Evolution of Modifier Genes and Biological Systems
Sarah P. Otto
OUTLINE
1. Evolution of biological systems
2. Evolution of dominance
3. Direct versus indirect selection
4. The evolution of genetic transmission
5. The evolution of the mutation rate
6. The evolution of sex and recombination
7. The evolution of haploidy versus diploidy
8. On evolution and optimization
The features that define how an organism lives and reproduces—how its genes are transmitted over time and space—have been molded by evolution. Scientists study this process by tracking changes over time at genes that alter the biological system (so-called modifier genes). Genes that modify a particular feature evolve in response to both direct and indirect selection, where the former depends on which modifier allele(s) an individual carries, and the latter depends on genetic associations that develop between the modifier and other genes affecting fitness. This chapter reviews the philosophy of modifier models and how they are used to study the evolution of biological systems.
GLOSSARY
Dominance. The degree to which phenotype is affected more by one allele than another at a gene. In population genetics, the phenotype of interest is often fitness. Here, if an allele is dominant, the fitness of a heterozygote is closer to the homozygote carrying this allele than to the fitness of the opposite homozygote.
Genetic Transmission. Processes associated with the inheritance of genes from parents to offspring, including mutation, segregation distortion, and recombination.
Modifier Gene. A gene whose alleles alter a feature of interest in a species, such as its mating system, mutation or recombination rate, or life history characteristics.
Ploidy Level. The number of homologous copies of each chromosome carried by a cell (excluding sex chromosomes). A haploid carries one copy, a diploid two, and a polyploid more than two.
1. EVOLUTION OF BIOLOGICAL SYSTEMS
Arguably the most fundamental equations in evolutionary biology describe the changes that occur within a population under natural selection. For example, as described in chapter III.3, selection favoring one version of a gene (say, the “A” allele) over another version of the gene (the “a” allele) causes the frequency, p, of the A allele to rise over time. This rise can be predicted using mathematical models under a particular set of assumptions, for example, that the population is diploid (with two copies of every gene), completely sexual, and randomly mating, which in turn implies that organisms and the genes they carry are well mixed across the species range. In other words, such models make a series of assumptions about the “biological system”: how the organism lives out its life, how it reproduces, and how it moves over space.
Yet one might wonder how the biological system itself evolves. Why do some species live their lives as diploids, whereas other species are predominantly haploid (with one copy of every gene), and yet others alternate between haploid and diploid phases? Why do some species reproduce sexually, whereas other species eschew sex and reproduce clonally? Why do some species move freely over the landscape, while others choose to stay put? That is, how do the features that define how an organism lives and reproduces—how its genes are transmitted over time and space—evolve? Of course, these features may themselves be directly subject to natural selection (e.g., to avoid costs of dispersal), but they may also evolve because they alter the genetic constitution and environmental context of an individual’s descendants. Because most introductions to the field of evolutionary biology focus on the direct action of natural selection (see chapter III.3) and sexual selection (see chapter VII.6), assuming a particular biological system, most biologists and laypeople have little idea how evolutionary biologists would address these broader questions about how and why a species lives and reproduces the way it does (see chapters III.3 and VII.4).
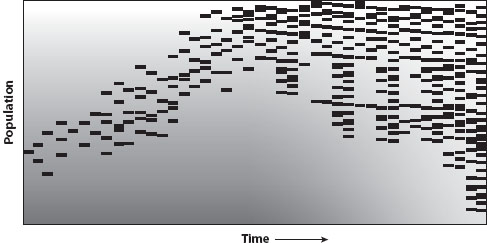
This chapter provides an introduction to the evolution of biological systems. The focus is on a body of theory that takes some feature of a biological system and examines how genes that shape this feature are expected to evolve. For example, whether yeast reproduce sexually depends on the activity of a series of genes that regulate entry into meiosis. Variants of these genes (called modifier genes) make it more or less likely for the cell to enter meiosis. These variants thus modify the transmission of the parents’ genes to offspring, determining whether they are inherited as a direct copy of the parental genome (asexual reproduction) or a mixture of two parental cells (sexual reproduction via meiosis). To study how the mode of reproduction and inheritance might evolve, evolutionary biologists develop mathematical models that track variants at such modifier genes. As described below, a particular modifier allele can rise or decline in frequency either because it directly affects the fitness of its carriers and/or because it alters the array of offspring and future descendants that are produced (figure 1). Modifier theory integrates these effects by tracking a modifier allele across generations to determine how the biological system is expected to evolve over the long term (see examples and links to other chapters in Table 1).
