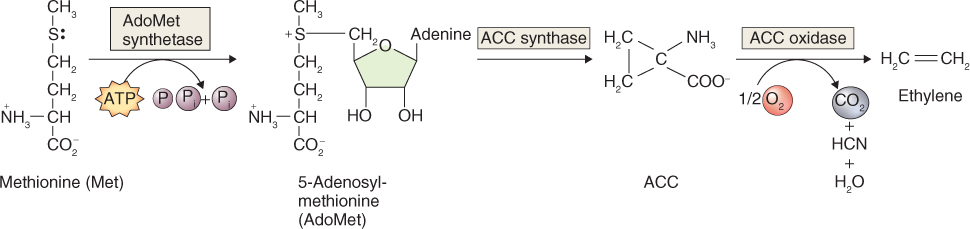
Ethylene is synthesized from methionine in three enzyme steps: adenosyl methionine synthetase which produces S-adenosyl methionine (SAM) from methionine; ACC synthase (ACS) which produces ACC (1-aminocyclopropane-1-carboxylic acid); and ACC oxidase (ACO) which produces ethylene as well as CO2 and HCN from ACC (Figure 10.25). ACC synthase and ACC oxidase are highly regulated enzymes in vivo. The expression of ACS genes is stimulated by auxin, fruit ripening, wounding and several abiotic stresses, especially flooding, and ACO genes are strongly upregulated in ripening climacteric fruits.
Figure 10.25 Biosynthesis of ethylene from the amino acid methionine. ACC, 1-aminocyclopropane-1-carboxylic acid.
ACS and ACO are represented by large multigene families. Tomato has at least ten ACS genes that can be subdivided into groups depending on whether their expression is regulated by auxin, ripening, wounding, abiotic stress, etc. ACS activity can also be regulated by alterations in its stability, for example by phosphorylation at the carboxyl terminus of the protein, which reduces its turnover by the 26S proteasome.
Whereas specific mechanisms exist for the break-down of the major plant hormones, ethylene is a notable exception. Ethylene quickly diffuses from sites of synthesis, meaning that specific catabolic reactions are not required to break it down. Nevertheless, catabolic reactions that can breakdown ethylene are found in plant tissues; however inhibition of these reactions does not affect the response of tissues to ethylene. These experiments support the conclusion that catabolism does not play a role in ethylene homeostasis. Rather, diffusion of the gas from its source is sufficient to remove the signal.
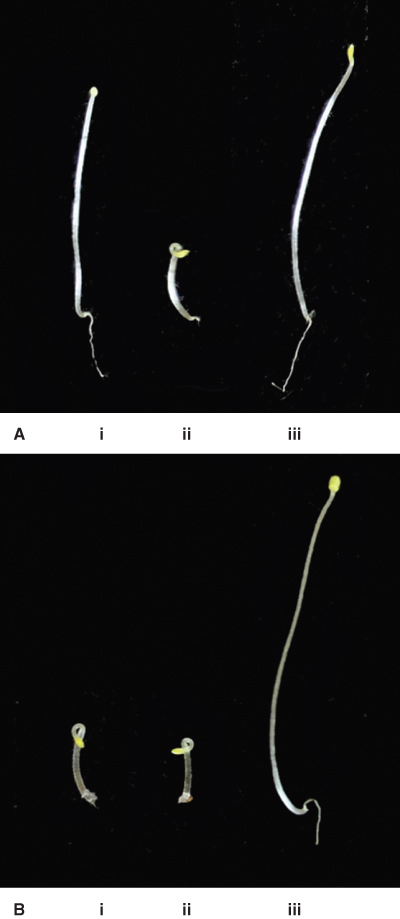
Ethylene receptors were identified in Arabidopsis plants screened for their insensitivity to the gas. A mutant isolated in this screen grew normally in the presence of ethylene and did not show the triple response; it was designated etr1 (ethylene response1; Figure 10.26). Isolation and sequencing of the etr1 gene showed it had similarities to the bacterial histidine kinase two-component response regulator; it was the first of this type of hormone receptor to be identified in plants. We now know that the CK receptor (see Figures 10.22 and 10.23) is also a member of this class of receptor. Other ethylene receptor genes have now been identified including ERS2 (ethylene response sensor2), which is a homolog of ETR1, ETR2 and EIN4 (ethylene insensitive4). All of these ethylene receptors have an amino-terminal membrane-spanning domain that binds ethylene and a histidine kinase domain in the carboxyl half of the molecule. ETR1 and ERS1 have functional histidine kinase domains, whereas EIN4, ETR2 and ERS2 lack histidine kinase activity.
Figure 10.26 Wild-type and mutant Arabidopsis seedlings grown in the dark in the absence or presence of ethylene. (A) In the absence of ethylene, wild-type seedlings (i) and those of the ethylene-insensitive mutant ein2-1 (iii) grow normally, but the ctr1-1 mutant (ii) exhibits a constitutive triple response in the absence of ethylene, including reduced hypocotyl elongation, swelling of the hypocotyl and pronounced curling of the apical hook. (B) When seedlings of these three genotypes are grown in the presence of ethylene, both wild-type (i) and ctr1-1 (ii) seedlings show the triple response but the ethylene insensitive ein2-1 mutant (iii) grows as if ethylene were not present. Mutants such as these led to the identification of the ethylene receptor and the key steps in the ethylene signal transduction pathway.
Ethylene receptors in Arabidopsis are thought to interact with each other in large complexes located in the membrane of the ER. The binding of copper is required for ethylene receptor function. In the absence of ethylene, ethylene receptors inhibit the ethylene response pathway. When ethylene receptors bind ethylene, they are inactivated and the response pathway is no longer inhibited. In other words ethylene receptors are ‘on’ in the absence of ethylene and ‘off’ in its presence.
Mitogen-activated protein kinases (MAP kinases) are a family of highly conserved eukaryotic proteins that transmit a cascade of signals, often from receptors, protein kinases or GTPases at the plasma membrane, to intracellular targets. While signals from receptors are short-lived, the phosphorylation cascade via MAP kinases is long-lived and widely broadcast throughout the cell by the many homologs of the proteins that belong to this protein kinase family. MAP kinases are serine/threonine protein kinases. Arabidopsis has 90 genes that encode this class of kinase. MAP kinases belong to three functionally distinct families that operate in series. MAPKKK (MAP kinase kinase kinase, for which there are 60 genes in Arabidopsis) becomes phosphorylated by upstream kinases such as the RAF-like kinase CTR1 in ethylene signaling; then MAPKKK phosphorylates MAPKK (MAP kinase kinase, ten Arabidopsis genes). MAPKK phosphorylates MAPK (MAP kinase, 20 Arabidopsis genes) on both threonine and tyrosine. MAPK is thought to be the last kinase in the phosphorylation cascade, as shown in Figure 10.27. Examples of the targets for this signaling cascade are transcription factors that are involved in stomatal guard cell functioning, the regulation of cell division, defense against pathogen attack and in responses to various abiotic stresses.
Figure 10.27 MAP kinase (MAPK) cascades are involved in regulating many aspects of plant growth and development, including cytokinesis and the response to ethylene in Arabidopsis. During regulation of cytokinesis (left) a kinesin-like protein is thought to be upstream of a MAPK cascade that targets protein kinases such as MAPK11, a kinase that inhibits cell division. For ethylene signaling (right), the ethylene receptor ETR1 and another kinase, CTR1, are thought to be upstream of the MAPK cascade and EIN2, a transmembrane protein in the ethylene signal transduction pathway, is downstream of this cascade. MAPKK, MAP kinase kinase; MAPKKK, MAP kinase kinase kinase.