Figure 1. Tracking a modifier gene. In this schematic, each individual carrying a new modifier is represented by a line within a population (vertical axis) at a particular point in time (horizontal axis). Fitness is represented by background shading (lighter implying fitter). By altering the biological system, modifiers can become associated over time with particularly fit genotypes, as shown here. Initially, the modifier appears in an individual of relatively low fitness, but over time it becomes associated with fitter individuals (e.g., if a mutation modifier, a carrier might have produced a new beneficial mutation). Over time, survival and proliferation of the fittest individuals allows the associated modifier to spread throughout the population. Individuals who would have survived but die because of the direct costs of the modifier (e.g., costs of error correction) are represented as dotted lines. Alternatively, had the modifier allele stayed associated with low-fitness individuals, it would have been eliminated with the deaths of those individuals (not shown).
Table 1. Examples of direct and indirect selection shaping the evolution of biological systems
Feature |
Direct selection |
Indirect selection |
Mating preferences (chapter VII.4) |
Costs involved in searching for a preferred mate or rejecting a mate |
Modifier increasing preference becomes associated with sons that are attractive to females with a similar preference |
Migration rate (chapter IV.3) |
Energetic costs and associated risks of moving, e.g., risk of predation |
Modifier increasing migration rate becomes associated with alleles that were selectively favored in other habitats and with reduced competition among relatives |
Mutation rate (chapter IV.2) |
Costs involved in repairing DNA damage and in failing to do so |
Modifier increasing mutation rate becomes associated with novel deleterious and beneficial alleles |
Sexual versus asexual reproduction (chapter IV.4) |
Transmission advantage of asexuality; offspring inherit 100% of genes from an asexual parent rather than only 50% from a sexual parent |
Modifier increasing frequency of sex becomes associated with different genetic combinations |
Selfing rate (chapter IV.6) |
Transmission advantage of selfing; selfing individuals can gain extra fitness by fertilizing their own ovules |
Modifier increasing frequency of selfing becomes associated with more homozygous gene combinations |
The results of modifier theory are particularly interesting in cases where it is hard to predict ahead of time which of several possible paths evolution is likely to take. For example, take the case of haploid versus diploid life cycles. Diploids have two sets of chromosomes and so have twice the amount of DNA as haploids. As a result, they suffer twice as many new mutations each generation, resulting in diploids suffering twice the deleterious mutation load compared to haploids (i.e., twice the reduction in fitness due to deleterious mutations; see chapter IV.5). Thus, from the perspective of the species, it would be optimal to be haploid. On the other hand, any particular individual is more fit if it is diploid, because the functioning of the second copy of the gene can mask any deleterious mutations the individual happens to carry. Thus, from the perspective of the individual, it would be optimal to be diploid. Which is the right perspective?
Modifier theory sidesteps this question, asking not what is optimal but instead what evolves. As we shall see, models that track genes that modify the alternation of generations between haploid and diploid phases find that haploidy is expected to evolve under some circumstances and diploidy under others. Interestingly, modifier models can also be used to understand how the direct effects on an individual’s fitness and the long-term effects on the mean fitness of a lineage both play a role in the evolution of the biological system.
The goal of this chapter is to illustrate some of the insights provided by modifier models about the evolution of biological systems.
2. EVOLUTION OF DOMINANCE
Working in the 1900s, R. A. Fisher, Sewall Wright, and J.B.S. Haldane developed the mathematical underpinnings of our understanding of evolution. They were also the first to develop and analyze modifier models. Indeed, a modifier model of dominance played a central role in one of the most infamous early debates in evolutionary biology. Data from an increasing number of species indicated that wild-type alleles tend to be dominant over deleterious mutations, partially to fully masking the effects of mutations when both alleles are present in a heterozygote. To explain this phenomenon, Fisher considered a scenario with two genes: one gene under selection with a wild-type allele (A) subject to recurrent mutation to a less fit allele (a), and one gene that alters the degree of dominance, h, of the mutant allele with fitnesses given by:

where ij describes the genotype at the modifier locus (MM, Mm, or mm). Figure 2 shows how to derive equations for this two-gene model. Fisher then argued that modifier alleles that increased the fitness of heterozygotes (decreasing hij) would leave more descendants and hence should rise in frequency. Fisher concluded that biological systems would evolve by successive modification to the point where wild-type alleles were more dominant over mutant alleles.
Figure 2. Model for the evolution of dominance. The evolution of dominance can be explored using a model to track changes at two genes: a modifier (M) and a gene under selection (A). We start (top) by censusing the frequency, pkl, of each gamete (for kl equal to MA, Ma, mA, or ma). These gametes come together to form diploids, which are then subject to selection (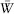 represents the mean fitness in the population). The surviving adults undergo meiosis (r is the recombination rate between M and A), with each gamete subject to mutation (at rate μ from A to a), producing the next generation of gametes. This cycle is repeated over and over to determine whether a modifier that alters the degree of dominance rises or falls in frequency. (Only a few examples of the equations for selection, meiosis, and mutation are shown.)
represents the mean fitness in the population). The surviving adults undergo meiosis (r is the recombination rate between M and A), with each gamete subject to mutation (at rate μ from A to a), producing the next generation of gametes. This cycle is repeated over and over to determine whether a modifier that alters the degree of dominance rises or falls in frequency. (Only a few examples of the equations for selection, meiosis, and mutation are shown.)
Wright did not dispute Fisher’s reasoning; indeed, Wright explored a similar model to reach the opposite conclusion. Wright observed that while the model predicts that modifiers of dominance would spread, the rate at which they spread is extremely low, being proportional to the mutant frequency at the A gene. Wright concluded that such a weak evolutionary force would likely be overwhelmed by other forces, including mutation at the modifier gene, side effects of the modifier gene, or random genetic drift. Instead, Wright argued that the fitness benefits of many genes diminish as their function increases, because the gene product becomes less a limiting resource; consequently, fitness is less affected in heterozygotes that still have one functioning gene copy than in homozygote mutants, whose gene product is much more limiting.
The interchange between Fisher and Wright over the power and efficacy of evolution is fascinating, and it eventually led to the breakdown of communication between them (Provine 1986). Subsequent authors, including Haldane, painted a less black-and-white picture, pointing out that selection on modifiers of dominance becomes strong when both alleles at the A gene are common (e.g., during the initial spread of wild-type alleles over previous alleles or when different alleles are favored over different parts of the species’ range) or if modifiers can affect dominance at multiple genes simultaneously (e.g., by promoting the degradation of misfolded proteins).
While it is sometimes said that Fisher’s modifier theory of dominance ultimately lost out to Wright’s physiological theory, this view is misleading. Both sides relied on insights obtained from modifier theory, with Fisher focusing on the direction of selection acting on dominance and Wright focusing on the strength of this selection.
3. DIRECT VERSUS INDIRECT SELECTION
In modifier models of dominance, the fitness of an individual depends on its modifier genotype (through hij). The same is true of models of epistasis, where modifiers alter the fitness of individuals carrying specific gene combinations at different loci (Liberman and Feldman 2005). In such cases, we say that the modifier gene is under direct selection.