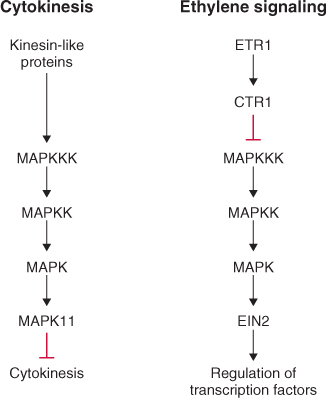
MAP kinases are thought to function downstream of the ethylene receptor ETR1 (Figure 10.28) and a second protein kinase, called CTR1 (Constitutive Triple Response1). The ctr1 gene was identified in a genetic screen and, as the name suggests, plants carrying this mutation display a triple response in the absence of ethylene (see Figure 10.26). Because the ctr1 mutation gave rise to the ethylene response phenotype in the absence of ethylene, the wild-type gene was proposed to be a negative regulator of ethylene responses. CTR1 is thought to interact directly with ETR1.
Figure 10.28 Ethylene signal transduction pathway in Arabidopsis. (1) Ethylene receptor ETR1 is localized in the ER membrane. In the absence of ethylene it activates a phosphorylation cascade that begins with CTR1, and includes members of the MAP kinase cascade. (2) EIN2 is a membrane-localized protein with 12 transmembrane spanning domains and is downstream of the MAP kinase cascade. (3) EIN2 brings about activation of a set of transcription factors, including EIN3 and ERF1, that regulate the expression of ethylene-responsive genes.
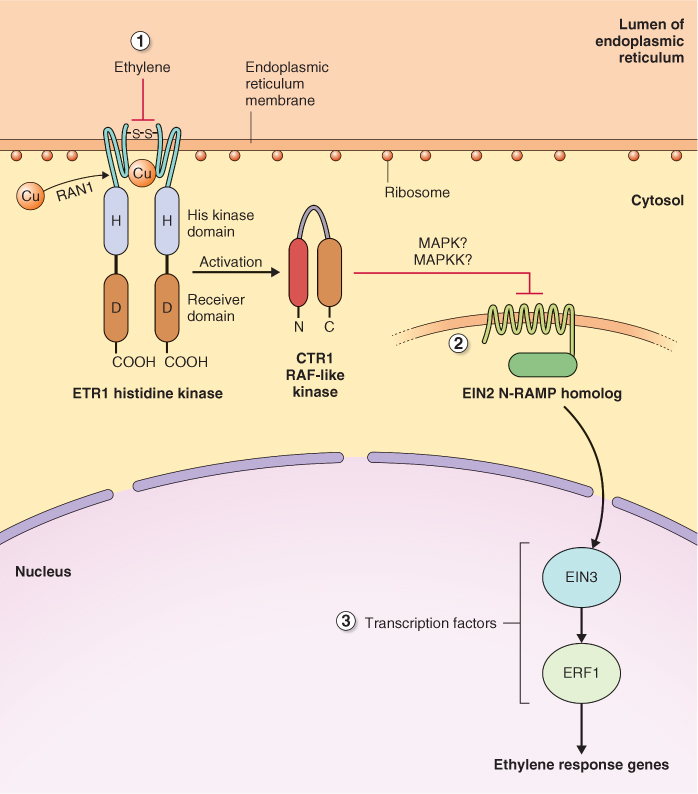
Ethylene brings about a change in the expression of a suite of genes that contain ethylene response elements, including those that regulate fruit ripening. It does this by affecting at least two sets of transcription factors, EIN3 and ERF1. The link between CTR1 and the transcription factors that recognize ethylene response elements is EIN2, another protein with transmembrane motifs whose localization and function is unknown. Ethylene receptors are localized on the ER membrane with their receiver domains directed to the ER lumen. How information is transferred from CTR1 to EIN2, another membrane-localized protein, and then to transcription factors in the nucleus, remains to be resolved.
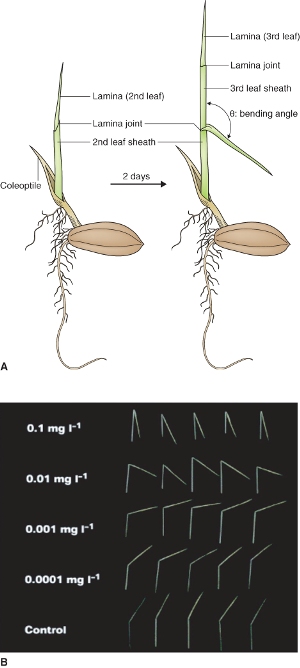
Brassinosteroids (BRs), triterpene molecules with distinctive chemical structures containing from 27 to 29 carbon atoms, share biosynthetic origins with several other plant hormones (see Figure 10.14). Brassinolide (BL), the most active of the native BRs in plants, has 28 carbon atoms; its structure with all its carbon atoms numbered is shown in Figure 10.29. The biological activity of BRs is dependent on the presence of hydroxyl groups at carbons 2 and 3 of ring A and positions 22 and 23 of the side-chain, and a seven-membered lactone B ring. BL was first isolated from germinating pollen of Brassica rapa. Later, castasterone, another BR, was isolated from insect galls of chestnut (Castanea spp.). BRs elicit a response in the rice lamina inclination bioassay, a test used to measure the biological activity of BR-like molecules (Figure 10.30).
Figure 10.29 Highly simplified biosynthetic pathway of brassinosteroids (BRs). Campesterol and campestanol are inactive precursors of the biologically active compounds castasterone and brassinolide. Brassinolide is the most active naturally occurring BR and is metabolized to inactive 26-hydroxybrassinolide by a cytochrome P450 enzyme encoded by the gene BAS1. The biological activity of BRs is dependent on the presence of hydroxyl groups at carbons 2 and 3 of ring A and positions 22 and 23 of the side-chain, and a seven-membered lactone B ring.
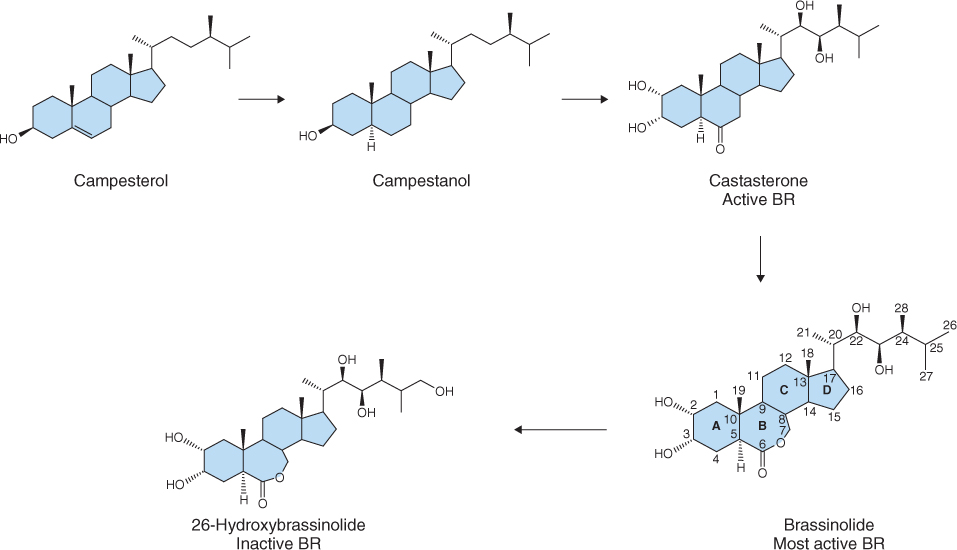
Figure 10.30 Brassinosteroid bioassay. (A) Diagram of rice seedling showing the lamina and leaf sheath. Brassinosteroids to be bioassayed are applied at the leaf sheath–lamina joint and the angle of the lamina is measured after 2 days of growth. (B) Photographs showing the effect of increasing BR on the lamina angle of rice seedlings.
Genetic analysis in Arabidopsis has shown that BRs are required for normal plant development. Mutations in BR biosynthesis and signaling pathways show a variety of phenotypes including extreme dwarfism as a result of Bri1 (BR insensitive1), a mutation in the BR receptor, and de-etiolation in mutants such as det2 (de-etiolated2) and cpd (constitutive photomorphogenesis and dwarfism). The de-etiolation phenotype is most pronounced in dark-grown Arabidopsis seedlings, where mutants exhibit short, fat hypocotyls, expanded cotyledons and the presence of anthocyanin pigments (Figure 10.31).