Not all modifiers affect fitness directly. They can nevertheless evolve because they alter the types of descendants produced, experiencing indirect selection according to the fitness of these descendants. For example, a modifier that alters the frequency of recombination may have little direct effect on the fitness of its carrier, but it will change the genetic makeup of any offspring produced. If these offspring happen to have high fitness, on average, then the modifier allele will rise to higher frequency (figure 1). In essence, the modifier allele hitchhikes along with the successes (and failures) of the descendants it produces. To determine whether a modifier spreads or not requires that we track each descendant carrying the allele and the type of offspring it in turn produces. It can take several generations before the fate of the modifier becomes clear, in which case various mathematical techniques (such as local stability analyses and quasi-linkage equilibrium methods) are used to determine the direction of modifier evolution over longer periods of time.
One of the earliest examples of a modifier experiencing indirect selection was used by Fisher in a verbal account of the widespread observation that approximately equal numbers of males and females are found in many species. Whether an individual produces sons or daughters may have little consequence for the fitness of the parent, especially if parental investment ends after offspring are produced. Thus the sex ratio of an individual is often not under direct selection, yet indirect selection does act. Specifically, if an individual carries a gene that causes it to produce more offspring of the rarer sex, those offspring would make up a larger proportion of the mating pool of that sex and would contribute more genes, on average, to the grandoffspring and the great-grandoffspring, etc. Consequently, modifier alleles promoting the production of the currently rare sex would rise in frequency, causing the sex ratio to evolve toward 50:50. This argument was initially verbal. Indeed, the roots of the idea trace back to Darwin’s The Descent of Man and Selection in Relation to Sex before genetic inheritance was understood, but it has subsequently been modeled and verified mathematically (e.g., Bodmer and Edwards 1960).
Of course, many biological systems evolve under the influence of both direct and indirect selection, as illustrated in Table 1, so that both must be considered when predicting the evolution of the system. It can be difficult, however, to intuit exactly how direct and indirect selection combine to influence the frequency of modifier alleles as they are transmitted from generation to generation, particularly because we must account for associations that develop between modifier alleles and other genes within the genome. Mathematical models are particularly illuminating in such cases, because they help guide and correct our evolutionary reasoning. These models are developed as illustrated in the case of dominance (figure 2), considering each step within the life cycle of an organism, so that modelers need not guess ahead of time what the net result of direct and indirect selection will be.
We turn next to a few examples of the application of modifier theory and the insights they have provided.
4. THE EVOLUTION OF GENETIC TRANSMISSION
One early and general result from modifier theory investigated the transmission of genes from parents to offspring. Assuming that a single randomly mating population has reached equilibrium under constant selection, modifier alleles that cause perfect transmission—where offspring are accurate copies of their parents—are always able to spread. This result was shown to apply to a number of processes that alter transmission, including mutation, migration, recombination, and segregation distortion, each of which is predicted to evolve toward zero. This general result is known as the reduction principle (for further information, see Altenberg and Feldman 1987).
While initially surprising, the reduction principle does make sense: if the world weren’t changing and if selection favored certain alleles or combinations of alleles, then biological systems should evolve to exactly replicate parental genotypes, given that these parents survived to reproduce and so have genotypes that work well in the current environment.
The reduction principle tells us that, all else being equal, the simplest form of transmitting genes by copying them perfectly across generations should evolve. The real world is, however, much more complex; all else is not equal. Direct costs of perfect transmission make it impossible to replicate DNA without mutation and, in many species, to segregate chromosomes properly without recombination. Furthermore, the world is changing. Environmental changes, mutation, and drift can all push biological systems away from their equilibrium points and cause the reduction principle to fail. Research in this area thus attempts to figure out exactly when and why organisms transmit their genes the way they do, imperfectly.
5. THE EVOLUTION OF THE MUTATION RATE
All biological systems are subject to errors during the replication of their genetic material; mutations thus represent a universal example of imperfect transmission. The frequency of mutational errors is influenced by a variety of genes, including those involved directly in DNA replication (e.g., polymerases) as well as genes involved in the detection and repair of errors (e.g., excision repair and mismatch repair genes) (see chapter IV.2). Both direct and indirect selection are thought to shape the evolution of the resulting mutation rate.
Direct selection on the mutation rate results from the costs and benefits of detecting and repairing DNA damage and replication errors. These include energetic costs of producing error-correcting proteins and reduced growth rates resulting from high-fidelity replication. The benefits include avoiding mutations that arise during development and directly reduce the fitness of an individual (e.g., cancer-causing mutations).
Indirect selection results from transmitting different numbers of mutations to future generations. On the one hand, alleles increasing the mutation rate produce offspring that suffer a higher load of deleterious mutations. On the other hand, these offspring are also more likely to carry beneficial mutations that improve fitness in the current environment.
What is the net result of these selective forces? The proof of the reduction principle allowed only deleterious mutations and assumed no costs, and so it cannot tell us. Subsequent work has shown, interestingly, that the answer depends on the way the organism reproduces.
If the organism reproduces asexually, clones evolve a mutation rate that maximizes the lineage’s long-term mean fitness. Consequently, higher mutation rates are expected to evolve in a changing environment where there is an advantage to producing beneficial mutations, as observed in a number of empirical studies (see review by Sniegowski et al. 2000).
With sex and recombination, however, genes that modify mutation rates evolve toward lower rates than would be optimal for the population. To understand this result, one again must think about the genes that influence the mutation rate and what happens to them from generation to generation. Carriers of a modifier that increases the mutation rate produce descendants that are more likely to carry deleterious as well as advantageous mutations. Deleterious mutations do not, on average, persist for long within the population, killing off their carriers as well as copies of the mutation modifier. Advantageous mutations persist longer, but sex and recombination separate these advantageous mutations from the mutation modifier that initially produced them. Thus, individuals carrying modifiers that increase the mutation rate soon bare the costs of producing more deleterious mutations, but because of sex, they share the benefits of their advantageous mutations with noncarriers. The more sex and recombination within a population, the more diluted the benefits become, hindering the evolution of higher mutation rates. Thus, mutation rates are expected to evolve to a lower level than what would maximize the mean fitness of the species.
6. THE EVOLUTION OF SEX AND RECOMBINATION
One of the most puzzling aspects of biological systems is that the vast majority of species engage in sexual reproduction, at least occasionally. Given that parents have survived to reproduce and so are “tried-and-true,” why should a parent shuffle its genome with the genome of another? What makes this question even more puzzling is that sexual reproduction generally entails many direct costs, including the cost of finding a mate, the risk of remaining unmated, and the dangers of disease transmission and predation during mating, not to mention the fact that transmitting only half of one’s genes to offspring automatically halves a parent’s fitness (the twofold cost of sex).
Theoretical models have confirmed that genomic shuffling is generally a bad idea. Only if the environment changes very rapidly over time (changing direction every two to five generations) or over space is the mean fitness of offspring higher with genomic shuffling than without. Thus, under most circumstances, the short-term effect of a modifier that increases the frequency of sexual reproduction is to produce less fit descendants. The same holds for a modifier that increases the frequency of recombination. Basically, by breaking apart the gene combinations that have been favored by past generations of selection, sex and recombination tend to produce less fit offspring. That is, these offspring suffer from two types of fitness load: a segregation load and a recombination load (see chapter IV.5).
Why then is sex so prevalent? The key is thought to lie not in the mean fitness of offspring but in the variability of their fitness. Genetic variation is the fuel of evolutionary change, and if sex increases genetic variation, the response to selection will be faster among sexually produced offspring, promoting the spread of modifiers that increase the frequency of sex. Put another way, if the fittest individuals of a generation are produced sexually, their survival and reproduction spread modifier genes that enhance the rate of sexual reproduction. Lutz Becks and Aneil Agrawal have recently provided experimental evidence for this phenomenon in rotifers.
There are some big “ifs” in the preceding paragraph, however: “if sex increases genetic variation” and “if the fittest individuals of a generation are produced sexually.” These conditions are not necessarily true. The problem is that while a single sexual individual can produce more variable offspring than a single asexual, a group of asexuals can be remarkably diverse, and altogether produce more variable offspring than a group of sexual individuals.