Figure 10.31 Effect of mutations in the brassinosteroid biosynthesis pathway on plant morphology, showing dark-grown wild-type and de-etiolated2 (det2) mutant Arabidopsis plants defective in brassinosteroid synthesis.

Brassinosteriods are synthesized from simpler sterol precursors such as sitosterol (C29), stigmasterol (C29), campesterol (C28) and cholesterol (C26) (see Figure 15.21). Although sitosterol is the most abundant sterol in plants, campesterol is the favored substrate for BR biosynthesis. Campesterol is converted to castasterone through a series of reactions that involve oxidation at carbon 6, reduction at carbon 5 and hydroxylation by enzymes of the cytochrome P450 class that add hydroxyl groups at carbons 2, 3, 22 and 23 (Figure 10.29). There are many intermediates between campestanol and castasterone and research indicates that the order of hydroxylation reactions is random. Formation of a seven-carbon lactone B ring results in the conversion of castasterone to BL.
As with other hormones, maintenance of BL homeostasis is central to its effectiveness as a hormone. BL biosynthesis is regulated by feedback loops that transcriptionally regulate the expression of many of the genes that encode the cytochrome P450 enzymes involved in hydroxylation of campestanol. Thus when BL concentration is high, its biosynthesis is reduced. Catabolism of BL is less well understood but hydroxylation, epimerization, oxidation and sulfonation, as well as conjugation to sugars and lipids, are all known to lead to a reduction in BL activity. The best understood reaction is hydroxylation at carbon 26 by a cytochrome P450 enzyme encoded by the gene BAS1 (Figure 10.29), which results in lowering of BL levels and produces dwarfism in Arabidopsis.
The BR receptor is a leucine-rich repeat (LRR) receptor serine/threonine kinase localized on the plasma membrane (Figure 10.32). This receptor was identified in a screen for Arabidopsis mutants that show normal root growth at high BL concentration. Mutants that grew normally in the presence of high BL were called bri1 (brassinolide insensitve1). The extracellular domain of BRI1 consists of multiple LRR repeats and a BL-binding domain, while the cytoplasmic domain consists of a serine/threonine protein kinase domain. An inhibitor of BRI1 phosphorylation called BKI1 (BRI1 kinase inhibitor1) prevents phosphorylation of the cytoplasmic domain of BRI1 and this inhibition is relieved as a consequence of BL binding. BL attaches to the binding site located in the extracellular portion of the receptor and initiates BRI1 activation. Activation involves autophosphorylation at multiple sites and the formation of homodimers. Activated BRI1 then forms heterodimers with BAK1, another LRR receptor kinase. Subsequently the activated heterodimer phosphorylates soluble cytoplasmic BR signaling kinases (BSKs).
Figure 10.32 Brassinosteroid (BR) signal transduction pathway. (1) The BR receptor BRI1 is located on the plasma membrane where it forms homo-oligmers. Upon binding BR, BRI1 becomes phosphorylated and releases BRI1 kinase inhibitor (BKI1) into the cytosol. (2) Activated BRI1 forms a hetero-oligomer with BAK1, which becomes phosphorylated. (3) It in turn phosphorylates BSK proteins. (4) BSK then activates the protein phosphatase BSU. (5) BSU dephosphorylates BIN2, causing it to be degraded by the ubiquitin–proteasome system (UbPS). (6) In the absence of BR, phosphorylated BIN2 phosphorylates the transcriptional regulators BES1 and BZR1, thereby abolishing their activity and targeting them for degradation by the UbPS, or by enhancing their transport to the cytoplasm when they bind 14-3-3 proteins. (7) In the presence of BR, BES1 and BZR1 are not phosphorylated and are active as transcriptional regulators leading to brassinosteroid responses.
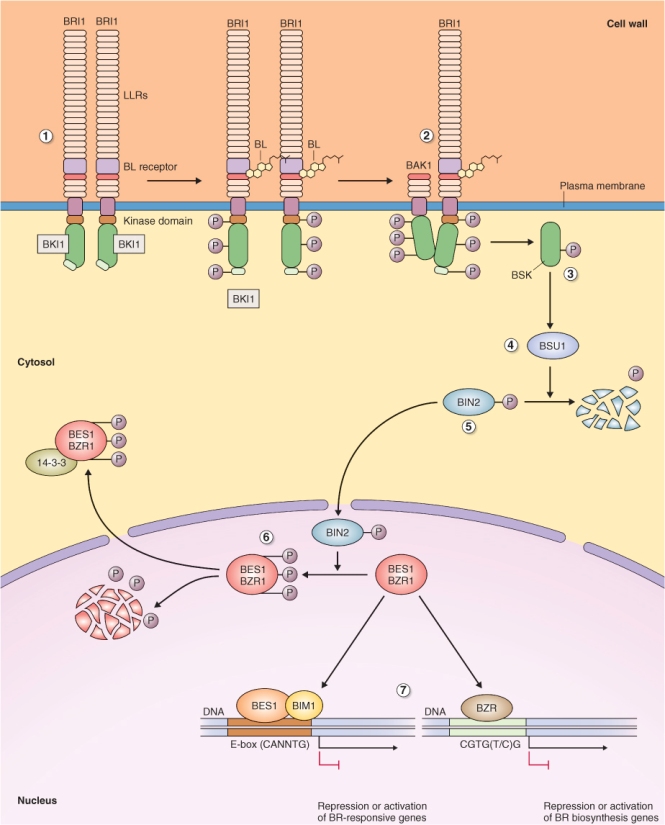
The role of BSK proteins is to bind to and activate a protein phosphatase known as BSU1 (BRI1 suppressor1). Activated BSU1 dephosphorylates BIN2 (Br insensitive2), a repressor of BR-induced gene expression, leading to its degradation by the UbPS. In the absence of BL, the phosphorylated form of BIN2 phosphorylates two DNA-binding proteins BES and BZR1, which cannot bind DNA when phosphorylated. BIN2 may participate in either activation or inhibition of transcription, because once BES and BZR are dephosphorylated they can bind DNA and act to either activate or repress transcription. Phosphorylated BES and BZR1 are rapidly turned over by the 26S proteasome or exported to the cytoplasm with the aid of 14-3-3 proteins (see Chapter 5).
Abscisic acid (ABA) was isolated and its structure was elucidated in the early 1960s. At that time, one of the bioassays used to test for the presence of ABA in plant extracts measured abscission of petioles from cotton seedling explants in sealed containers, and this bioassay is the origin of the trivial name abscisic acid. It is now known that the effect of ABA on abscission in this assay was indirect. ABA stimulates ethylene production by cotton seedling explants, and enhanced ethylene synthesis in the sealed containers was responsible for the abscission observed. ABA has been shown to be involved in responses of plants to abiotic stress including drought, and one of its effects on plant function is to bring about rapid closure of stomatal guard cells (see Chapter 14). Other roles for ABA include imposing bud and seed dormancy, suppressing germination and accelerating leaf senescence (see Chapters 6, 17 and 18). As leaves senesce they produce more ethylene which brings about abscission in some species.
Abscisic acid is a tetraterpene synthesized in the plastid from intermediates in the isoprenoid pathway (see Figures 10.2 and 10.14). Steps in the biosynthesis of ABA are similar to those for carotenoids, and the pigment β-carotene is an intermediate in its synthesis (Figure 10.33). The phenotypes of ABA-deficient mutants include those that wilt readily because they have poor control of stomatal closure, and many that exhibit precocious seed germination or vivipary as in the case of some Zea mays mutants (see Figure 17.20). β-Carotene is converted to zeaxanthin in several steps, and zeaxanthin is converted in a single reaction to trans-violaxanthin. Violaxanthin can follow two routes to ABA. Under conditions of abiotic stress, trans-violaxanthin is converted to cis-neoxanthin via the intermediate trans-neoxanthin. Alternatively, trans-violaxanthin can be converted directly to cis-neoxanthin and thence to xanthoxin, the first committed step in ABA biosynthesis, in a reaction catalyzed by the enzyme NCED (9-cis-epoxycarotenoid dioxygenase), a product of the Arabidopsis NCED gene. Xanthoxin has biological activity similar to that of ABA. Several AtNCED genes have been isolated; AtNCED3 catalyzes xanthoxin synthesis during abiotic stress, whereas AtNCED6 and AtNCED9 catalyze xanthoxin synthesis in seeds.
Figure 10.33 Abscisic acid (ABA) biosynthesis from isopentenyl diphosphate begins in the plastid. Xanthoxin conversion to ABA occurs in the cytosol. AAO, ABA-oxidase; NCED, 9-cis-epoxycarotenoid dioxygenase; SDR, short chain dehydrogenase/reductase.
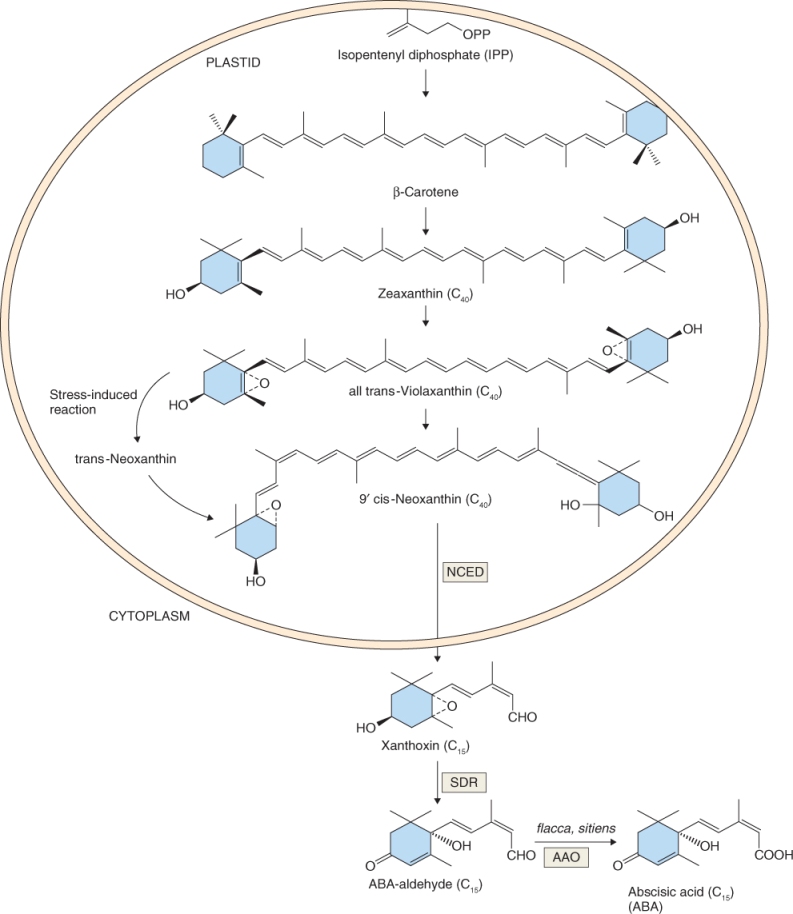
Synthesis of ABA from xanthoxin occurs in the cytosol in two steps, beginning with the formation of ABA-aldehyde. In Arabidopsis the ABA2 gene encodes an enzyme that catalyzes conversion of xanthoxin to ABA-aldehyde. The conversion of ABA-aldehyde to ABA is catalyzed by ABA-oxidase (AAO). Many mutants have been isolated that have defects in the conversion of ABA-aldehyde to ABA including flacca and sitiens in tomato (Solanum lycopersicum), nar2a in barley (Hordeum vulgare) and aba3 and aao3 in Arabidopsis.
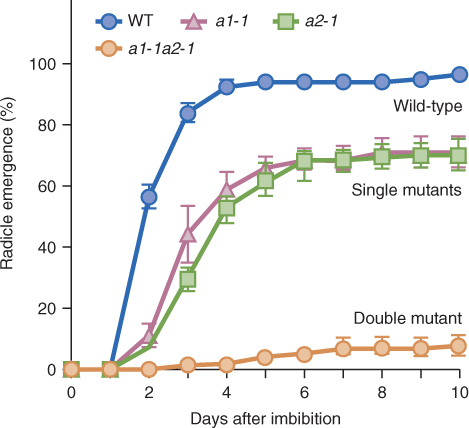
Catabolism of ABA and its conjugation to small molecules such as sugars are both mechanisms that ensure that ABA is quickly removed from cells, which allows it to function as a signaling molecule. The main pathway for ABA catabolism is its oxidation to biologically inactive 8′-hydroxy-ABA, phaseic acid and dihydrophaseic acid (Figure 10.34). ABA 8′-hydroxylase catalyzes 8′-hydroxy-ABA synthesis and, like the enzymes that inactivate BRs and GAs, it is a cytochrome P450 monooxygenase, in this case CYP707A. Loss-of-function mutations in CYP707A cause a drastic reduction in germination of Arabidopsis seeds, emphasizing the importance of ABA in germination and of CYP707A in ABA inactivation (Figure 10.35). Conjugation of ABA to sugars is another route for removing active ABA from cells. ABA β-glucosyl ester is an important catabolite that is thought to be stored in the vacuole. Enzymes that can hydrolyze ABA conjugates have been isolated and it has been proposed that conjugates can provide a supply of active ABA.
Figure 10.34 Steps in the inactivation of abscisic acid (ABA). ABA inactivation by esterification with glucose is reversible. ABA is irreversibly inactivated when converted to more oxidized derivatives such as 8'-hydroxy-ABA, phaseic acid and dihydrophaseic acid. The step from ABA to phaseic acid is catalyzed by a member of the CYP450 enzyme family.
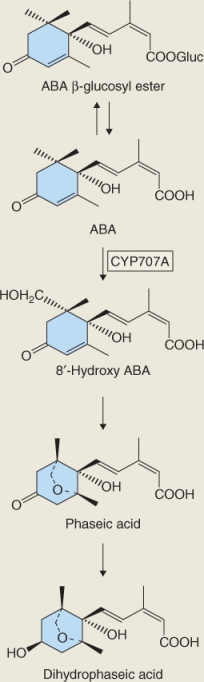
Figure 10.35 Mutations in the abscisic acid (ABA) 8'-hydroxylase gene CYP707A lead to an accumulation of active ABA, hence reduced germination of Arabidopsis seeds. All seeds of wild-type (WT) plants germinate by 4 days but single (a1-1, a2-1) and double (a1-1a2-1) mutants show decreasing levels of germination.
Abscisic acid elicits two distinct response types in plants: rapid responses such as the loss of turgor in guard cells to close stomata; and more gradual responses including effects on seed dormancy and germination and reactions to abiotic stresses. It was proposed that these relatively fast and slow responses use different receptors, making their isolation in a mutant screen more difficult. The likelihood that there are multiple ABA receptors is supported by evidence that indicates the existence of both soluble and membrane-bound receptors. Three classes of ABA receptors have been identified to date. These include the plasma membrane-localized G proteins GTG1 and GTG2; a plastid-localized enzyme that coordinates nucleus to plastid signaling; and cytosolic ligand-binding proteins of the START domain superfamily. Recent research shows that soluble START domain proteins are the principal ABA receptors that function in responses as diverse as stomatal closing, germination and abiotic stress.
The identification of START domain proteins as ABA receptors arose from two different experimental strategies. One employed pyrabactin (PY), a synthetic compound that mimics ABA, and searched for Arabidopsis mutants that were PY-insensitive. The gene that conferred insensitivity to PY was cloned and called Pyrabactin Resistance1 (PYR1). PYR1, and its homologs, which in Arabidopsis are called PYL (PYR-like), are START domain proteins and have been shown to be ABA-dependent inhibitors of one class of protein phosphatase type 2C (PP2C) proteins (Figure 10.36). What made this discovery exciting was the knowledge that the products of genes corresponding to several types of ABA-insensitive Arabidopsis mutants, including ABI1 and ABI2, were members of the PP2C class of proteins previously shown to be involved in ABA signaling. A different experimental approach, based on protein–protein interaction, also found a link between PYR1/PYL and PP2Cs. Using ABI1 and ABI2 as bait in the yeast two-hybrid system (see Figure 8.14) resulted in the isolation of additional PYR1/PYL homologs, called regulatory component of ABA receptors (RCARs).