Evolutionary theory has thus sought conditions where (1) genetic variation is lacking, (2) sexual reproduction can increase this variation, and (3) the long-term advantages of increasing genetic variation outweigh the short-term segregation and recombination loads. Two main conditions have been found that satisfy these requirements and allow the spread of genes that increase the rate of sexual reproduction. First, genetic variation may be lacking because of past selection; this explanation requires that individuals at the extremes in fitness (high and low) are less fit, on average, compared with intermediate individuals, which in turn requires negative fitness interactions either within a gene (dominance) or among genes (epistasis). Second, genetic variation may be lacking because populations contain only a finite number of genetic combinations, and it is unlikely that the fittest alleles all occur within the same individual. Indeed, after a period of selection, a finite population tends to become composed of individuals that have similar fitness but carry different mixtures of good and bad genes. Sex and recombination can recover some of this hidden genetic variation by combining genes into different configurations, producing some particularly sick offspring and some particularly healthy offspring. With the survival and spread of the extremely fit individuals, genes promoting sex and recombination hitchhike along in frequency.
The jury is still out regarding whether the puzzle of sex is solved by the need for variation depleted by past selection and/or past drift in finite populations, but there is no doubt that modifier theory has clarified which evolutionary explanations can work and the conditions needed for them to do so.
7. THE EVOLUTION OF HAPLOIDY VERSUS DIPLOIDY
As one final example, let us return to the question of ploidy evolution. All sexual organisms pass through a stage with a reduced genome following meiosis (haploid) and a doubled genome following the union of gametes (diploid). Some organisms, like ourselves, spend little time in the haploid stage, whereas others, including a variety of fungi, algae, and mosses, spend little time in the diploid stage.
Evolutionary biologists can explore the conditions favoring the evolution of haploidy or diploidy by tracking changes at genes that alter the timing of the life cycle (figure 3A). Genetic changes that promote meiosis soon after gametes unite produce a predominantly haploid species, and vice versa for a predominantly diploid species.
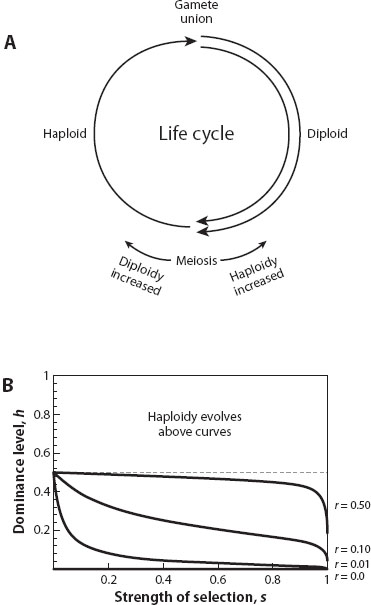
Figure 3. The evolution of haploidy versus diploidy. (A) With an alternation of generations, the extent of the haploid or diploid phase can be altered by genes that control the relative timing of meiosis and gamete union. (B) Modifier alleles that slightly expand the haploid phase are favored above the curves, with diploidy favored below the curves. The size of the region in which haploidy is favored expands when there is less genetic mixing (here shown as less recombination, r, between M and A). Selection acts against mutations with strength 1 – hs in heterozygous diploids and strength 1 – s in haploids and homozygous diploids.
Tracking such modifier genes, we find another interesting result. Haploid life cycles are favored in species that have low effective rates of recombination—for example, when reproduction is typically asexual or often involves inbreeding. In addition, haploid life cycles are favored when heterozygous diploids have a lower fitness than homozygous diploids, on average (i.e., when beneficial alleles are partially recessive and/or deleterious mutations are partially dominant). In contrast, diploidy evolves in highly sexual populations as long as heterozygotes have relatively high fitness (figure 3B). Additionally, in large multicellular organisms, modifiers that promote diploidy are more likely to spread because of the direct benefits of protecting an individual from the cancer-causing mutations that arise during development. Otto and Gerstein (2008) review empirical work testing these predictions.