Figure 10.36 Simplified model of abscisic acid (ABA) signal transduction via the soluble PYR receptor. PYR binds ABA in the cytosol and then binds protein phosphatase PP2C, inhibiting its activity. Protein kinases of the SNRK class are downstream of PP2C. When phosphorylated, these kinases modulate the activity of ion channels, such as the potassium channel KAT1, and of transcription factors, such as the ABFs (ABRE binding factors) that regulate the activity of genes containing ABA response elements (ABREs).

Proteins of the PYR1/PYL/RCAR group form dimers, specifically bind ABA and function as ABA receptors. When one of these dimers binds ABA, it can, in turn, bind members of the PP2C class of protein phosphatase, inhibiting the activity of PP2Cs. The target of PP2Cs is a class of SNF1-related protein kinase 2 (SNRK2). Analysis of Arabidopsis mutants has shown that protein kinases of the SNRK class, such as OST (Open Stomata), are positive regulators of the ABA signaling pathway. In other words, SNRK kinases are active in the presence of ABA and are required for ABA signal transduction. ABA exerts its physiological effects by regulating the activity of SNRK2 kinases, which in turn phosphorylate target proteins. In the absence of ABA, PP2Cs such as ABI1 and ABI2 are active and dephosphorylate SNRK2, inactivating these kinases. Conversely, in the presence of ABA, PP2C is inhibited and SNRK kinases are active. Many downstream targets of the PYR/PYL/RCAR-PP2C-SNRK2 pathway have now been identified. These include ion channels (e.g. the Arabidopsis potassium channel KAT1—a member of the same family of channels at AKT1 which is discussed in Chapter 13), the reactive oxygen-generating enzyme RbohF, an NADPH oxidase (see Chapter 18), and ABA-responsive transcription factors such as ABF2. Transcription factors of the ABF2 class interact with ABA response elements (ABREs) in the promoters of ABA-responsive genes (see Chapter 3).
Strigolactones (SLs) were initially isolated from plant root exudates as compounds that stimulate the germination of seeds of several parasitic plants from the family Orobanchaceae, such as Orobanche spp. (broom rape) and Striga spp. (witch weed) (Figure 10.37; see also Figure 15.42), and the branching of mycorrhizal hyphae. Striga asiatica is a common parasite of grasses and can cause total crop losses in some instances. Orobanche species, on the other hand, are highly specific parasites of many eudicot crops such as Helianthus annuus (sunflower). The severity of parasitism by Striga and Orobanche is increased by the fact that each plant produces thousands of very small seeds that persist in the soil.
Figure 10.37 Orobanche hederae (brown stalks) growing on ivy (Hedera helix).

In addition to their involvement in promoting invasion by harmful parasites, SLs have been shown to act as host recognition signals in arbuscular mycorrhizal fungi (see Chapter 12). These fungi form beneficial symbiotic associations with the roots of most (>80%) plants. SLs released by the roots of host plants serve to attract the arbuscular mycorrhiza (AM) to the root surface and are also thought to stimulate branching of the fungal hyphae. SLs are synthesized not only by plants that are hosts to AM but by non-host species too, such as Arabidopsis and white lupine (Lupinus albus). Taking these observations together with the fact that roots of plants that are not hosts to parasitic weeds also produce SLs, it became clear that these novel plant compounds were likely to play roles in normal plant growth and development.
Strigol was the first SL to be purified. It has a characteristic chemical structure that includes two lactone rings (rings C and D) linked via an enol ether bridge (Figure 10.38). Strigol was first isolated from root exudates of Gossypium hirsutum (cotton), a plant that is not parasitized by members of the Orobanchaceae. G. hirsutum is therefore one of many crops that is used as a ‘Striga trap’, that is, it produces strigol which induces germination of Striga seeds in the absence of an appropriate host plant, thus reducing the abundance of viable seeds in soil.
Figure 10.38 Chemical structures of strigolactones. Orobanchol and strigol are biologically active strigolactones isolated from plants, while 5-deoxystrigol is thought to be a precursor of strigol. GR24 is a synthetic strigol analog.
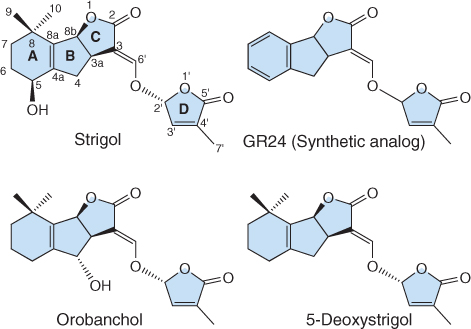
More than ten SLs have now been isolated from plants and all stimulate the germination of weedy parasites. Orobanchol, isolated from red clover (Trifolium pratense) root exudates, stimulates the germination of seeds of many Orobanche species. 5-Deoxystrigol has been found in root exudates of almost all plants that produce SLs, leading to the hypothesis that strigol, orobanchol and other SLs are products of its hydroxylation (Figure 10.38). SLs are present in plants at low concentration, and their chemical synthesis has only recently been achieved. Therefore, research on SL function has relied on the use of a synthetic SL analog known as GR24 (Figure 10.38). Note the similarity between GR24 and naturally occurring SLs, especially the presence of C and D rings and the enol ether bridge.
As we have discussed earlier in this chapter, auxins and CKs have profound effects on branching in plants. The analysis of branching mutants of several species, especially max (more axillary growth) of Arabidopsis, rms (ramosus) of garden pea (Pisum sativum) and various dwarf (d17 and d10) mutants of rice (Oryza sativa), led to the recognition that SLs are potent regulators of branching (Figure 10.39). MAX1, MAX3, MAX4, RMS1, RMS5, D10 and D17 are all members of the family of plastid localized enzymes known as carotenoid cleavage dioxygenases (CCDs). Mutations in CCD-encoding genes result both in increased branching, also called tillering in O. sativa, and in much lower endogenous SL concentrations. These mutations can be rescued by the addition of the synthetic SL GR24 (Figure 10.39A). These and other results indicate that branching in O. sativa as well as in Arabidopsis and P. sativum is regulated by SL, whose synthesis is dependent on the activity of plastid-localized CCDs. Grafting experiments in Arabidopsis showed that SLs are produced by the root system and are translocated upward in the transpiration stream where they act to prevent the outgrowth of lateral branches (Figure 10.39B). In these experiments mutant scions were grafted on to wild-type (WT) rootstocks and in the case of max3 and max4, which lack enzymes required for SL biosynthesis, the mutant phenotype was rescued. The max1 mutant of Arabidopsis encodes a CYP450 enzyme and can also be rescued by application of GR24 and by grafting the max1 scions to WT rootstock. The results of these experiments suggest that SLs are produced in roots by a pathway that involves CCD and CYP450 enzymes. The Arabidopsis max2 mutant cannot be rescued by root grafting to WT plants. In this case the mutation is in an F-box protein gene that is involved in SL signaling, not synthesis, which means that no amount of added SL can rescue the phenotype.
Figure 10.39 Branching mutants in Arabidopsis and rice indicate a role for strigolactones in regulating axillary bud growth. (A) Wild-type and d3 and d10 mutants of rice showing that the tillering (branch growth) phenotype of the mutant can be suppressed by treatment with the synthetic strigolactone GR24. (B) Drawings of wild-type (WT) and several max mutants of Arabidopsis (top row) and the corresponding mutant scion grafted to a WT rootstock (bottom row). Mutants in strigolactone synthesis (max1, max3, max4) can be rescued by grafting to WT but plants with a mutation in the strigolactone signaling gene max2 cannot be rescued by grafting.

An SL biosynthetic scheme that incorporates work with the various branching mutants discussed above is shown in Figure 10.40. Carotenoids are cleaved by CCDs to a product that is converted to SL by the action of CYP450 enzymes. Signaling from SLs leading to inhibition of lateral bud growth involves an F-box protein, which is part of an SCF-type E3 ubiquitin ligase and is a product of the MAX2 gene. The role of SLs in branching is discussed further in Chapter 12.
Figure 10.40 Steps in the synthesis and signal transduction pathway of strigolactones (SLs). SLs are synthesized in plastids from carotenoids and converted to active SLs in the cytosol by enzymes of the cytochrome P450 (CYP450) class. SL signaling involves the participation of the UbPS. Mutations affecting steps leading to SL synthesis in Arabidopsis, Petunia, Pisum and rice and an F-box protein in the SL signaling pathway are shown.

Jasmonates are a group of naturally occurring compounds, similar to mammalian prostaglandins, that are synthesized from the fatty acid linolenic acid (Figure 10.41). Jasmonic acid (JA) and methyl jasmonate (MeJA) are the most commonly occurring jasmonates in plants. MeJA is also one of the principal components of fragrance produced by jasmine (Jasminum grandiflorum). The roles of jasmonates as plant hormones were discovered in the 1970s, when JA was reported to inhibit the growth of rice, wheat and lettuce seedlings. It was subsequently shown to inhibit seed and pollen germination, retard root growth and stimulate the curling of tendrils. JA also promotes tuber formation and the accumulation of storage proteins in seeds.
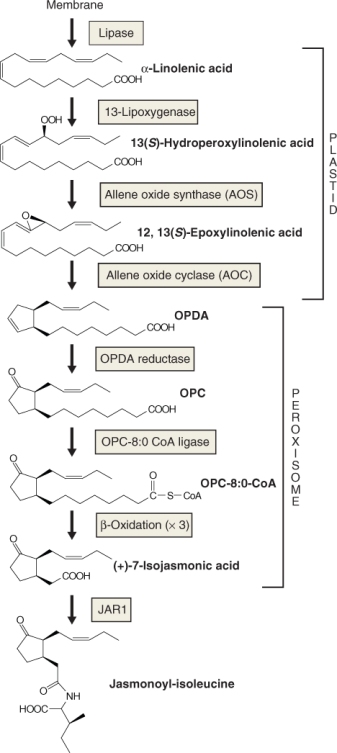
Figure 10.41 Biosynthesis of jasmonic acid. α-Linolenic acid released from membranes by lipase activity is converted to oxo-phytodienoic acid (OPDA) in the plastid followed by several steps, including three cycles of β-oxidation that occur in the peroxisome. JA conjugated to isoleucine is the active form of jasmonate in the cytosol. JAR1, product of the JASMONATE RESISTANT 1 locus; OPC, oxo-cyclopentane octanoic acid.
Arabidopsis mutants that are defective in JA biosynthesis or response are male-sterile because anther filaments do not elongate to allow dispersal of pollen to the stigma. Furthermore, anther dehiscence does not occur when the flowers open, and the majority of mutant pollen grains are not viable. Fertility of these mutants can be rescued by JA treatment. Jasmonates also play an important role in plant defense as they are produced when herbivores damage plant tissue (see Chapter 15).
Two organelles, the plastid and peroxisome, are involved in the synthesis of JA from linolenic acid. The first step in JA biosynthesis is catalyzed by a lipase that releases α-linolenic acid, a C18 polyunsaturated fatty acid, from plastid membrane lipid. Linolenic acid is converted to oxo-phytodienoic acid (OPDA) in the plastid by three enzymes, lipoxygenase, allene oxide synthase and allene oxide cyclase. It is not understood how OPDA exits the plastid, but it is transported into the peroxisome by an ABC class transporter (see Chapter 5). OPDA is then converted to JA in the peroxisome in five further steps catalyzed by OPDA reductase and oxo-phytoenoic acid CoA ligase followed by three rounds of β-oxidation (Figure 10.41; see Chapter 6).
JA is conjugated via an amide link to isoleucine (Ile) and other amino acids to yield jasmonoyl-isoleucine (JA-Ile) and other jasmonoyl–amino acid conjugates, respectively (Figure 10.41). JA-Ile is thought to be the functionally active form of JA. In Arabidopsis, conjugation of JA to amino acids is catalyzed by an enzyme encoded by the JAR1 (JASMONATE RESISTANT 1) locus. The JAR1 gene was isolated in a screen for plants that were resistant to high JA concentrations, a phenotype that suggests that conversion of JA to JA-Ile is required to activate the JA signaling pathway. Unlike the synthesis pathways of other plant hormones, JA-Ile synthesis is under positive feedback regulation. JA treatment results in elevation of the synthesis of many JA biosynthetic genes including lipoxygenase, AOS, AOC, the gene encoding OPDA reductase (OPR3) and JAR1.
In addition to being converted to JA-Ile, JA is also the precursor of volatile methyl jasmonate, the product of a SAM-dependent carboxyl methyltransferase. Methyl jasmonate is biologically active and is thought to act as a volatile signal in plant defenses against herbivory (see Chapter 15). There are many other catabolites of JA that accumulate in plants and which have biological activity, including tuberonic acid which, as the name suggests, promotes tuber formation. Surprisingly little is known about the formation of inactive JA catabolites. As we have emphasized, mechanisms for removing hormones from cells are as important as those that bring about their synthesis. Perhaps, as is the case with ethylene, the volatility of JA and its derivatives makes chemical inactivation redundant.
Jasmonic acid regulates the transcription of a large number of genes, especially those that are related to defense responses in plants. The JA signaling pathway resembles the pathways of several other plant hormones in that the UbPS is involved in the degradation of a member of the JAZ family of transcriptional repressor proteins (Figure 10.42). JAZ functions to repress the activity of transcription factors that regulate JA-responsive genes. Proteolysis of JAZ results in relief of repression and JA-responsive genes are transcribed. JA targets JAZ proteins for ubiquitination by binding to the F-box protein COI1, a component of the SCFCOI1 ubiquitin ligase complex.
Figure 10.42 A simplified model of the jasmonic acid (JA) signal transduction pathway. Jasmonate forms a complex with the F-box protein COL1 that targets the transcription repressor JAZ for degradation by the UbPS. Release of inhibition of JAZ repression allows the transcription of JA-responsive genes by transcription factors such as MYC2. Transcribed genes include defense proteins.

JAZ proteins repress transcription by binding to MYC2, a transcription factor required for the expression of JA-responsive genes. When JA-Ile binds the SCFCOI1 complex it facilitates the binding of JAZ to the complex, which leads to its ubiquitination and eventual destruction by the proteasome. Repression of MYC2 is thus relieved and transcription of JA-responsive genes is initiated. Interestingly, genes that encode JAZ are among those upregulated by JA-Ile, giving rise to a negative feedback loop.
Polyamines (PAs) are small organic compounds that have two or more amino groups and range in molecular mass from 88 Da for putrescine to 202 Da for spermine. Putrescine, spermidine and spermine are the most abundant PAs in plants, but in legumes cadaverine is also abundant (Figure 10.43). Other less abundant PAs such as thermospermine are very important physiologically. Unlike the other hormones discussed so far, PAs are found in plants at millimolar concentrations. They elicit a range of responses, including cell differentiation and division, embryogenesis, flower development, fruit ripening, root initiation, abiotic stress tolerance and tuber formation.
Figure 10.43 Chemical structures of polyamines (PAs) that occur naturally in plants. Putrescine, spermidine and spermine are the most common PAs but cadaverine is abundant in legumes.
PAs have a high affinity for anionic constituents such as DNA, RNA, phospholipids and acidic proteins, as well as anionic groups in membranes and cell walls, including hydroxycinnamic acids. In addition to being present as free amines, a significant fraction of the PA pool is found as amide conjugates with p-coumaric, ferulic and caffeic acids. Conjugates between PAs and hydroxycinnamic acids play roles in development including flowering and seed and fruit development, and in hypersensitive responses to viral and fungal infections.