8. ON EVOLUTION AND OPTIMIZATION
In the introduction, we pointed out that haploidy is optimal from the perspective of a species: haploids carry, on average, half as many mutations, and these mutations are immediately exposed to selection and purged. From the perspective of the individual, however, it is best to be diploid: deleterious mutations carried by a diploid can be masked by the second gene copy, allowing that individual to survive and reproduce. Which perspective is correct?
In some ways, the answer is neither. Considering what is best for the group would predict a world without diploids. Considering what is best for the individual would predict a world in which haploids are rare.
In another sense, however, the answer is that both perspectives provide clues about the evolutionary forces acting on a biological system. Because we track modifier genes over generations, whether those genes spread or disappear is influenced by what is best for the individuals that carry them in any particular generation and also what is best for the lineage (or group) of descendants that inherit the gene.
In many of the examples highlighted in this chapter, the way the biological system evolves depends on the amount of genetic mixing occurring within a species. When reproduction is primarily sexual, modifier alleles are more likely to evolve in ways that are best for the individual (e.g., lower mutation rates even when beneficial mutations are needed, more diploid life cycles even though deleterious mutations accumulate), because genetic mixing via sex prevents genetic differences from building up between the group of descendants that carry a particular modifier gene and those that do not. With little sex, modifier alleles are more likely to evolve in ways that are best for the group (e.g., higher mutation rates when beneficial mutations are needed, more haploid life cycles), because the modifier tends to be inherited for several generations alongside whatever genetic changes it causes, allowing the modifier to reap the long-term benefits of these changes.
What modifier theory allows us to do is to look at the biological world and to change the question from “What is best?” to “What will evolve?” What will evolve is sometimes best for the individual, sometimes best for the species, and sometimes neither. To understand how complex biological systems evolve and how this evolution has led to the remarkable diversity of life requires us to pay attention to how the genes that mold an organism change over time and how this evolution depends on the features of the organism and its environment.
FURTHER READING
Altenberg, L., and M. W. Feldman. 1987. Selection, generalized transmission and the evolution of modifier genes. I. The reduction principle. Genetics 117: 559–572. A technical proof of the reduction principle, showing when and why evolution favors perfect transmission.
Barton, N. H. 1995. A general model for the evolution of recombination. Genetical Research 65: 123–145. A tour de force analysis of the conditions under which increased recombination evolves.
Bodmer, W. F., and A. W. Edwards. 1960. Natural selection and the sex ratio. Annals of Human Genetics 24: 239–244. A mathematical treatment demonstrating that the sex ratio evolves toward equal parental investment in daughters and sons.
Liberman, U., and M. W. Feldman. 2005 On the evolution of epistasis I: Diploids under selection. Theoretical Population Biology 67: 141–160. A mathematical study exploring, for the first time, the evolution of epistasis using a modifier model.
Mayo, O., and R. Bürger. 1997. The evolution of dominance: A theory whose time has passed? Biological Reviews 72: 97–110. An examination of the evolution of dominance, providing a strong historical overview.
M’Gonigle, L. K., and S. P. Otto. 2011. Ploidy and the evolution of parasitism. Proceedings of the Royal Society B 278: 2814–2822. An exploration of genetic factors that could influence the transition between free-living and parasitic life cycles.
Otto, S. P. 2009. The evolutionary enigma of sex. American Naturalist 174: S1–S14. A review of the evolutionary forces acting on rates of sex and recombination.
Otto, S. P., and A. C. Gerstein. 2008. The evolution of haploidy and diploidy. Current Biology 18: R1121–R1124. A primer describing theoretical and empirical results on the evolutionary transitions between haploidy and diploidy, as well as open questions.
Sniegowski, P. D., P. J. Gerrish, T. Johnson, and A. Shaver. 2000. The evolution of mutation rates: Separating causes from consequences. Bioessays 22: 1057–1066. An excellent overview of the evolution of the mutation rate.