Putrescine, the simplest of the PAs, can serve as the precursor of spermidine and spermine synthesis (Figure 10.44). In some species putrescine can be synthesized from L-ornithine (Orn) in one step catalyzed by Orn decarboxylase (ODC), or from L-arginine (Arg) in a two-step process initiated by Arg decarboxylase (ADC). ODC is usually less active than ADC, suggesting that Arg decarboxylation is the major pathway for putrescine synthesis. Support for the importance of Arg in PA synthesis comes from the observation that Arabidopsis has two ADC genes but lacks an ODC gene. Furthermore, Arabidopsis mutants that lack both ADC genes produce abnormal seeds that fail to germinate. In many species putrescine production is increased by environmental stress as a consequence of upregulation of ADC activity. Exposing Arabidopsis to low temperature, high salt or dehydration, as well as ABA treatment, induces the expression of ADC1 or ADC2.
Figure 10.44 Biosynthesis of polyamines from arginine, ornithine and S-adenosyl methionine.
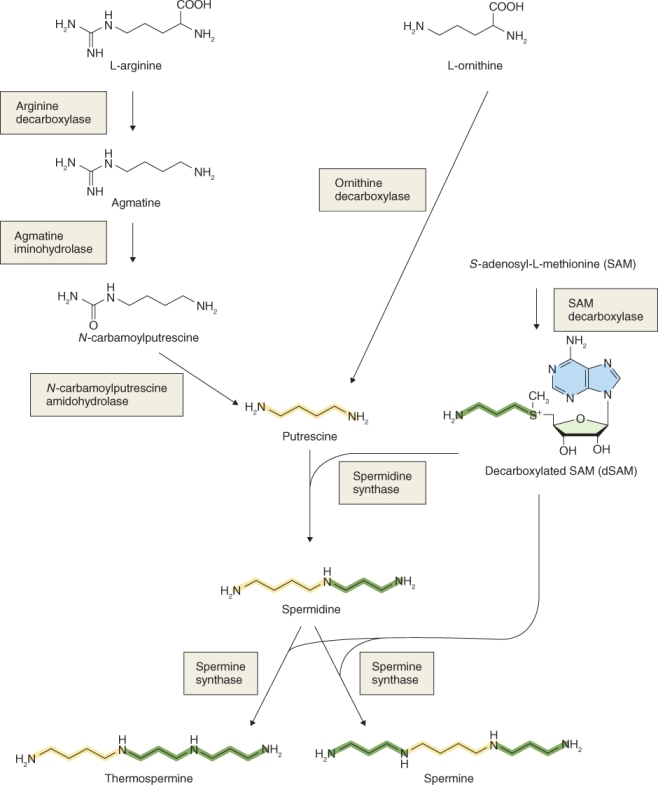
SAM and putrescine are precursors of spermidine, spermine and thermospermine synthesis (Figure 10.44). As we have seen in Section 10.5.1, SAM is also a precursor of ethylene; consequently PAs and ethylene act as competitive inhibitors of each other's biosynthetic pathways. SAM is first decarboxylated to dSAM, which plays a role in the synthesis of spermidine from putrescine catalyzed by spermidine synthase. Spermine and thermospermine are synthesized from spermidine in reactions catalyzed by spermine synthases, in which dSAM also participates. The synthesis of dSAM is under feed-forward and feedback control. Its synthesis increases as putrescine levels rise but it is inhibited by spermidine.
There are two Arabidopsis genes encoding spermine synthase, SDMS and ACL5. SDMS catalyzes the synthesis of spermine and ACL5 catalyzes the synthesis of thermospermine. ACL5 loss-of-function mutants have defects in shoot xylem differentiation and elongation (Figure 10.45). The phenotype can be rescued by the application of thermospermine, but not spermine, indicating that thermospermine is required for normal stem elongation in Arabidopsis.
Figure 10.45 Mutations in the acl5 gene, that encodes the enzyme for thermospermine synthesis from spermidine, cause defects in xylem (X) development in Arabidopsis seedling hypocotyls. Light micrographs of stem cross-sections sampled after 5, 13 and 35 days' growth of wild-type (A, C, E) and acl5 (B, D, F) seedlings. Note the very aberrant xylem development in the acl5 mutant.
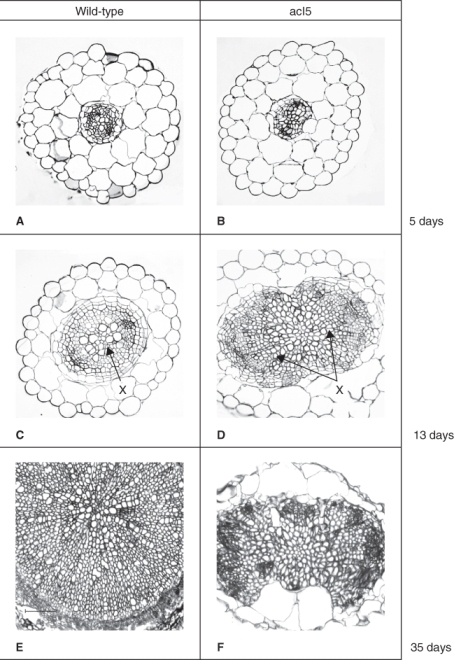
PA catabolism is catalyzed by two classes of enzymes, copper-containing diamine oxidases and flavin-linked polyamine oxidases. Both classes of enzyme generate H2O2 while PAs are broken down eventually to inactive compounds. Plants contain many forms of polyamine oxidase and some are localized to the apoplast where the H2O2 they release plays a role in lignification by cross-linking hydroxycinnamic acids. Three of the five polyamine oxidases of Arabidopsis are localized to peroxisomes, but H2O2 released by these enzymes is unlikely to be developmentally important since the abundant catalase in this organelle will convert H2O2 to H2O.
The effects of PAs on tracheary element differentiation have been extensively studied in Arabidopsis and in the model system Zinnia elegans. Arabidopsis acl5 mutants have reduced thermospermine levels and defective xylem differentiation in hypocotyls (Figure 10.45). Compared with WT, vessel elements in acl5 mutants are small, have only spiral thickenings and lack pits in their walls. Xylem fibers are also absent. The observations that spermine application can reverse the effects of acl5 on xylem differentiation, and that ACL5 is expressed specifically in developing xylem elements in WT Arabidopsis, lend strong support to the idea that PA synthesis is required for normal xylem development.
Further evidence for a role of PAs in xylem differentiation comes from work with Zinnia elegans, in which tracheary element differentiation can be studied at the cellular level in isolated cell cultures. The addition of spermine to cultured Z. elegans cells that have been induced to form tracheary elements dramatically increases vessel size, and wall thickenings are more elaborate. It is likely that in both Arabidopsis and Z. elegans PAs play a role in delaying programmed cell death of tracheary elements, the final step in the differentiation of functional vessels and tracheids (see Chapter 18).
Salicylic acid (SA) has long been recognized as a biologically important molecule produced by plants. Until recently its biological importance was related to the medicinal role of its derivative acetylsalicylic acid, more commonly known as aspirin. SA was initially isolated from the bark of Salix alba or willow, where it is abundant, hence its name. Its formal chemical name is 2-hydroxybenzenecarboxcylic acid and it has a molecular mass of 138 Da.
There are two possible routes for the synthesis of SA in plants, one from trans-cinnamic acid (tCA), the other from chorismic acid (Figure 10.46). In the tCA pathway trans-cinnamic acid is synthesized from phenylalanine in a reaction catalyzed by phenylalanine ammonia lyase (PAL). tCA is converted to benzoic acid by the reactions of β-oxidation in which CoA is added to tCA; then trans-cinnamoyl-CoA undergoes three rounds of β-oxidation to produce benzoyl-CoA which is converted to benzoic acid. Benzoic acid is hydroxylated to form SA by a soluble cytochrome P450 enzyme. Support for the role of this pathway in SA synthesis comes from experiments that show that when Nicotiana tabacum plants are inoculated with tobacco mosaic virus (TMV), PAL activity is suppressed and there are much lower levels of SA.
Figure 10.46 Biosynthesis of salicylic acid from phenylalanine and chorismate. SA can be converted to methyl salicylate or be conjugated to glucose.
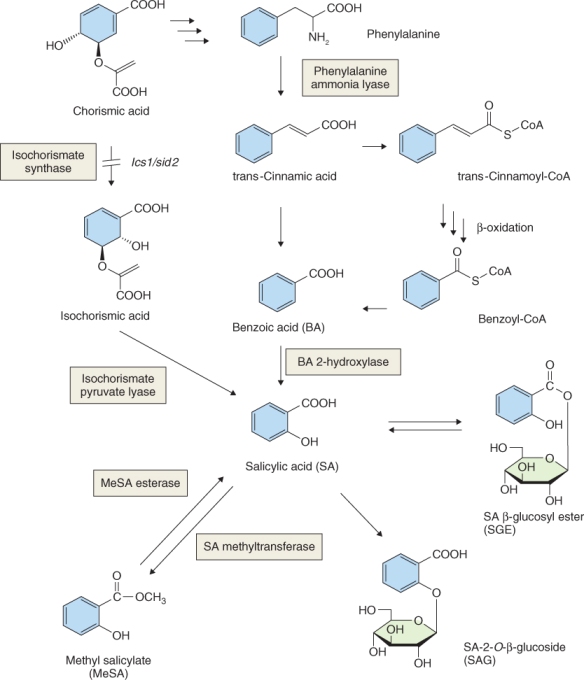
The chorismate pathway for SA synthesis occurs in two steps (Figure 10.46). The first step is catalyzed by isochorismate synthase, encoded by the ICS1/SID2 gene; the second is catalyzed by isochorismate pyruvate lyase (IPL). The ics1/sid2 mutant of Arabidopsis fails to accumulate SA under various inductive conditions, suggesting that the chorismate pathway predominates in this species. Observations on other species, however, show that SA is synthesized via the tCA pathway. Tobacco plants experiencing abiotic stress from ozone exposure synthesize SA from benzoic acid in the absence of ICS expression. In Arabidopsis, on the other hand, ICS is expressed in response to ozone and SA accumulates. These data suggest that the pathway for SA biosynthesis in plants is species-specific and perhaps treatment-specific.
SA is converted in vivo to methyl salicylate (MeSA), and glucosyl esters and glucosides can be produced (Figure 10.46). Methylation of SA on the carboxyl group to form MeSA is catalyzed by a carboxyl methyltransferase that uses SAM as the methyl donor. MeSA can be converted back to SA by MeSA esterase.
Salicylic acid can retard senescence of petals, induce flowering in duckweeds and bring about thermogenesis (heat production) in arum lilies from the family Araceae (see Figure 7.18). Adding aspirin to a vase of cut flowers is a widely used remedy to delay their senescence. In this case, the mechanism of SA action is a consequence of its effects on the conversion of ACC to ethylene. SA inhibits ACO, which lowers ethylene levels and thus delays senescence. Duckweeds (small aquatic angiosperms of the family Araceae that includes Lemna gibba, Spirodela polyrrhiza and Wolffia microscopia) require a series of long days to flower but can be induced to flower in short days if treated with SA. It is unlikely that SA is an endogenous flowering hormone in duckweeds, since it is relatively abundant in plants grown under both long and short days.
Arum spadix thermogenesis is an interesting biological phenomenon and SA is thought to be the signal that induces this process. The spadix produces amines that become volatile as the temperature of the spadix tissue increases by 10 °C or more, the result of thermal energy released by uncoupled electron flow through the alternative oxidase pathway in mitochondria (see Chapter 7). The pungent volatilized amines attract pollinators to the flower. The thermogenic-inducing principle was initially called calorigen and was identified as SA in the late1980s. SA application to the spadix can bring about thermogenicity, accompanied by an increase in endogenous SA levels—an example of positive feedback.
In response to pathogen infection many plants develop resistance to the disease by limiting the spread of the pathogen (see Chapter 15). Not only is localized spread of the pathogen limited but parts of the plant remote from the site of infection also develop resistance to the pathogen, a phenomenon called systemic acquired resistance (SAR). SAR is also associated with the production of a suite of proteins called pathogenesis-related (PR) proteins. SA is an important participant in this mechanism of resistance to pathogenic infection.
Evidence that SA was involved in SAR came from experiments showing that SA treatment of leaves of Nicotiana tabacum cv. Xanthi, which are susceptible to TMV, induces the synthesis of PA proteins and resistance to TMV infection. Exposure to SA has now been shown to induce SAR in a variety of pathogen-infected plants. Further proof that SA was involved in SAR came from experiments showing that infection of TMV-resistant N. tabacum plants with TMV causes a 40-fold increase in SA levels in inoculated leaves and a ten-fold increase in leaves remote from the infection site. The increase in SA concentrations in TMV-resistant plants is accompanied by enhanced expression of genes that encode PA proteins in inoculated and uninoculated leaves. N. tabacum plants that are susceptible to TMV do not show an increase in SA levels or in PA protein gene expression when infected with the virus. Results from these and similar experiments led to the conclusion that SA plays a key role in signal transduction leading to the development of SAR.
MeSA has a function in SAR that appears to differ between species. Experiments show that it is essential for SAR in N. tabacum, where it plays a role in regulating the pool of available SA. There is, however no evidence that MeSA participates in SA homeostasis or SAR in Arabidopsis.
Nitric oxide (NO) is a gas produced by all living organisms. Investigations in the 1980s into the biochemistry of nitrate (NO3−) metabolism by plants led to the discovery that NO can be synthesized during nitrate assimilation when NO3− is reduced to NH4+. Bacteria, on the other hand, generate NO during the process of denitrification when NH4+ ions are oxidized to N2 (see Chapter 13). Animals produce NO from arginine using the enzyme NO synthase. NO synthases have not been found definitively in plants and, as we will discuss below, plants can use a variety of other enzymes and pathways to produce NO, including the enzyme nitrate reductase that plays a key role in the conversion of NO3− to NH4.
In mammals, NO is a regulator of vasodilation, a property that has generated a great deal of research interest and led to the development of drugs such as Viagra. NO also elicits a range of responses in plants. These include regulating stomatal closure, lateral root initiation, flowering, seed germination and responses to a wide variety of abiotic stresses including osmotic stress, salinity, flooding and temperature extremes. NO is also involved in plant defense against pathogen attack by stimulating the production of H2O2. Treatment of plant tissues with ABA and IAA results in increased NO production, and in stomatal guard cells H2O2 stimulates the synthesis of NO.
In contrast to mammals, there has been no unequivocal demonstration of a plant NO synthase activity that uses arginine as the substrate for the synthesis of NO. In plants the synthesis of NO from NO3− or nitrite (NO2−) catalyzed by nitrate reductase is now well established (see Chapter 13). During normoxic conditions, i.e. in the presence of normal atmospheric oxygen concentrations, nitrate reductase is the source of NO produced enzymatically by plants. Arabidopsis has two genes encoding nitrate reductase, NIA1 and NIA2, and nia1/nia2 double mutants have a dramatic reduction in the synthesis of NO by stomatal guard cells and in the ability of these guard cells to close stomata in response to ABA. While NIA2 is the more abundant enzyme in Arabidopsis, mutations in NIA1 show that production of NO by NIA1 is more important in regulating stomatal closure.
Other biosynthetic pathways in plants can give rise to significant amounts of NO, especially under conditions of limited oxygen availability, when substrates such as NO2− accept electrons from cytochrome oxidase, cytochrome c, cytochrome P450 or other donors (Equation 10.1).
Non-enzymatic mechanisms for NO synthesis are also believed to operate in animals and plants. The chemistry of NO production from NO2− is well established. Under mildly acidic conditions nitrite is protonated to form nitrous acid (Equation 10.2). Nitrous acid can then be reduced to NO by a range of biological reductants, including ascorbate and phenolic components of the plant cell wall as shown in Equation 10.3.
Plants have several acidic compartments that would support the production of NO from nitrite, especially the cell wall where the activity of the plasma membrane H+-ATPase acidifies the apoplast and where phenolic components of the wall can provide the reductant to reduce nitrite to NO.